Genome-Wide Association Study of Breast Density among Women of African Ancestry
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Data
2.2. Breast Density
2.3. Genome-Wide Association Study in Penn Medicine BioBank
2.4. Functional Mapping
2.5. Transcriptome-Wide Association Study (TWAS)
2.6. GWAS-Catalog Lookup
2.7. Correlation Analyses
3. Results
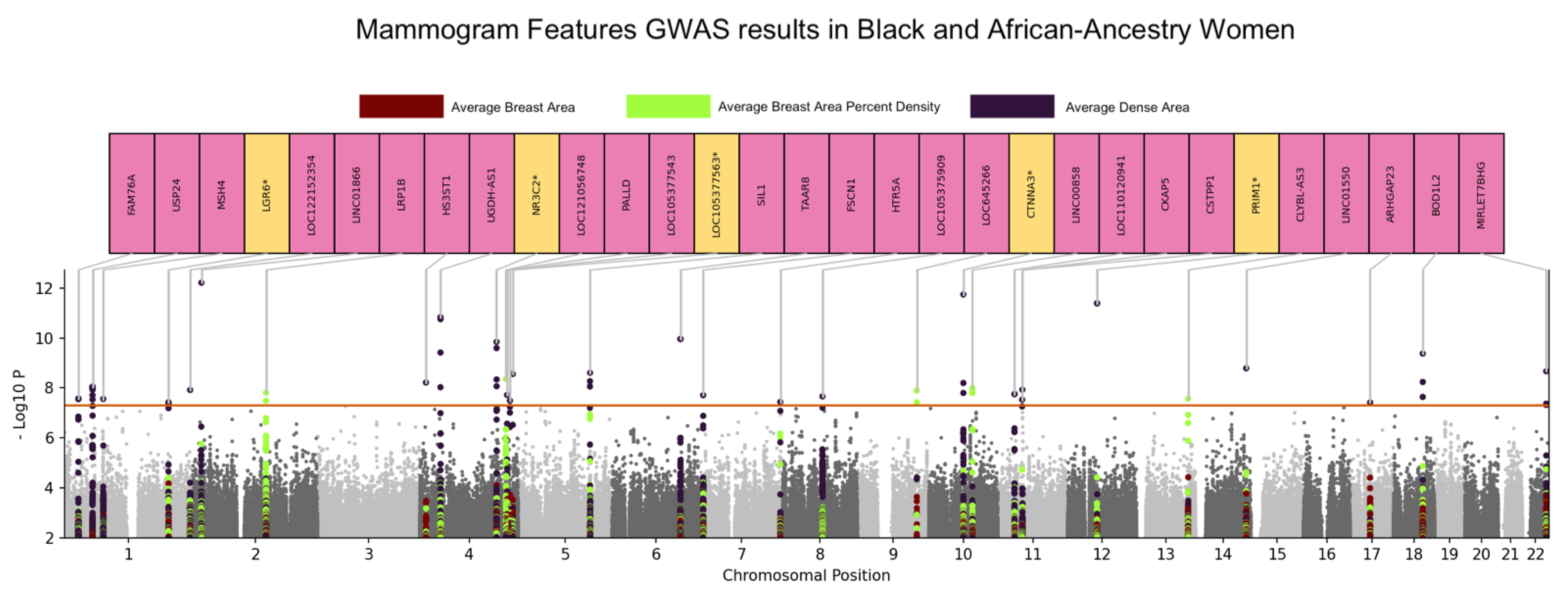
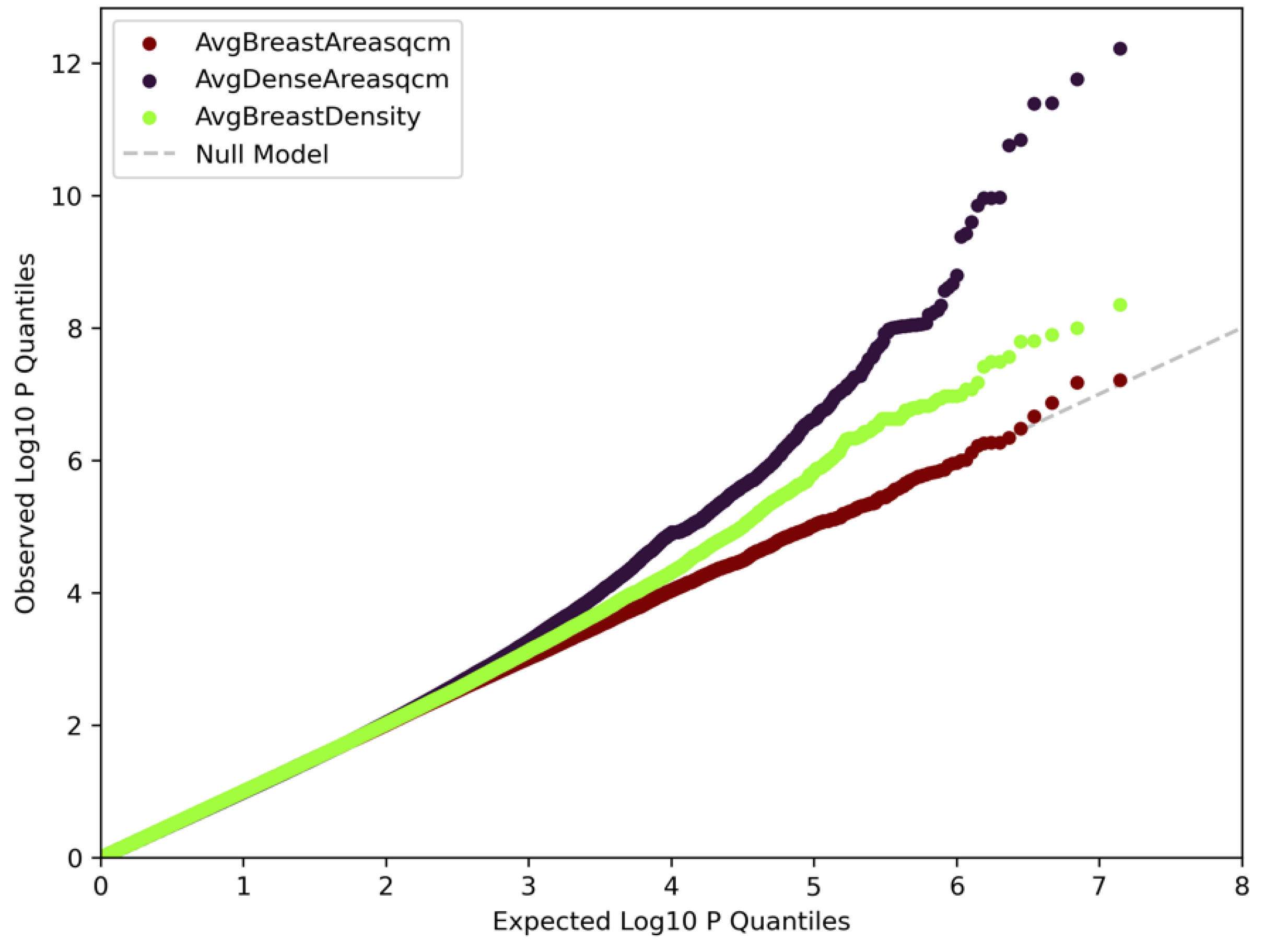
3.1. Genome-Wide Association Analyses
3.2. Functional Mapping
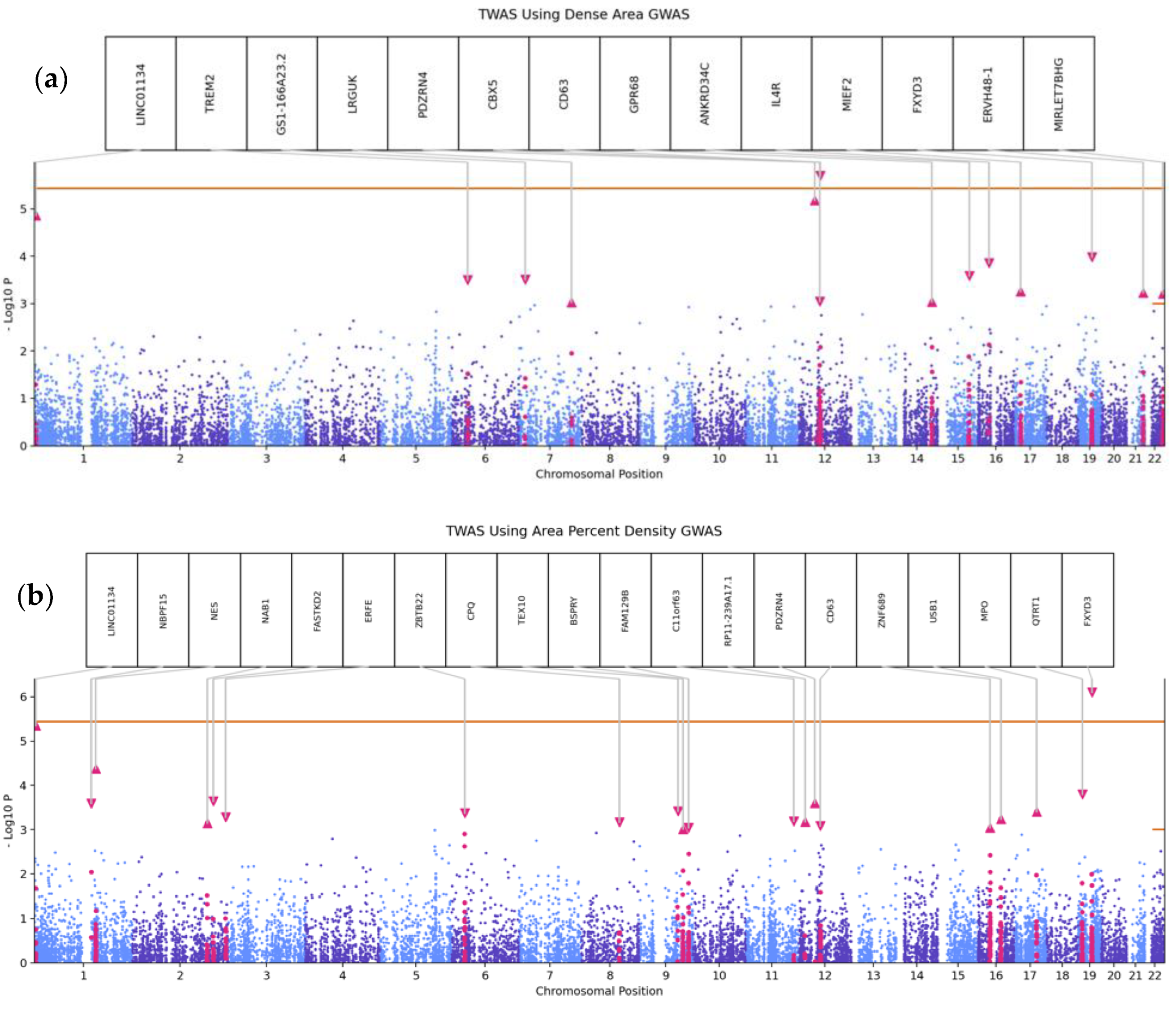
3.3. Transcriptome-Wide Association Analyses
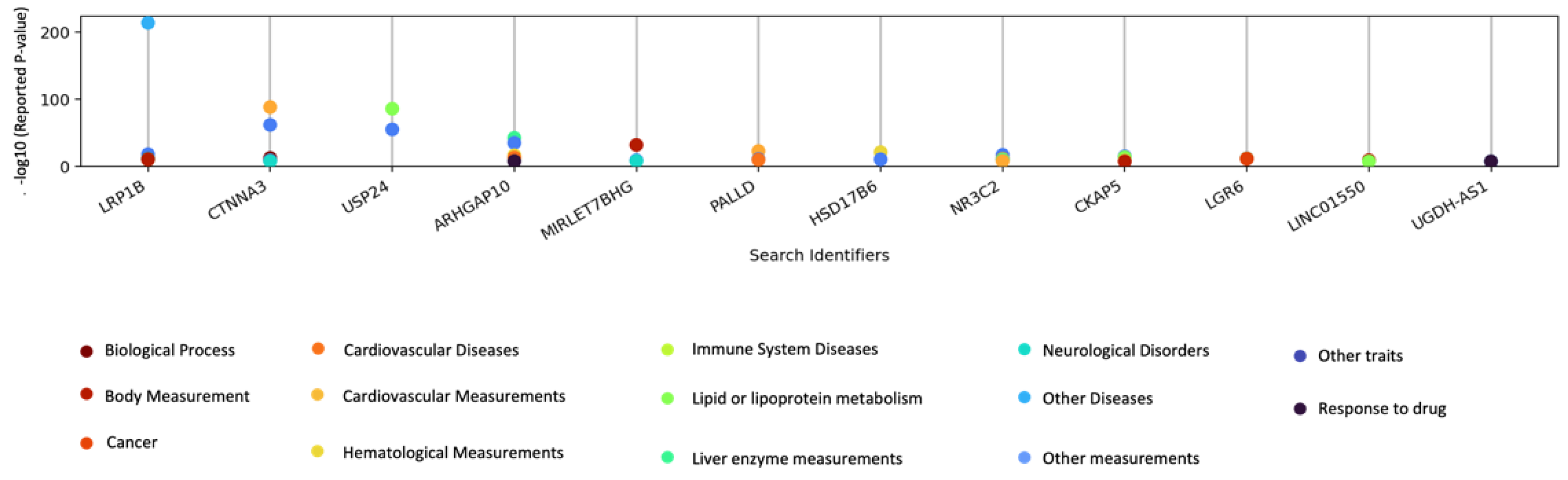
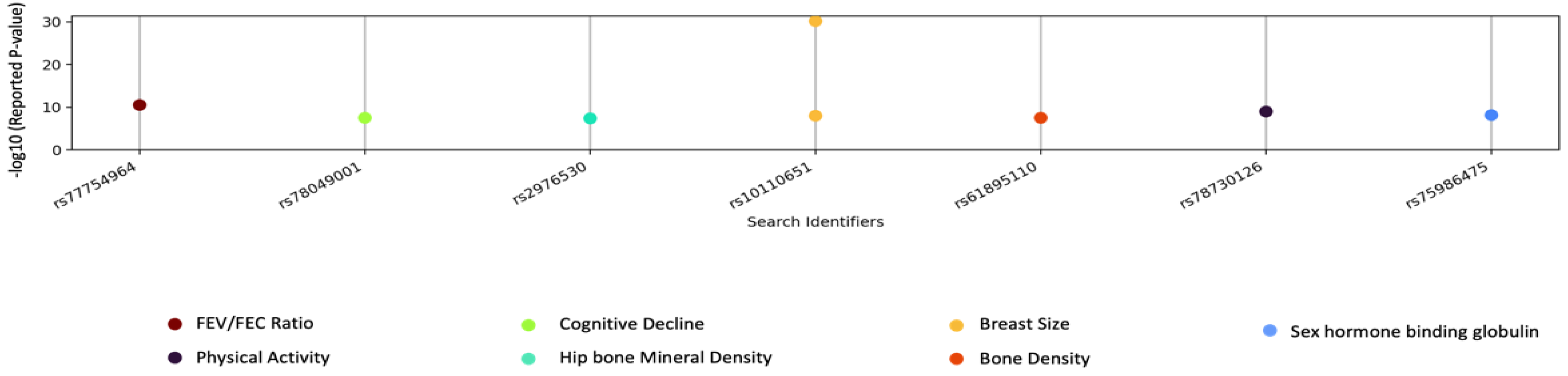
3.4. GWAS Catalog Lookups
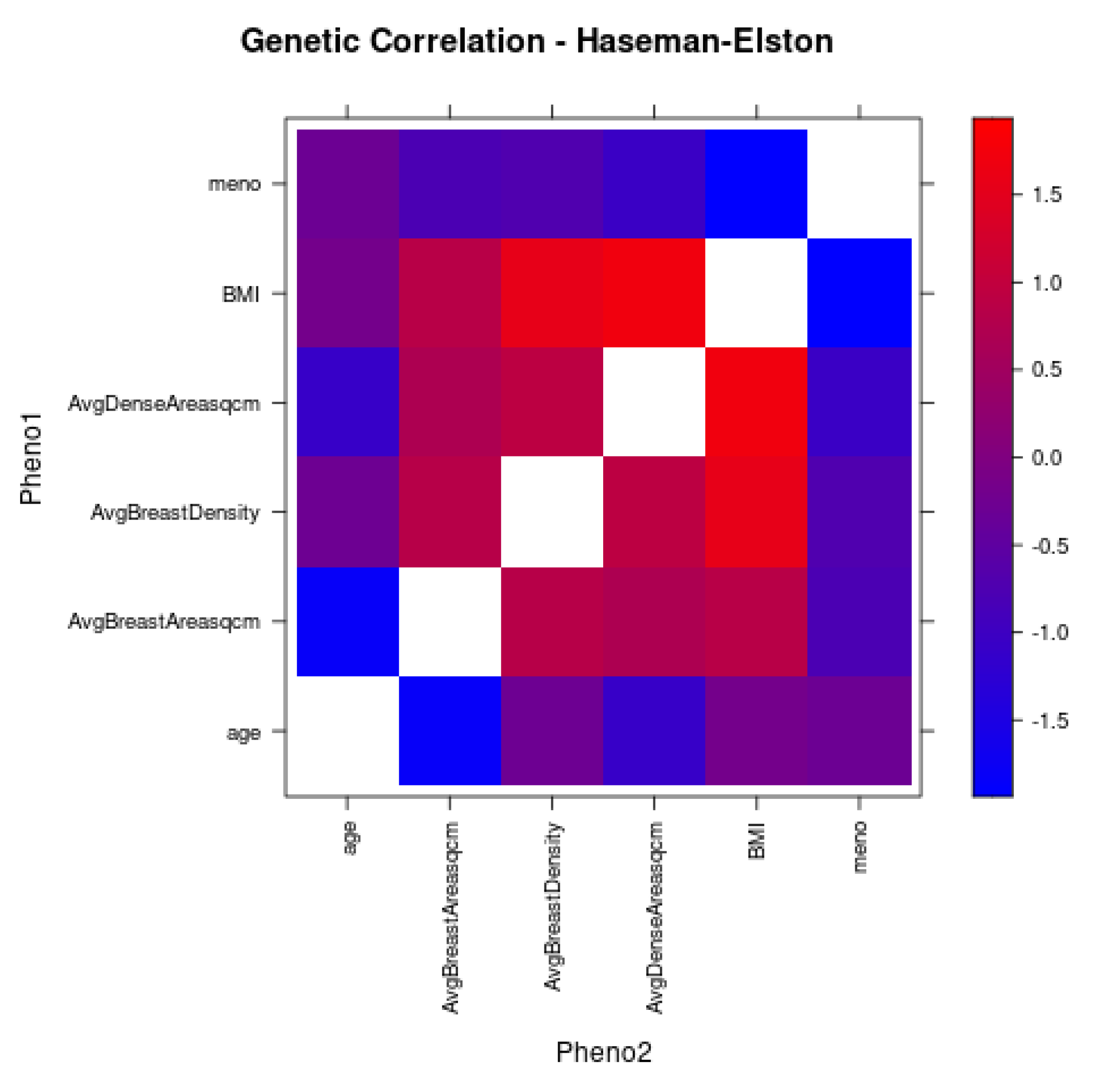
3.5. Correlation Analyses
3.6. Comparison among EUR and AFR Breast Density GWAS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- DeSantis, C.E.; Miller, K.D.; Goding Sauer, A.; Jemal, A.; Siegel, R.L. Cancer statistics for African Americans. CA Cancer J. Clin. 2019, 69, 211–233. [Google Scholar] [CrossRef] [PubMed]
- Howlader, N.; Altekruse, S.F.; Li, C.I.; Chen, V.W.; Clarke, C.A.; Ries, L.A.; Cronin, K.A. US incidence of breast cancer subtypes defined by joint hormone receptor and HER2 status. J. Natl. Cancer Inst. 2014, 106, dju055. [Google Scholar] [CrossRef] [PubMed]
- Kohler, B.A.; Sherman, R.L.; Howlader, N.; Jemal, A.; Ryerson, A.B.; Henry, K.A.; Boscoe, F.P.; Cronin, K.A.; Lake, A.; Noone, A.M.; et al. Annual Report to the Nation on the Status of Cancer, 1975–2011, Featuring Incidence of Breast Cancer Subtypes by Race/Ethnicity, Poverty, and State. J. Natl. Cancer Inst. 2015, 107, djv048. [Google Scholar] [CrossRef] [PubMed]
- Pettersson, A.; Graff, R.E.; Ursin, G.; Santos Silva, I.D.; McCormack, V.; Baglietto, L.; Vachon, C.; Bakker, M.F.; Giles, G.G.; Chia, K.S.; et al. Mammographic density phenotypes and risk of breast cancer: A meta-analysis. J. Natl. Cancer Inst. 2014, 106, dju078. [Google Scholar] [CrossRef] [PubMed]
- Bond-Smith, D.; Stone, J. Methodological Challenges and Updated Findings from a Meta-analysis of the Association between Mammographic Density and Breast Cancer. Cancer Epidemiol. Biomark. Prev. 2019, 28, 22–31. [Google Scholar] [CrossRef]
- McCarthy, A.M.; Keller, B.M.; Pantalone, L.M.; Hsieh, M.K.; Synnestvedt, M.; Conant, E.F.; Armstrong, K.; Kontos, D. Racial Differences in Quantitative Measures of Area and Volumetric Breast Density. J. Natl. Cancer Inst. 2016, 108, djw104. [Google Scholar] [CrossRef] [PubMed]
- Bissell, M.C.S.; Kerlikowske, K.; Sprague, B.L.; Tice, J.A.; Gard, C.C.; Tossas, K.Y.; Rauscher, G.H.; Trentham-Dietz, A.; Henderson, L.M.; Onega, T.; et al. Breast Cancer Population Attributable Risk Proportions Associated with Body Mass Index and Breast Density by Race/Ethnicity and Menopausal Status. Cancer Epidemiol. Biomark. Prev. 2020, 29, 2048–2056. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Gong, W.; Wang, S.; Li, Z. Trends in body mass index, overweight and obesity among adults in the USA, the NHANES from 2003 to 2018: A repeat cross-sectional survey. BMJ Open 2022, 12, e065425. [Google Scholar] [CrossRef]
- Gastounioti, A.; Kasi, C.D.; Scott, C.G.; Brandt, K.R.; Jensen, M.R.; Hruska, C.B.; Wu, F.F.; Norman, A.D.; Conant, E.F.; Winham, S.J.; et al. Evaluation of LIBRA Software for Fully Automated Mammographic Density Assessment in Breast Cancer Risk Prediction. Radiology 2020, 296, 24–31. [Google Scholar] [CrossRef]
- Keller, B.M.; Chen, J.; Daye, D.; Conant, E.F.; Kontos, D. Preliminary evaluation of the publicly available Laboratory for Breast Radiodensity Assessment (LIBRA) software tool: Comparison of fully automated area and volumetric density measures in a case-control study with digital mammography. Breast Cancer Res. 2015, 17, 117. [Google Scholar] [CrossRef]
- Sprague, B.L.; Conant, E.F.; Onega, T.; Garcia, M.P.; Beaber, E.F.; Herschorn, S.D.; Lehman, C.D.; Tosteson, A.N.; Lacson, R.; Schnall, M.D.; et al. Variation in Mammographic Breast Density Assessments Among Radiologists in Clinical Practice: A Multicenter Observational Study. Ann. Intern. Med. 2016, 165, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Brand, J.S.; Humphreys, K.; Li, J.; Karlsson, R.; Hall, P.; Czene, K. Common genetic variation and novel loci associated with volumetric mammographic density. Breast Cancer Res. 2018, 20, 30. [Google Scholar] [CrossRef] [PubMed]
- Holowko, N.; Eriksson, M.; Kuja-Halkola, R.; Azam, S.; He, W.; Hall, P.; Czene, K. Heritability of Mammographic Breast Density, Density Change, Microcalcifications, and Masses. Cancer Res. 2020, 80, 1590–1600. [Google Scholar] [CrossRef]
- Chen, H.; Fan, S.; Stone, J.; Thompson, D.J.; Douglas, J.; Li, S.; Scott, C.; Bolla, M.K.; Wang, Q.; Dennis, J.; et al. Genome-wide and transcriptome-wide association studies of mammographic density phenotypes reveal novel loci. Breast Cancer Res. BCR 2022, 24, 27. [Google Scholar] [CrossRef] [PubMed]
- Sieh, W.; Rothstein, J.H.; Klein, R.J.; Alexeeff, S.E.; Sakoda, L.C.; Jorgenson, E.; McBride, R.B.; Graff, R.E.; McGuire, V.; Achacoso, N.; et al. Identification of 31 loci for mammographic density phenotypes and their associations with breast cancer risk. Nat. Commun. 2020, 11, 5116. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. National Health and Nutrition Examination Survey: 2011–2012 Data Documentation, Codebook, and Frequencies. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, 2013; Updated 2015. Available online: https://wwwn.cdc.gov/nchs/nhanes/search/datapage.aspx?Component=Examination&CycleBeginYear=2011 (accessed on 22 June 2021).
- Keller, B.M.; Nathan, D.L.; Wang, Y.; Zheng, Y.; Gee, J.C.; Conant, E.F.; Kontos, D. Estimation of breast percent density in raw and processed full field digital mammography images via adaptive fuzzy c-means clustering and support vector machine segmentation. Med. Phys. 2012, 39, 4903–4917. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Wigginton, J.E.; Cutler, D.J.; Abecasis, G.R. A note on exact tests of Hardy-Weinberg equilibrium. Am. J. Hum. Genet. 2005, 76, 887–893. [Google Scholar] [CrossRef]
- Truong, V.Q.; Woerner, J.A.; Cherlin, T.A.; Bradford, Y.; Lucas, A.M.; Okeh, C.C.; Shivakumar, M.K.; Hui, D.H.; Kumar, R.; Pividori, M.; et al. Quality Control Procedures for Genome-Wide Association Studies. Curr. Protoc. 2022, 2, e603. [Google Scholar] [CrossRef]
- Southern, J.A.; Young, D.F.; Heaney, F.; Baumgartner, W.K.; Randall, R.E. Identification of an epitope on the P and V proteins of simian virus 5 that distinguishes between two isolates with different biological characteristics. J. Gen. Virol. 1991, 72, 1551–1557. [Google Scholar] [CrossRef]
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef] [PubMed]
- Galinsky, K.J.; Bhatia, G.; Loh, P.R.; Georgiev, S.; Mukherjee, S.; Patterson, N.J.; Price, A.L. Fast Principal-Component Analysis Reveals Convergent Evolution of ADH1B in Europe and East Asia. Am. J. Hum. Genet. 2016, 98, 456–472. [Google Scholar] [CrossRef]
- Bush, W.S.; Dudek, S.M.; Ritchie, M.D. Biofilter: A knowledge-integration system for the multi-locus analysis of genome-wide association studies. Pac. Symp. Biocomput. 2009, 2009, 368–379. [Google Scholar]
- Pendergrass, S.A.; Frase, A.; Wallace, J.; Wolfe, D.; Katiyar, N.; Moore, C.; Ritchie, M.D. Genomic analyses with biofilter 2.0: Knowledge driven filtering, annotation, and model development. BioData Min. 2013, 6, 25. [Google Scholar] [CrossRef] [PubMed]
- Weissbrod, O.; Hormozdiari, F.; Benner, C.; Cui, R.; Ulirsch, J.; Gazal, S.; Schoech, A.P.; van de Geijn, B.; Reshef, Y.; Marquez-Luna, C.; et al. Functionally informed fine-mapping and polygenic localization of complex trait heritability. Nat. Genet. 2020, 52, 1355–1363. [Google Scholar] [CrossRef]
- Boix, C.A.; James, B.T.; Park, Y.P.; Meuleman, W.; Kellis, M. Regulatory genomic circuitry of human disease loci by integrative epigenomics. Nature 2021, 590, 300–307. [Google Scholar] [CrossRef]
- Barbeira, A.N.; Dickinson, S.P.; Bonazzola, R.; Zheng, J.; Wheeler, H.E.; Torres, J.M.; Torstenson, E.S.; Shah, K.P.; Garcia, T.; Edwards, T.L.; et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 2018, 9, 1825. [Google Scholar] [CrossRef]
- Consortium, G.T. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar]
- Sollis, E.; Mosaku, A.; Abid, A.; Buniello, A.; Cerezo, M.; Gil, L.; Groza, T.; Gunes, O.; Hall, P.; Hayhurst, J.; et al. The NHGRI-EBI GWAS Catalog: Knowledgebase and deposition resource. Nucleic. Acids Res. 2023, 51, D977–D985. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Yang, J.; Zeng, J.; Goddard, M.E.; Wray, N.R.; Visscher, P.M. Concepts, estimation and interpretation of SNP-based heritability. Nat. Genet. 2017, 49, 1304–1310. [Google Scholar] [CrossRef] [PubMed]
- Michailidou, K.; Lindstrom, S.; Dennis, J.; Beesley, J.; Hui, S.; Kar, S.; Lemacon, A.; Soucy, P.; Glubb, D.; Rostamianfar, A.; et al. Association analysis identifies 65 new breast cancer risk loci. Nature 2017, 551, 92–94. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.H.; Chen, L.C.; Lee, C.J.; Huang, C.C.; Ho, Y.S.; Yang, P.S.; Ho, C.T.; Chang, H.L.; Lin, I.H.; Chang, H.W.; et al. DNA primase polypeptide 1 (PRIM1) involves in estrogen-induced breast cancer formation through activation of the G2/M cell cycle checkpoint. Int. J. Cancer 2019, 144, 615–630. [Google Scholar] [CrossRef]
- Tominaga, N.; Hagiwara, K.; Kosaka, N.; Honma, K.; Nakagama, H.; Ochiya, T. RPN2-mediated glycosylation of tetraspanin CD63 regulates breast cancer cell malignancy. Mol. Cancer 2014, 13, 134. [Google Scholar] [CrossRef]
- Justo, B.L.; Jasiulionis, M.G. Characteristics of TIMP1, CD63, and beta1-Integrin and the Functional Impact of Their Interaction in Cancer. Int. J. Mol. Sci. 2021, 22, 9319. [Google Scholar] [CrossRef] [PubMed]
- Terävä, J.; Verhassel, A.; Botti, O.; Islam, M.K.; Leivo, J.; Wittfooth, S.; Härkönen, P.; Pettersson, K.; Gidwani, K. Primary breast cancer biomarkers based on glycosylation and extracellular vesicles detected from human serum. Cancer Rep. 2022, 5, e1540. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Mao, J.; Barrero, R.A.; Wang, P.; Zhang, F.; Wang, T. Development of a CD63 Aptamer for Efficient Cancer Immunochemistry and Immunoaffinity-Based Exosome Isolation. Molecules 2020, 25, 5585. [Google Scholar] [CrossRef]
- Lee, S.; Liang, X.; Woods, M.; Reiner, A.S.; Concannon, P.; Bernstein, L.; Lynch, C.F.; Boice, J.D.; Deasy, J.O.; Bernstein, J.L.; et al. Machine learning on genome-wide association studies to predict the risk of radiation-associated contralateral breast cancer in the WECARE Study. PLoS ONE 2020, 15, e0226157. [Google Scholar] [CrossRef]
- Yamamoto, H.; Okumura, K.; Toshima, S.; Mukaisho, K.; Sugihara, H.; Hattori, T.; Kato, M.; Asano, S. FXYD3 protein involved in tumor cell proliferation is overproduced in human breast cancer tissues. Biol. Pharm. Bull. 2009, 32, 1148–1154. [Google Scholar] [CrossRef]
- Xue, Y.; Lai, L.; Lian, W.; Tu, X.; Zhou, J.; Dong, P.; Su, D.; Wang, X.; Cao, X.; Chen, Y.; et al. SOX9/FXYD3/Src Axis Is Critical for ER(+) Breast Cancer Stem Cell Function. Mol. Cancer Res. 2019, 17, 238–249. [Google Scholar] [CrossRef]
- Mavaddat, N.; Pharoah, P.D.; Michailidou, K.; Tyrer, J.; Brook, M.N.; Bolla, M.K.; Wang, Q.; Dennis, J.; Dunning, A.M.; Shah, M.; et al. Prediction of breast cancer risk based on profiling with common genetic variants. J. Natl. Cancer Inst. 2015, 107, djv036. [Google Scholar] [CrossRef] [PubMed]
- Adedokun, B.; Du, Z.; Gao, G.; Ahearn, T.U.; Lunetta, K.L.; Zirpoli, G.; Figueroa, J.; John, E.M.; Bernstein, L.; Zheng, W.; et al. Cross-ancestry GWAS meta-analysis identifies six breast cancer loci in African and European ancestry women. Nat. Commun. 2021, 12, 4198. [Google Scholar] [CrossRef] [PubMed]
- Zou, W.; Hu, C.H.; Zhou, J.P. Relationship between the expression of E-cadherin-catenins and alpha-, beta-, gamma-catenin and the metastasis and prognosis of breast cancer. Hunan Yi Ke Da Xue Xue Bao 2002, 27, 499–502. [Google Scholar] [PubMed]
- Lobo, S.; Benusiglio, P.R.; Coulet, F.; Boussemart, L.; Golmard, L.; Spier, I.; Huneburg, R.; Aretz, S.; Colas, C.; Oliveira, C. Cancer predisposition and germline CTNNA1 variants. Eur. J. Med. Genet. 2021, 64, 104316. [Google Scholar] [CrossRef]
- Clark, D.F.; Michalski, S.T.; Tondon, R.; Nehoray, B.; Ebrahimzadeh, J.; Hughes, S.K.; Soper, E.R.; Domchek, S.M.; Rustgi, A.K.; Pineda-Alvarez, D.; et al. Loss-of-function variants in CTNNA1 detected on multigene panel testing in individuals with gastric or breast cancer. Genet. Med. 2020, 22, 840–846. [Google Scholar] [CrossRef]
- Ju, R.; Wu, W.; Fei, J.; Qin, Y.; Tang, Q.; Wu, D.; Xia, Y.; Wu, J.; Wang, X. Association analysis between the polymorphisms of HSD17B5 and HSD17B6 and risk of polycystic ovary syndrome in Chinese population. Eur. J. Endocrinol. 2015, 172, 227–233. [Google Scholar] [CrossRef]
- Jones, M.R.; Mathur, R.; Cui, J.; Guo, X.; Azziz, R.; Goodarzi, M.O. Independent confirmation of association between metabolic phenotypes of polycystic ovary syndrome and variation in the type 6 17beta-hydroxysteroid dehydrogenase gene. J. Clin. Endocrinol. Metab. 2009, 94, 5034–5038. [Google Scholar] [CrossRef]
- Burgdorf, K.S.; Gjesing, A.P.; Grarup, N.; Justesen, J.M.; Sandholt, C.H.; Witte, D.R.; Jorgensen, T.; Madsbad, S.; Hansen, T.; Pedersen, O. Association studies of novel obesity-related gene variants with quantitative metabolic phenotypes in a population-based sample of 6,039 Danish individuals. Diabetologia 2012, 55, 105–113. [Google Scholar] [CrossRef]
- Principe, C.; Dionisio de Sousa, I.J.; Prazeres, H.; Soares, P.; Lima, R.T. LRP1B: A Giant Lost in Cancer Translation. Pharmaceuticals 2021, 14, 836. [Google Scholar] [CrossRef]
- Hu, H.; Wang, Y.; Zhang, T.; Zhang, C.; Liu, Y.; Li, G.; Zhou, D.; Lu, S. Association of LncRNA-GACAT3 with MRI features of breast cancer and its molecular mechanism. J. BUON 2019, 24, 2377–2384. [Google Scholar]
- Zhong, H.; Yang, J.; Zhang, B.; Wang, X.; Pei, L.; Zhang, L.; Lin, Z.; Wang, Y.; Wang, C. LncRNA GACAT3 predicts poor prognosis and promotes cell proliferation in breast cancer through regulation of miR-497/CCND2. Cancer Biomark. 2018, 22, 787–797. [Google Scholar] [CrossRef] [PubMed]
- Borneman, R.M.; Gavin, E.; Musiyenko, A.; Richter, W.; Lee, K.J.; Crossman, D.K.; Andrews, J.F.; Wilhite, A.M.; McClellan, S.; Aragon, I.; et al. Phosphodiesterase 10A (PDE10A) as a novel target to suppress beta-catenin and RAS signaling in epithelial ovarian cancer. J. Ovarian Res. 2022, 15, 120. [Google Scholar] [CrossRef] [PubMed]
- De, S.; Cipriano, R.; Jackson, M.W.; Stark, G.R. Overexpression of kinesins mediates docetaxel resistance in breast cancer cells. Cancer Res. 2009, 69, 8035–8042. [Google Scholar] [CrossRef]
- Fang, W.J.; Zheng, Y.; Wu, L.M.; Ke, Q.H.; Shen, H.; Yuan, Y.; Zheng, S.S. Genome-wide analysis of aberrant DNA methylation for identification of potential biomarkers in colorectal cancer patients. Asian Pac. J. Cancer Prev. 2012, 13, 1917–1921. [Google Scholar] [CrossRef] [PubMed]
- Homer, M.V.; Charo, L.M.; Natarajan, L.; Haunschild, C.; Chung, K.; Mao, J.J.; DeMichele, A.M.; Su, H.I. Genetic variants of age at menopause arhe not related to timing of ovarian failure in breast cancer survivors. Menopause 2017, 24, 663–668. [Google Scholar] [CrossRef]
| Characteristic | All Self-Reported Black or African American Women, N = 10,090 1 | Study Cohort, N = 1333 |
|---|---|---|
| Breast Area (cm2), Mean (SD) | 206.71 (81.80) | 217.63 (85.08) |
| Area Percent Density (%), Mean (SD) | 13.89 (11.40) | 11.52 (6.65) |
| Dense Area (cm2), Mean (SD) | 26.30 (28.28) | 22.52 (11.06) |
| Age, Mean (SD) | 56.45 (10.57) | 56.65 (9.80) |
| Atypical Hyperplasia, n (%) | 26 (0.26) | 4 (0.3) |
| Body Mass Index (Kg/m2), Mean (SD) | 32.44 (7.40) | 33.87 (7.85) |
| Menopausal Status, n (%) | ||
| Postmenopausal | 6926 (68.64) | 963 (72.2) |
| Premenopausal | 3164 (31.36) | 370 (27.8) |
| Breast Density BI-RADS Categories, n (%) | ||
| Almost entirely fat | 1810 (17.94) | 269 (20.2) |
| Scattered fibroglandular tissue | 6053 (59.99) | 814 (61.2) |
| Heterogeneously dense | 2111 (20.92) | 240 (18.0) |
| Extremely dense | 91 (0.90) | 7 (0.5) |
| Missing/Unknown | 25 (0.25) | 3 (0.2) |
| Cancer Diagnosis After Screening, n (%) | 171 (1.69) | 33 (2.5) |
| Number of Biopsies, n (%) | ||
| None | 8206 (81.33) | 1053 (79.0) |
| One | 1489 (14.76) | 217 (16.3) |
| Two or more | 395 (3.91) | 63 (4.7) |
| Age at Menarche, n (%) | ||
| <12 years | 2129 (21.10) | 318 (23.9) |
| 12–13 years | 4358 (43.19) | 550 (41.3) |
| 14+ years | 2239 (22.19) | 306 (23.0) |
| Unknown | 1364 (13.52) | 159 (11.9) |
| Age at First Live Birth, n (%) | ||
| No Births | 1495 (14.82) | 187 (14.0) |
| <20 | 3536 (35.04) | 524 (39.3) |
| 20–24 | 2504 (24.82) | 354 (26.6) |
| 25–29 | 1272 (12.61) | 145 (10.9) |
| 30+ | 650 (6.44) | 72 (5.4) |
| Birth age missing | 633 (6.27) | 51 (3.8) |
| Family History of Breast Cancer, n (%) | ||
| None | 8788 (87.10) | 1171 (87.8) |
| One relative | 1147 (11.37) | 145 (10.9) |
| Two or more relatives | 155 (1.54) | 17 (1.3) |
| Snpid | Variant_ID | Trait | Additional #SNPS in Loci | SNP2 | Gene | p-Value |
|---|---|---|---|---|---|---|
| RS75413938 | chr1:55151613:T:G | Dense Area | 29 | 1:55066599(1),1:55069787(1),1:55078520(1),1:55079038(1),1:55079364(1),1:55091687(1),1:55096857(1),1:55099538(1),1:55103811(1),1:55108626(1),1:55120773(1),1:55126542(1),1:55128390(1),1:55128925(1),1:55128926(1),1:55134044(1),1:55155440(1),1:55161852(1),1:55167712(1),1:55177218(1),1:55183862(1),1:55189932(1),1:55193134(1),1:55194898(1),1:55195273(1),1:55195399(1),1:55203473(1),1:55217682(1),1:55226938(1) | USP24 | 8.92 × 10−9 |
| RS79314581 | chr1:244097544:G:C | Dense Area | 0 | NONE | LOC122152354/ RN7SL148P | 1.22 × 10−8 |
| RS190395094 | chr1:27735235:C:T | Dense Area | 8 | 1:27646562(1),1:27649415(1),1:27657130(1),1:27674922(1),1:27694043(1),1:27725469(1),1:27820501(1),1:27823924(1) | FAM76A | 2.64 × 10−8 |
| RS60005977 | chr1:75838225:C:T | Dense Area | 0 | NONE | MSH4 | 2.77 × 10−8 |
| RS78168242 | chr1:202232716:C:T | Dense Area | 2 | 1:202217643(1),1:202220169(1) | LGR6 | 3.73 × 10−8 |
| RS142447005 | chr2:16941818:G:T | Dense Area | 0 | NONE | LINC01866 | 6.06 × 10−13 |
| RS139721819 | chr4:39529250:C:G | Dense Area | 6 | 4:39487863(1),4:39494788(1),4:39497063(1),4:39523173(1),4:39528094(1),4:39608882(1) | UGDH-AS1 | 1.45 × 10−11 |
| RS191367039 | chr4:148052195:G:A | Dense Area | 4 | 4:148066298(1),4:148070999(1),4:148125334(1),4:148134899(1) | ARHGAP10 | 1.40 × 10−10 |
| RS147570646 | chr4:179504076:C:T | Dense Area | 1 | 4:179560038(1) | LOC105377563/ LOC124900818 | 2.73 × 10−9 |
| RS58470658 | chr4:148160004:C:T | Dense Area | 1 | 4:148166598(1) | NR3C2 | 4.57 × 10−9 |
| RS116045382 | chr4:11398948:C:T | Dense Area | 0 | NONE | HS3ST1 | 6.09 × 10−9 |
| RS143507397 | chr4:168554153:G:A | Dense Area | 0 | NONE | PALLD | 1.89 × 10−8 |
| RS186021694 | chr4:173988596:A:G | Dense Area | 3 | 4:173962614(1),4:173969459(1),4:173982430(1) | LOC105377543 | 3.22 × 10−8 |
| RS113187843 | chr5:138964719:T:A | Dense Area | 0 | NONE | SIL1 | 2.46 × 10−9 |
| RS112818595 | chr5:138863360:C:T | Dense Area | 2 | 5:138813316(1),5:138850667(1) | CTNNA1 | 5.39 × 10−9 |
| RS6912620 | chr6:132552803:G:A | Dense Area | 6 | 6:132460453(1),6:132479524(1),6:132480587(1),6:132551032(1),6:132551150(1),6:132592333(1) | TAAR8 | 1.08 × 10−10 |
| RS59162058 | chr7:5603217:C:T | Dense Area | 2 | 7:5644533(1),7:5651116(1) | FSCN1 | 1.97 × 10−8 |
| RS1657248 | chr7:155112424:G:A | Dense Area | 2 | 7:155112360(1),7:155113278(1) | HTR5A | 3.64 × 10−8 |
| RS114517045 | chr8:77390103:T:G | Dense Area | 5 | 8:77439758(1),8:77447273(1),8:77460647(1),8:77474685(1),8:77485921(1) | LOC105375909 | 2.20 × 10−8 |
| RS189070945 | chr10:66699780:C:T | Dense Area | 11 | 10:66669847(1),10:66681835(1),10:66694622(1),10:66716457(1),10:66717173(1),10:66720546(1),10:66721909(1),10:66733490(1),10:66739808(1),10:66742213(1),10:66745040(1) | CTNNA3 | 1.76 × 10−12 |
| RS59522962 | chr10:66806298:T:C | Dense Area | 1 | 10:66790114(1) | CTNNA3 | 1.58 × 10−8 |
| RS79232684 | chr11:47150163:G:A | Dense Area | 0 | NONE | CSTPP1 | 1.18 × 10−8 |
| RS114694584 | chr11:31973064:A:G | Dense Area | 6 | 11:31930445(1),11:31935750(1),11:31939541(1),11:31948997(1),11:31955129(1),11:31986978(1) | LOC110120941/ LOC107984420 | 1.72 × 10−8 |
| RS75439556 | chr11:46775991:C:A | Dense Area | 1 | 11:46802307(1) | CKAP5 | 2.95 × 10−8 |
| RS184938993 | chr12:56780836:G:A | Dense Area | 1 | 12:56780646(1) | HSD17B6 | 4.02 × 10−12 |
| RS150208861 | chr14:97883545:C:T | Dense Area | 0 | NONE | LINC01550 | 1.61 × 10−9 |
| RS150796751 | chr17:38494521:G:A | Dense Area | 0 | NONE | ARHGAP23 | 3.82 × 10−8 |
| RS143065709 | chr18:57155409:C:T | Dense Area | 2 | 18:57148687(1),18:57168501(1) | BOD1L2/LINC02565 | 4.17 × 10−10 |
| RS73886707 | chr22:46112492:C:T | Dense Area | 3 | 22:46105565(1),22:46109178(1),22:46109365(1) | MIRLET7BHG | 2.15 × 10−9 |
| RS148811443 | chr2:142145550:G:A | Percent Density | 7 | 2:142055838(1),2:142158131(1),2:142161710(1),2:142167799(1),2:142215347(1),2:142218937(1),2:142242824(1) | LRP1B | 1.56 × 10−8 |
| RS79331071 | chr4:166343624:C:T | Percent Density | 26 | 4:166296011(1),4:166296276(1),4:166296612(1),4:166308772(1),4:166311509(1),4:166322829(1),4:166331896(1),4:166337499(1),4:166340457(1),4:166342254(1),4:166343104(1),4:166343888(1),4:166344382(1),4:166345601(1),4:166345825(1),4:166349179(1),4:166350417(1),4:166350715(1),4:166351544(1),4:166351573(1),4:166352076(1),4:166353507(1),4:166355475(1),4:166376134(1),4:166409358(1),4:166415758(1) | LOC121056748/ LOC121056749 | 4.41 × 10−9 |
| RS138783664 | chr9:114879160:C:T | Percent Density | 1 | 9:114917870(1) | LOC645266/ LOC124310630 | 1.27 × 10−8 |
| RS143877555 | chr10:84298935:C:T | Percent Density | 5 | 10:84294546(1),10:84344702(1),10:84348787(1),10:84352282(1),10:84366026(1) | LINC00858/CCSER2 | 1.01 × 10−8 |
| RS145826214 | chr13:99921286:C:T | Percent Density | 3 | 13:99908944(1),13:99916943(1),13:99920000(1) | CLYBL-AS3 | 2.71 × 10−8 |
| Trait | Gene | Annotation | SNPID | PIP |
|---|---|---|---|---|
| Dense Area | PDE10A | Het | rs480268 | 0.98738 |
| PDE10A | Het | rs6907588 | 0.98244 | |
| PDE10A | Het | rs576853 | 0.99956 | |
| PDE10A | Het | rs481701 | 1 | |
| LOC101927404 | rs9967157 | 1 | ||
| LOC101927404 | rs716961 | 1 | ||
| LOC101927404/LOC105372168 | rs34217531 | 1 | ||
| LOC101927404/LOC105372168 | rs1306871 | 1 | ||
| LOC105372168 | rs9954012 | 1 | ||
| LOC105372168 | ncRNA_gene | rs611750 | 1 | |
| LOC105372310 | Het | rs10412042 | 1 | |
| LOC105372310 | Het | rs12462802 | 1 | |
| LOC105372310 | Het | rs7253843 | 1 | |
| LOC105372310 | Het | rs4277458 | 1 | |
| LOC100129265/BNIP3P19 | Het | rs28493283 | 1 | |
| BNIP3P19/BNIP3P20 | Het | rs10426611 | 1 | |
| BNIP3P20/BNIP3P21 | pseudogene | rs9989730 | 1 | |
| BNIP3P20/BNIP3P21 | pseudogene | rs28786195 | 1 | |
| Percent Density | SH3GL3 | ncRNA_gene | rs10906974 | 1 |
| SH3GL3 | ncRNA_gene | rs301847 | 1 | |
| SH3GL3 | ncRNA_gene | rs6602974 | 1 | |
| SH3GL3 | lnc_RNA | rs55641568 | 1 | |
| SH3GL3 | lnc_RNA | rs11853676 | 1 | |
| SH3GL3 | lnc_RNA | rs7350762 | 1 | |
| SH3GL3 | lnc_RNA | rs12905964 | 1 | |
| KIFC3 | mRNA | rs1582594 | 1 | |
| KIFC3 | mRNA | rs2967139 | 1 | |
| KIFC3 | EnhA1 | rs4784864 | 1 | |
| KIFC3 | EnhA1 | rs2911348 | 1 | |
| KIFC3 | EnhA2 | rs2967137 | 1 | |
| KIFC3 | EnhA2 | rs59350294 | 1 | |
| KIFC3 | mRNA | rs140234666 | 1 | |
| KIFC3 | Het | rs9938048 | 0.99999 | |
| KIFC3/CNGB1 | EnhA1 | rs838583 | 1 | |
| CNGB1 | mRNA | rs691656 | 1 | |
| Breast Area | Percent Density | Dense Area | |||||||
|---|---|---|---|---|---|---|---|---|---|
| A1_FREQ | BETA | p | A1_FREQ | BETA | p | A1_FREQ | BETA | p | |
| rs11205303 | 0.078 | −0.047 | 0.447 | 0.077 | −0.053 | 0.481 | 0.077 | −0.083 | 0.310 |
| rs1868992 | 0.412 | 0.058 | 0.160 | 0.413 | 0.030 | 0.547 | 0.412 | 0.048 | 0.370 |
| rs17625845 | 0.069 | 0.154 | 0.019 | 0.068 | 0.055 | 0.486 | 0.068 | 0.103 | 0.229 |
| rs6851733 | 0.093 | −0.038 | 0.496 | 0.095 | 0.018 | 0.786 | 0.094 | −0.016 | 0.824 |
| rs413472 | 0.266 | 0.057 | 0.123 | 0.266 | −0.045 | 0.311 | 0.266 | −0.028 | 0.557 |
| rs335189 | 0.167 | −0.004 | 0.927 | 0.167 | 0.032 | 0.533 | 0.168 | 0.093 | 0.093 |
| rs11745230 | 0.402 | 0.021 | 0.521 | 0.401 | −0.016 | 0.696 | 0.402 | 0.008 | 0.859 |
| rs2112670 | 0.171 | −0.035 | 0.410 | 0.171 | −0.026 | 0.612 | 0.171 | −0.054 | 0.327 |
| rs2042239 | 0.154 | −0.013 | 0.766 | 0.152 | −0.074 | 0.168 | 0.154 | −0.061 | 0.289 |
| rs3819405 | 0.460 | −0.003 | 0.936 | 0.456 | −0.125 | 0.002 | 0.456 | −0.106 | 0.015 |
| rs4897107 | 0.159 | −0.049 | 0.257 | 0.160 | −0.055 | 0.287 | 0.160 | −0.153 | 0.007 |
| rs9397436 | 0.069 | 0.043 | 0.490 | 0.068 | −0.048 | 0.524 | 0.068 | 0.077 | 0.344 |
| rs16885613 | 0.356 | −0.115 | 0.001 | 0.356 | 0.197 | 8.07 × 10−7 | 0.357 | 0.117 | 0.007 |
| rs10087804 | 0.296 | −0.113 | 0.001 | 0.296 | 0.209 | 9.17 × 10−7 | 0.297 | 0.146 | 0.002 |
| rs58847541 | 0.290 | −0.009 | 0.792 | 0.288 | −0.036 | 0.384 | 0.290 | −0.017 | 0.700 |
| rs2138555 | 0.315 | −0.032 | 0.355 | 0.318 | −0.034 | 0.416 | 0.318 | −0.047 | 0.295 |
| rs10995187 | 0.071 | −0.018 | 0.773 | 0.070 | −0.012 | 0.876 | 0.070 | 0.014 | 0.864 |
| rs4980383 | 0.274 | −0.002 | 0.956 | 0.275 | 0.055 | 0.194 | 0.275 | 0.020 | 0.671 |
| rs11836164 | 0.221 | 0.026 | 0.510 | 0.221 | −0.003 | 0.953 | 0.221 | 0.011 | 0.835 |
| rs7297051 | 0.141 | −0.004 | 0.929 | 0.141 | 0.010 | 0.852 | 0.142 | 0.014 | 0.802 |
| rs61938093 | 0.209 | 0.132 | 0.001 | 0.211 | −0.107 | 0.022 | 0.210 | −0.038 | 0.451 |
| rs4499190 | 0.150 | −0.070 | 0.137 | 0.151 | −0.050 | 0.377 | 0.151 | −0.089 | 0.145 |
| rs11646715 | 0.166 | −0.051 | 0.234 | 0.168 | 0.029 | 0.573 | 0.168 | −0.005 | 0.931 |
| rs12462111 | 0.169 | 0.063 | 0.139 | 0.169 | −0.022 | 0.658 | 0.169 | 0.033 | 0.549 |
| rs1231281 | 0.263 | 0.038 | 0.306 | 0.263 | 0.025 | 0.580 | 0.263 | 0.040 | 0.412 |
| rs17789629 | 0.040 | −0.039 | 0.639 | 0.041 | 0.132 | 0.185 | 0.040 | −0.022 | 0.836 |
| rs34066050 | 0.209 | −0.018 | 0.661 | 0.208 | 0.012 | 0.799 | 0.209 | 0.037 | 0.476 |
| rs73169097 | 0.056 | −0.104 | 0.132 | 0.057 | −0.016 | 0.843 | 0.057 | −0.045 | 0.615 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verma, S.S.; Guare, L.; Ehsan, S.; Gastounioti, A.; Scales, G.; Ritchie, M.D.; Kontos, D.; McCarthy, A.M.; Penn Medicine Biobank. Genome-Wide Association Study of Breast Density among Women of African Ancestry. Cancers 2023, 15, 2776. https://doi.org/10.3390/cancers15102776
Verma SS, Guare L, Ehsan S, Gastounioti A, Scales G, Ritchie MD, Kontos D, McCarthy AM, Penn Medicine Biobank. Genome-Wide Association Study of Breast Density among Women of African Ancestry. Cancers. 2023; 15(10):2776. https://doi.org/10.3390/cancers15102776
Chicago/Turabian StyleVerma, Shefali Setia, Lindsay Guare, Sarah Ehsan, Aimilia Gastounioti, Gabrielle Scales, Marylyn D. Ritchie, Despina Kontos, Anne Marie McCarthy, and Penn Medicine Biobank. 2023. "Genome-Wide Association Study of Breast Density among Women of African Ancestry" Cancers 15, no. 10: 2776. https://doi.org/10.3390/cancers15102776
APA StyleVerma, S. S., Guare, L., Ehsan, S., Gastounioti, A., Scales, G., Ritchie, M. D., Kontos, D., McCarthy, A. M., & Penn Medicine Biobank. (2023). Genome-Wide Association Study of Breast Density among Women of African Ancestry. Cancers, 15(10), 2776. https://doi.org/10.3390/cancers15102776








