Development of a Machine Learning Model to Predict Recurrence of Oral Tongue Squamous Cell Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Seer Database Query (Data Source)
2.2. Patient Grouping and Feature Extraction
- The oral tongue should be the primary site of the first case for each patient.
- All cases corresponding to patients with missing or unknown values for any variable critical for analysis, including the Total Number of Malignant Tumors, Sequence Number, Survival Months, and Year of Diagnosis, were filtered out.
2.3. ML Training & Validation (Balancing, under Sampling, Number of Runs and Distribution of Data)
3. Results
3.1. Study Population Characteristics and Cancer Recurrence Information
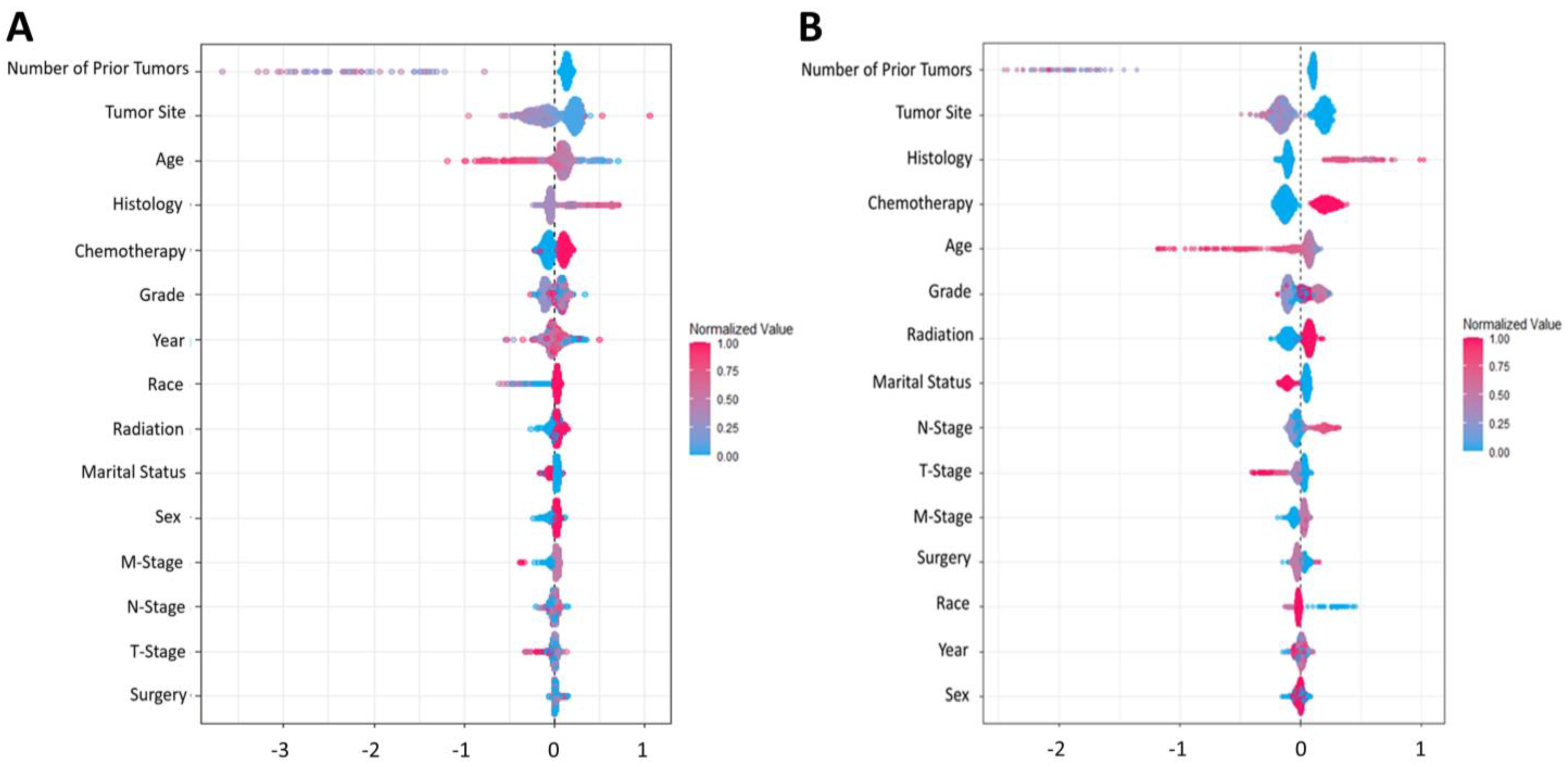
3.2. Model Prediction and Development (Performance Metric for the Algorithm)
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Noone, A.M.; Howlader, N.; Krapcho, M.; Miller, D.; Brest, A.; Yu, M.; Cronin, K.A. SEER Cancer Statistics Review; National Cancer Institute: Bethesda, MD, USA, 2018; pp. 1975–2015. [Google Scholar]
- Kim, Y.-J.; Kim, J.H. Increasing Incidence and Improving Survival of Oral Tongue Squamous Cell Carcinoma. Sci. Rep. 2020, 10, 7877. [Google Scholar] [CrossRef]
- Patel, S.C.; Carpenter, W.R.; Tyree, S.; Couch, M.E.; Weissler, M.; Hackman, T.; Hayes, D.N.; Shores, C.; Chera, B.S. Increasing Incidence of Oral Tongue Squamous Cell Carcinoma in Young White Women, Age 18 to 44 Years. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2011, 29, 1488–1494. [Google Scholar] [CrossRef] [PubMed]
- Warnakulasuriya, S. Global Epidemiology of Oral and Oropharyngeal Cancer. Oral Oncol. 2009, 45, 309–316. [Google Scholar] [CrossRef]
- Mukdad, L.; Heineman, T.E.; Alonso, J.; Badran, K.W.; Kuan, E.C.; St John, M.A. Oral Tongue Squamous Cell Carcinoma Survival as Stratified by Age and Sex: A Surveillance, Epidemiology, and End Results Analysis. Laryngoscope 2019, 129, 2076–2081. [Google Scholar] [CrossRef] [PubMed]
- Camisasca, D.R.; Silami, M.A.N.C.; Honorato, J.; Dias, F.L.; de Faria, P.A.S.; Lourenço, S. de Q.C. Oral Squamous Cell Carcinoma: Clinicopathological Features in Patients with and without Recurrence. ORL J. Oto-Rhino-Laryngol. Its Relat. Spec. 2011, 73, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Ermer, M.A.; Kirsch, K.; Bittermann, G.; Fretwurst, T.; Vach, K.; Metzger, M.C. Recurrence Rate and Shift in Histopathological Differentiation of Oral Squamous Cell Carcinoma—A Long-Term Retrospective Study over a Period of 13.5 Years. J. Cranio-Maxillo-Facial Surg. Off. Publ. Eur. Assoc. Cranio-Maxillo-Facial Surg. 2015, 43, 1309–1313. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhang, S.; Yue, K.; Wang, X.-D. The Recurrence and Survival of Oral Squamous Cell Carcinoma: A Report of 275 Cases. Chin. J. Cancer 2013, 32, 614–618. [Google Scholar] [CrossRef]
- Yuan, Q.; Cai, T.; Hong, C.; Du, M.; Johnson, B.E.; Lanuti, M.; Cai, T.; Christiani, D.C. Performance of a Machine Learning Algorithm Using Electronic Health Record Data to Identify and Estimate Survival in a Longitudinal Cohort of Patients with Lung Cancer. JAMA Netw. Open 2021, 4, e2114723. [Google Scholar] [CrossRef] [PubMed]
- Ko, C.; Brody, J.P. A Genetic Risk Score for Glioblastoma Multiforme Based on Copy Number Variations. Cancer Treat. Res. Commun. 2021, 27, 100352. [Google Scholar] [CrossRef]
- Karatza, P.; Dalakleidi, K.; Athanasiou, M.; Nikita, K.S. Interpretability Methods of Machine Learning Algorithms with Applications in Breast Cancer Diagnosis. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine & Biology Society, Virtual, 1–5 November 2021; Volume 2021, pp. 2310–2313. [Google Scholar] [CrossRef]
- Howard, F.M.; Kochanny, S.; Koshy, M.; Spiotto, M.; Pearson, A.T. Machine Learning-Guided Adjuvant Treatment of Head and Neck Cancer. JAMA Netw. Open 2020, 3, e2025881. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.Q.; Gardiner, L.; Wang, H.; Hueman, M.T.; Chen, D. Creating Prognostic Systems for Well-Differentiated Thyroid Cancer Using Machine Learning. Front. Endocrinol. 2019, 10, 288. [Google Scholar] [CrossRef] [PubMed]
- Toh, C.; Brody, J.P. Genetic Risk Score for Ovarian Cancer Based on Chromosomal-Scale Length Variation. BioData Min. 2021, 14, 18. [Google Scholar] [CrossRef]
- Zhou, C.; Hu, J.; Wang, Y.; Ji, M.-H.; Tong, J.; Yang, J.-J.; Xia, H. A Machine Learning-Based Predictor for the Identification of the Recurrence of Patients with Gastric Cancer after Operation. Sci. Rep. 2021, 11, 1571. [Google Scholar] [CrossRef]
- Zhao, M.; Tang, Y.; Kim, H.; Hasegawa, K. Machine Learning with K-Means Dimensional Reduction for Predicting Survival Outcomes in Patients with Breast Cancer. Cancer Inform. 2018, 17, 1176935118810215. [Google Scholar] [CrossRef]
- Lynch, C.M.; Abdollahi, B.; Fuqua, J.D.; de Carlo, A.R.; Bartholomai, J.A.; Balgemann, R.N.; van Berkel, V.H.; Frieboes, H.B. Prediction of Lung Cancer Patient Survival via Supervised Machine Learning Classification Techniques. Int. J. Med. Inform. 2017, 108, 1–8. [Google Scholar] [CrossRef]
- Chu, C.S.; Lee, N.P.; Adeoye, J.; Thomson, P.; Choi, S.-W. Machine Learning and Treatment Outcome Prediction for Oral Cancer. J. Oral Pathol. Med. Off. Publ. Int. Assoc. Oral Pathol. Am. Acad. Oral Pathol. 2020, 49, 977–985. [Google Scholar] [CrossRef]
- Sarkar, S.; Min, K.; Ikram, W.; Tatton, R.W.; Riaz, I.B.; Silva, A.C.; Bryce, A.H.; Moore, C.; Ho, T.H.; Sonpavde, G.; et al. Performing Automatic Identification and Staging of Urothelial Carcinoma in Bladder Cancer Patients Using a Hybrid Deep-Machine Learning Approach. Cancers 2023, 15, 1673. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, A.; Wolsztynski, E.; Cronin, M.; Moghaddam, S. Improving the Post-Operative Prediction of BCR-Free Survival Time with MRNA Variables and Machine Learning. Cancers 2023, 15, 1276. [Google Scholar] [CrossRef] [PubMed]
- Umer, M.; Naveed, M.; Alrowais, F.; Ishaq, A.; Hejaili, A.A.; Alsubai, S.; Eshmawi, A.; Mohamed, A.; Ashraf, I. Breast Cancer Detection Using Convoluted Features and Ensemble Machine Learning Algorithm. Cancers 2022, 14, 6015. [Google Scholar] [CrossRef]
- Alabi, R.O.; Elmusrati, M.; Sawazaki-Calone, I.; Kowalski, L.P.; Haglund, C.; Coletta, R.D.; Mäkitie, A.A.; Salo, T.; Almangush, A.; Leivo, I. Comparison of Supervised Machine Learning Classification Techniques in Prediction of Locoregional Recurrences in Early Oral Tongue Cancer. Int. J. Med. Inform. 2020, 136, 104068. [Google Scholar] [CrossRef]
- Karadaghy, O.A.; Shew, M.; New, J.; Bur, A.M. Development and Assessment of a Machine Learning Model to Help Predict Survival Among Patients with Oral Squamous Cell Carcinoma. JAMA Otolaryngol.–Head Neck Surg. 2019, 145, 1115–1120. [Google Scholar] [CrossRef] [PubMed]
- Chernock, R.D. Morphologic Features of Conventional Squamous Cell Carcinoma of the Oropharynx: “keratinizing” and “Nonkeratinizing” Histologic Types as the Basis for a Consistent Classification System. Head Neck Pathol. 2012, 6 (Suppl. S1), S41–S47. [Google Scholar] [CrossRef] [PubMed]
- Gijsbers, P.; LeDell, E.; Thomas, J.; Poirier, S.; Bischl, B.; Vanschoren, J. An Open Source AutoML Benchmark. arXiv 2019, arXiv:1907.00909. [Google Scholar]
- Caruana, R.; Niculescu-Mizil, A. An Empirical Comparison of Supervised Learning Algorithms. In Proceedings of the 23rd International Conference on Machine Learning, New York, NY, USA, 25–29 June 2006; pp. 161–168. [Google Scholar]
- H2O.ai Generalized Linear Model (GLM)–H2O Documentation. Available online: https://docs.h2o.ai/h2o/latest-stable/h2o-docs/data-science/glm.html (accessed on 12 February 2021).
- H2O.ai Distributed Random Forest (DRF)–H2O Documentation. Available online: https://docs.h2o.ai/h2o/latest-stable/h2o-docs/data-science/drf.html (accessed on 12 February 2021).
- H2O.ai Deep Learning Neural Networks–H2O Documentation. Available online: https://docs.h2o.ai/h2o/latest-stable/h2o-docs/data-science/deep-learning.html%0A (accessed on 12 February 2021).
- H2O.ai Gradient Boosting Machine (GBM)–H2O Documentation. Available online: https://docs.h2o.ai/h2o/latest-stable/h2o-docs/data-science/gbm.html (accessed on 12 February 2021).
- Friedman, J.H. Greedy Function Approximation: A Gradient Boosting Machine. Ann. Stat. 2001, 29, 1189–1232. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority over-Sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Garcia-Pedrajas, N.; Herrera, F.; Fyfe, C.; Sánchez, J.M.B.; Ali, M. Trends in Applied Intelligent Systems. In Proceedings of the 23rd International Conference on Industrial Engineering and Other Applications of Applied Intelligent Systems, IEA/AIE 2010, Cordoba, Spain, 1–4 June 2010; Volume 6097. [Google Scholar]
- Lundberg, S.M.; Lee, S.-I. A Unified Approach to Interpreting Model Predictions. In Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 4768–4777. [Google Scholar]
- An, S.-Y.; Jung, E.-J.; Lee, M.; Kwon, T.-K.; Sung, M.-W.; Jeon, Y.K.; Kim, K.H. Factors Related to Regional Recurrence in Early Stage Squamous Cell Carcinoma of the Oral Tongue. Clin. Exp. Otorhinolaryngol. 2008, 1, 166–170. [Google Scholar] [CrossRef]
- Sharma, P.; Shah, S.V.; Taneja, C.; Patel, A.M.; Patel, M.D. A Prospective Study of Prognostic Factors for Recurrence in Early Oral Tongue Cancer. J. Clin. Diagn. Res. JCDR 2013, 7, 2559–2562. [Google Scholar] [CrossRef]
- Berdugo, J.; Thompson, L.D.R.; Purgina, B.; Sturgis, C.D.; Tuluc, M.; Seethala, R.; Chiosea, S.I. Measuring Depth of Invasion in Early Squamous Cell Carcinoma of the Oral Tongue: Positive Deep Margin, Extratumoral Perineural Invasion, and Other Challenges. Head Neck Pathol. 2019, 13, 154–161. [Google Scholar] [CrossRef]
- Safi, A.-F.; Kauke, M.; Grandoch, A.; Nickenig, H.-J.; Zöller, J.E.; Kreppel, M. Analysis of Clinicopathological Risk Factors for Locoregional Recurrence of Oral Squamous Cell Carcinoma–Retrospective Analysis of 517 Patients. J. Cranio-Maxillo-Facial Surg. Off. Publ. Eur. Assoc. Cranio-Maxillo-Facial Surg. 2017, 45, 1749–1753. [Google Scholar] [CrossRef]
- Vázquez-Mahía, I.; Seoane, J.; Varela-Centelles, P.; Tomás, I.; Álvarez García, A.; López Cedrún, J.L. Predictors for Tumor Recurrence after Primary Definitive Surgery for Oral Cancer. J. Oral Maxillofac. Surg. Off. J. Am. Assoc. Oral Maxillofac. Surg. 2012, 70, 1724–1732. [Google Scholar] [CrossRef]
- Lacko, M.; Braakhuis, B.J.M.; Sturgis, E.M.; Boedeker, C.C.; Suárez, C.; Rinaldo, A.; Ferlito, A.; Takes, R.P. Genetic Susceptibility to Head and Neck Squamous Cell Carcinoma. Int. J. Radiat. Oncol. Biol. Phys. 2014, 89, 38–48. [Google Scholar] [CrossRef]
- Copper, M.P.; Jovanovic, A.; Nauta, J.J.P.; Braakhuis, B.J.M.; de Vries, N.; van der Waal, I.; Snow, G.B. Role of Genetic Factors in the Etiology of Squamous Cell Carcinoma of the Head and Neck. Arch. Otolaryngol.–Head Neck Surg. 1995, 121, 157–160. [Google Scholar] [CrossRef]
- Matthias, C.; Harréus, U.; Strange, R. Influential Factors on Tumor Recurrence in Head and Neck Cancer Patients. Eur. Arch. Oto-Rhino-Laryngol. Head Neck 2006, 263, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Heroiu Cataloiu, A.-D.; Danciu, C.E.; Popescu, C.R. Multiple Cancers of the Head and Neck. Maedica 2013, 8, 80–85. [Google Scholar] [PubMed]
- Jerjes, W.; Upile, T.; Petrie, A.; Riskalla, A.; Hamdoon, Z.; Vourvachis, M.; Karavidas, K.; Jay, A.; Sandison, A.; Thomas, G.J.; et al. Clinicopathological Parameters, Recurrence, Locoregional and Distant Metastasis in 115 T1-T2 Oral Squamous Cell Carcinoma Patients. Head Neck Oncol. 2010, 2, 9. [Google Scholar] [CrossRef]
- Wolfer, S.; Elstner, S.; Schultze-Mosgau, S. Degree of Keratinization Is an Independent Prognostic Factor in Oral Squamous Cell Carcinoma. J. Oral Maxillofac. Surg. 2018, 76, 444–454. [Google Scholar] [CrossRef]
- Sinha, N.; Rigby, M.H.; McNeil, M.L.; Taylor, S.M.; Trites, J.R.B.; Hart, R.D.; Bullock, M.J. The Histologic Risk Model Is a Useful and Inexpensive Tool to Assess Risk of Recurrence and Death in Stage I or II Squamous Cell Carcinoma of Tongue and Floor of Mouth. Mod. Pathol. 2018, 31, 772–779. [Google Scholar] [CrossRef]
- Brandwein-Gensler, M.; Teixeira, M.S.; Lewis, C.M.; Lee, B.; Rolnitzky, L.; Hille, J.J.; Genden, E.; Urken, M.L.; Wang, B.Y. Oral Squamous Cell Carcinoma: Histologic Risk Assessment, but Not Margin Status, Is Strongly Predictive of Local Disease-Free and Overall Survival. Am. J. Surg. Pathol. 2005, 29, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, A.; Husain, N.; Misra, S.; Kumar, V.; Gupta, S.; Akhtar, N.; Lakshmanan, M.; Garg, S.; Arora, A.; Jain, K. Validation of the Brandwein Gensler Risk Model in Patients of Oral Cavity Squamous Cell Carcinoma in North India. Head Neck Pathol. 2020, 14, 616–622. [Google Scholar] [CrossRef]
- El-Mofty, S.K. Histopathologic Risk Factors in Oral and Oropharyngeal Squamous Cell Carcinoma Variants: An Update with Special Reference to HPV-Related Carcinomas. Med. Oral Patol. Oral Y Cir. Bucal 2014, 19, e377–e385. [Google Scholar] [CrossRef]
- O-charoenrat, P.; Pillai, G.; Patel, S.; Fisher, C.; Archer, D.; Eccles, S.; Rhys-Evans, P. Tumour Thickness Predicts Cervical Nodal Metastases and Survival in Early Oral Tongue Cancer. Oral Oncol. 2003, 39, 386–390. [Google Scholar] [CrossRef] [PubMed]
- H2O.ai XGBoost–H2O Documentation. Available online: https://docs.h2o.ai/h2o/latest-stable/h2o-docs/data-science/xgboost.html (accessed on 12 February 2021).
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. arXiv 2016, arXiv:1603.02754. [Google Scholar]
- Wang, C.; Deng, C.; Wang, S. Imbalance-XGBoost: Leveraging Weighted and Focal Losses for Binary Label-Imbalanced Classification with XGBoost. Pattern Recognit. Lett. 2020, 136, 190–197. [Google Scholar] [CrossRef]


| Variable | 5-Year (N = 14,995) | 10-Year (N = 7342) |
|---|---|---|
| No. (%) | No. (%) | |
| Mean Age, years (SD) | 58.4 (11.5) | 56.2 (11.5) |
| Sex | ||
| Male | 10,636 (72.0) | 4075 (67.7) |
| Female | 4129 (28.0) | 1943 (32.3) |
| Race | ||
| White | 13,261 (89.8) | 5991 (90.1) |
| Black | 706 (4.8) | 270 (4.1) |
| Asian | 798 (5.4) | 387 (5.8) |
| Marital Status | ||
| Single | 5056 (34.2) | 2040 (30.7) |
| Married | 9709 (65.8) | 4608 (69.3) |
| Number of Prior Tumors | ||
| 0 | 14,051 (95.2) | 6324 (95.1) |
| 1 | 496 (3.4) | 232 (3.5) |
| 2 | 161 (1.1) | 77 (1.2) |
| 3 | 46 (0.3) | 14 (0.2) |
| 4+ | 11 (0.1) | 1 (0.0) |
| Histology | ||
| Nonkeratinizing SCC with maturation | 11,468 (77.7) | 5276 (79.4) |
| Undifferentiated nonkeratinizing SCC | 86 (0.6) | 39 (1.0) |
| Differentiated nonkeratinizing SCC | 824 (5.6) | 288 (4.3) |
| Keratinizing SCC | 2286 (15.5) | 993 (15.0) |
| SCC NOS | 101 (0.7) | 52 (1.0) |
| Tumor Grade | ||
| Well-differentiated | 2262 (18.8) | 1067 (19.7) |
| Moderately differentiated | 5752 (47.8) | 2585 (47.6) |
| Poorly differentiated | 3896 (32.4) | 1710 (31.5) |
| Undifferentiated | 117 (1.0) | 64 (1.2) |
| T-Stage | ||
| T1 | 4443 (46.7) | 1594 (50.0) |
| T2 | 3274 (34.4) | 1109 (34.8) |
| T3 | 1013 (10.6) | 262 (8.2) |
| T4 | 784 (8.2) | 221 (6.9) |
| N-Stage | ||
| N0 | 5110 (45.5) | 1918 (48.3) |
| N1 | 1968 (17.5) | 764 (19.2) |
| N2 | 3847 (34.3) | 1187 (29.9) |
| N3 | 296 (2.6) | 102 (2.6) |
| M-Stage | ||
| M0 | 11,200 (99.3) | 3913 (99.2) |
| M1 | 75 (0.7) | 30 (0.8) |
| Surgery | ||
| Yes | 6125 (41.8) | 2506 (37.5) |
| No | 8519 (58.2) | 4185 (62.5) |
| Radiation | ||
| Yes | 8965 (60.7) | 3811 (57.3) |
| No | 5800 (39.3) | 2837 (42.7) |
| Chemotherapy | ||
| Yes | 6598 (44.7) | 2632 (39.6) |
| No | 8167 (55.3) | 4016 (60.4) |
| Prediction Window | Classification Model | AUC (SD) | Accuracy % (95% CI) | Recall % (SD) | Precision % (SD) |
|---|---|---|---|---|---|
| 5 Years | GBM | 0.75 (0.01) | 81.8 (79.7–83.9) | 83.0 (0.02) | 97.7 (0.002) |
| GLM | 0.73 (0.02) | 77.4 (74.5–80.2) | 78.1 (0.03) | 98.0 (0.002) | |
| DRF | 0.73 (0.03) | 72.8 (69.8–75.7)) | 73.3 (0.02) | 97.8 (0.003) | |
| Deep Learning | 0.70 (0.04) | 82.1 (74.7–89.6) | 83.5 (0.06) | 97.6 (0.002) | |
| 10 Years | GBM | 0.74 (0.02) | 80.0 [75.3, 84.1] | 82.8 (0.04) | 94.0 (0.004) |
| GLM | 0.73 (0.02) | 78.4 [74.2, 82.7] | 81.0 (0.04) | 94.3 (0.002) | |
| Deep Learning | 0.71 (0.02) | 74.4 [70.1, 78.8] | 76.6 (0.04) | 94.0 (0.002) | |
| DRF | 0.69 (0.01) | 70.6 [68.0, 73.3] | 72.2 (0.02) | 93.8 (0.004) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fatapour, Y.; Abiri, A.; Kuan, E.C.; Brody, J.P. Development of a Machine Learning Model to Predict Recurrence of Oral Tongue Squamous Cell Carcinoma. Cancers 2023, 15, 2769. https://doi.org/10.3390/cancers15102769
Fatapour Y, Abiri A, Kuan EC, Brody JP. Development of a Machine Learning Model to Predict Recurrence of Oral Tongue Squamous Cell Carcinoma. Cancers. 2023; 15(10):2769. https://doi.org/10.3390/cancers15102769
Chicago/Turabian StyleFatapour, Yasaman, Arash Abiri, Edward C. Kuan, and James P. Brody. 2023. "Development of a Machine Learning Model to Predict Recurrence of Oral Tongue Squamous Cell Carcinoma" Cancers 15, no. 10: 2769. https://doi.org/10.3390/cancers15102769
APA StyleFatapour, Y., Abiri, A., Kuan, E. C., & Brody, J. P. (2023). Development of a Machine Learning Model to Predict Recurrence of Oral Tongue Squamous Cell Carcinoma. Cancers, 15(10), 2769. https://doi.org/10.3390/cancers15102769







