Non-Expressed Donor KIR3DL1 Alleles May Represent a Risk Factor for Relapse after T-Replete Haploidentical Hematopoietic Stem Cell Transplantation
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells (PBMCs and Cell Lines)
2.2. Cohort of T-Replete Haploidentical HSCT Patients
2.3. KIR Genotyping
2.4. KIR Allele Typing
2.5. KIR3DL1 Constructs
2.6. Site-Directed Mutagenesis of KIR3DL1
2.7. Obtention of KIR3DL1-eGFP Transfected Jurkat Cell Line
2.8. Flow Cytometry Analysis
2.9. Fluorescence Microscopy Imaging
2.10. Statistical Analyses
2.11. Quality Management System (QMS)
3. Results
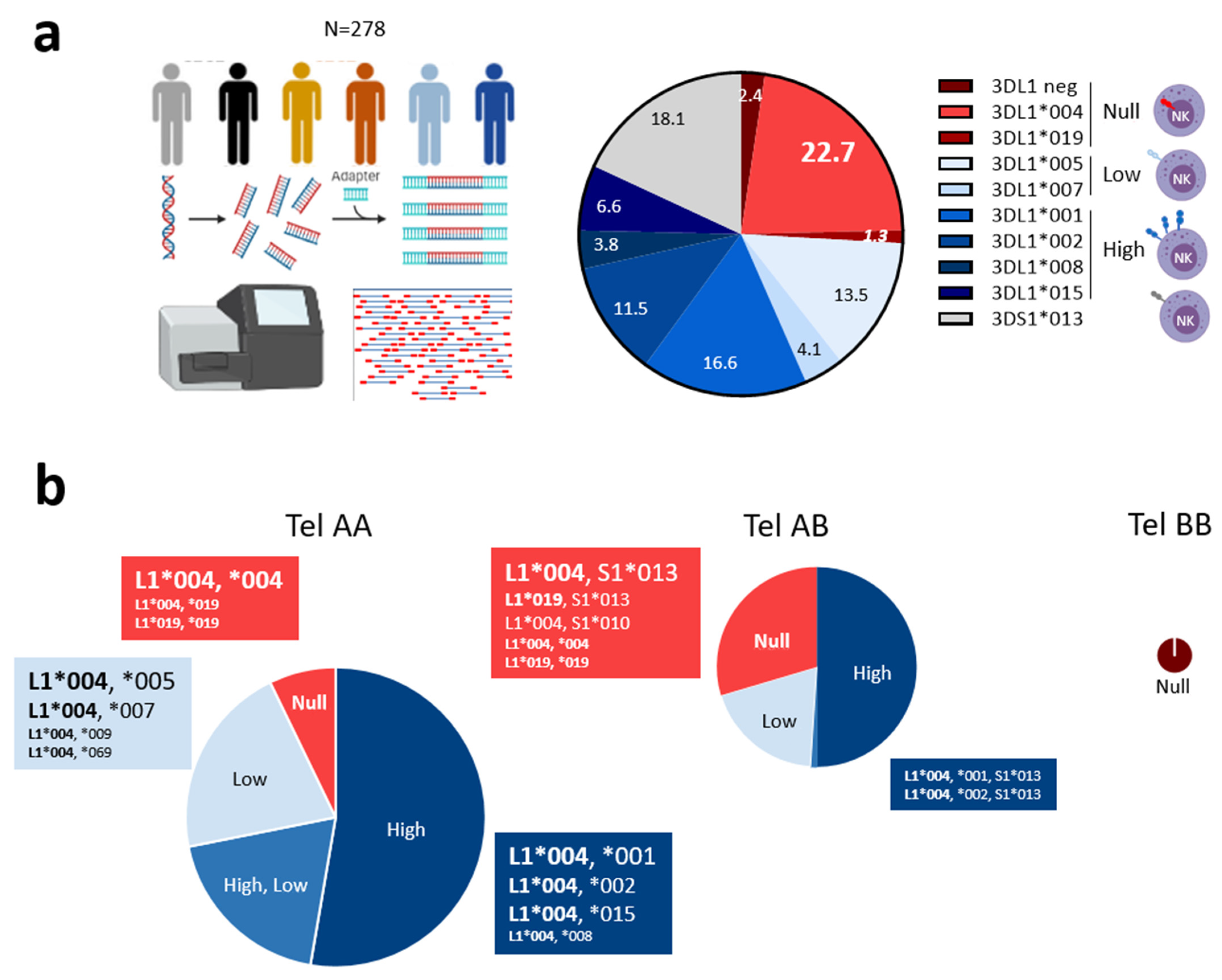
3.1. Predominant KIR3DL1*004 Allele and Unusual KIR3DL1*019 Allele Are Associated with KIR3DL1 Null Phenotype on NK Cells
3.2. Intracellular Localization of KIR3DL1*019
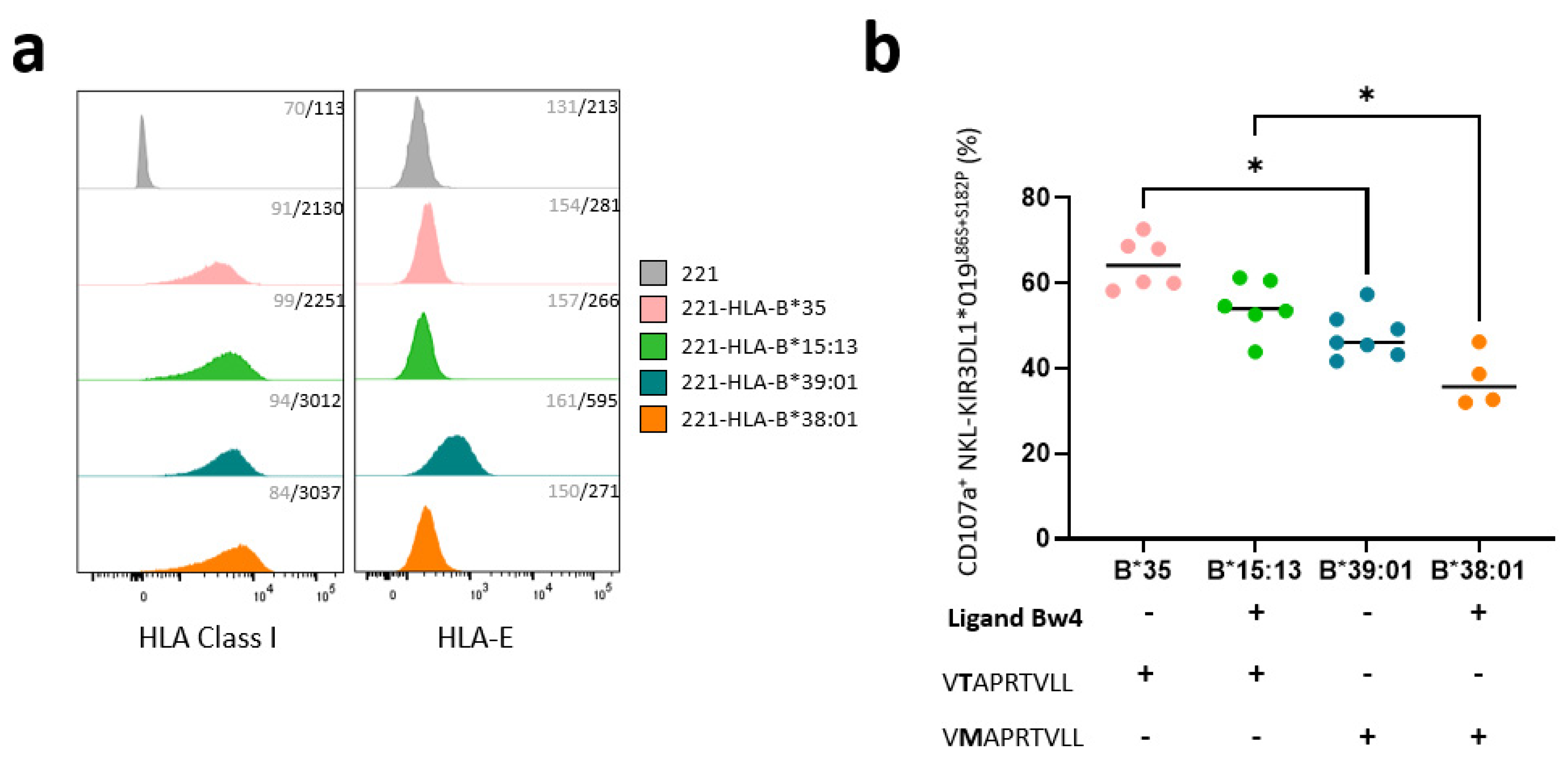
3.3. KIR3DL1*019L86S+S182P NKL Cell Line Recognizes HLA-B-Transfected 221 Target Cells Differently Depending on the Nature of HLA-B Allotypes
3.4. Non-Expressed KIR3DL1 Alleles Are a Risk Factor for Relapse Incidence after T-Replete Haploidentical HSCT in Myeloid Diseases
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anfossi, N.; André, P.; Guia, S.; Falk, C.S.; Roetynck, S.; Stewart, C.A.; Breso, V.; Frassati, C.; Reviron, D.; Middleton, D.; et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity 2006, 25, 331–342. [Google Scholar] [CrossRef]
- Rascle, P.; Woolley, G.; Jost, S.; Manickam, C.; Reeves, R.K. NK cell education: Physiological and pathological influences. Front. Immunol. 2023, 14, 1087155. [Google Scholar] [CrossRef]
- Kelly, A.; Trowsdale, J. Introduction: MHC/KIR and governance of specificity. Immunogenetics 2017, 69, 481–488. [Google Scholar] [CrossRef]
- Stern, M.; Ruggeri, L.; Capanni, M.; Mancusi, A.; Velardi, A. Human leukocyte antigens A23, A24, and A32 but not A25 are ligands for KIR3DL1. Blood 2008, 112, 708–710. [Google Scholar] [CrossRef]
- Béziat, V.; Hilton, H.G.; Norman, P.J.; Traherne, J.A. Deciphering the killer-cell immunoglobulin-like receptor system at super-resolution for natural killer and T-cell biology. Immunology 2017, 150, 248–264. [Google Scholar] [CrossRef]
- McErlean, C.; Gonzalez, A.A.; Cunningham, R.; Meenagh, A.; Shovlin, T.; Middleton, D. Differential RNA expression of KIR alleles. Immunogenetics 2010, 62, 431–440. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, G.M.; McVicar, D. The yin-yang of KIR3DL1/S1: Molecular mechanisms and cellular function. Crit. Rev. Immunol. 2013, 33, 203–218. [Google Scholar] [CrossRef]
- Cooley, S.; Weisdorf, D.J.; Guethlein, L.A.; Klein, J.P.; Wang, T.; Le, C.T.; Marsh, S.G.E.; Geraghty, D.; Spellman, S.; Haagenson, M.D.; et al. Donor selection for natural killer cell receptor genes leads to superior survival after unrelated transplantation for acute myelogenous leukemia. Blood 2010, 116, 2411–2419. [Google Scholar] [CrossRef]
- Erer, B.; Takeuchi, M.; Ustek, D.; Tugal-Tutkun, I.; Seyahi, E.; Özyazgan, Y.; Duymaz-Tozkir, J.; Gül, A.; Kastner, D.L.; Remmers, E.F.; et al. Evaluation of KIR3DL1/KIR3DS1 polymorphism in Behçet’s disease. Genes Immun. 2016, 17, 396–399. [Google Scholar] [CrossRef] [PubMed]
- Ahn, R.S.; Moslehi, H.; Martin, M.P.; Abad-Santos, M.; Bowcock, A.M.; Carrington, M.; Liao, W. Inhibitory KIR3DL1 alleles are associated with psoriasis. Br. J. Dermatol. 2016, 174, 449–451. [Google Scholar] [CrossRef] [PubMed]
- Castaño-Núñez, Á.; Montes-Cano, M.-A.; García-Lozano, J.-R.; Ortego-Centeno, N.; García-Hernández, F.-J.; Espinosa, G.; Graña-Gil, G.; Sánchez-Bursón, J.; Juliá, M.-R.; Solans, R.; et al. Association of Functional Polymorphisms of KIR3DL1/DS1 With Behçet’s Disease. Front. Immunol. 2019, 10, 2755. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Peña, R.; Vidal-Castiñeira, J.R.; Alonso-Arias, R.; Suarez-Alvarez, B.; Vicario, J.L.; Solana, R.; Collantes, E.; López-Vázquez, A.; Martínez-Borra, J.; López-Larrea, C. Association of the KIR3DS1*013 and KIR3DL1*004 alleles with susceptibility to ankylosing spondylitis. Arthritis Rheum. 2010, 62, 1000–1006. [Google Scholar] [CrossRef] [PubMed]
- Prakash, S.; Sarangi, A.N.; Alam, S.; Sonawane, A.; Sharma, R.K.; Agrawal, S. Putative role of KIR3DL1/3DS1 alleles and HLA-Bw4 ligands with end stage renal disease and long term renal allograft survival. Gene 2017, 637, 219–229. [Google Scholar] [CrossRef] [PubMed]
- Alicata, C.; Pende, D.; Meazza, R.; Canevali, P.; Loiacono, F.; Bertaina, A.; Locatelli, F.; Nemat-Gorgani, N.; Guethlein, L.A.; Parham, P.; et al. Hematopoietic stem cell transplantation: Improving alloreactive Bw4 donor selection by genotyping codon 86 of KIR3DL1/S1. Eur. J. Immunol. 2016, 46, 1511–1517. [Google Scholar] [CrossRef]
- Boudreau, J.E.; Giglio, F.; Gooley, T.A.; Stevenson, P.A.; Le Luduec, J.-B.; Shaffer, B.C.; Rajalingam, R.; Hou, L.; Hurley, C.K.; Noreen, H.; et al. KIR3DL1/HLA-B Subtypes Govern Acute Myelogenous Leukemia Relapse After Hematopoietic Cell Transplantation. J. Clin. Oncol. 2017, 35, 2268–2278. [Google Scholar] [CrossRef]
- Ureshino, H.; Shindo, T.; Sano, H.; Kubota, Y.; Ando, T.; Kidoguchi, K.; Kusaba, K.; Itamura, H.; Kojima, H.; Kusunoki, Y.; et al. Reconstitution of NK cells expressing KIR3DL1 is associated with reduced NK cell activity and relapse of CML after allogeneic hematopoietic stem cell transplantation. Int. J. Hematol. 2019, 111, 733–738. [Google Scholar] [CrossRef]
- Boulet, S.; Kleyman, M.; Kim, J.Y.; Kamya, P.; Sharafi, S.; Simic, N.; Bruneau, J.; Routy, J.-P.; Tsoukas, C.M.; Bernard, N.F. A combined genotype of KIR3DL1 high expressing alleles and HLA-B*57 is associated with a reduced risk of HIV infection. AIDS 2008, 22, 1487–1491. [Google Scholar] [CrossRef]
- Boudreau, J.E.; Mulrooney, T.J.; Le Luduec, J.-B.; Barker, E.; Hsu, K.C. KIR3DL1 and HLA-B Density and Binding Calibrate NK Education and Response to HIV. J. Immunol. 2016, 196, 3398–3410. [Google Scholar] [CrossRef]
- Luo, M.; Czarnecki, C.; Nebroski, M.; Kimani, J.; Bernard, N.; Plummer, F.A. KIR3DL1 alleles and their epistatic interactions with human leukocyte antigen class I influence resistance and susceptibility to HIV-1 acquisition in the Pumwani sex worker cohort. AIDS 2018, 32, 841–850. [Google Scholar] [CrossRef]
- Boulet, S.; Song, R.; Kamya, P.; Bruneau, J.; Shoukry, N.H.; Tsoukas, C.M.; Bernard, N.F. HIV protective KIR3DL1 and HLA-B genotypes influence NK cell function following stimulation with HLA-devoid cells. J. Immunol. 2010, 184, 2057–2064. [Google Scholar] [CrossRef]
- Hajeer, A.; Jawdat, D.; Massadeh, S.; Aljawini, N.; Abedalthagafi, M.S.; Arabi, Y.M.; Alaamery, M. Association of KIR gene polymorphisms with COVID-19 disease. Clin. Immunol. 2022, 234, 108911. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.; Yamada, E.; Alter, G.; Martin, M.P.; Bashirova, A.A.; Norman, P.J.; Altfeld, M.; Parham, P.; Anderson, S.K.; McVicar, D.W.; et al. Novel KIR3DL1 alleles and their expression levels on NK cells: Convergent evolution of KIR3DL1 phenotype variation? J. Immunol. 2008, 180, 6743–6750. [Google Scholar] [CrossRef]
- Carr, W.H.; Pando, M.J.; Parham, P. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J. Immunol. 2005, 175, 5222–5229. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, C.M.; Guethlein, L.A.; Shilling, H.G.; Pando, M.; Carr, W.H.; Rajalingam, R.; Vilches, C.; Parham, P. Different NK cell surface phenotypes defined by the DX9 antibody are due to KIR3DL1 gene polymorphism. J. Immunol. 2001, 166, 2992–3001. [Google Scholar] [CrossRef] [PubMed]
- Gumperz, J.E.; Barber, L.D.; Valiante, N.M.; Percival, L.; Phillips, J.H.; Lanier, L.L.; Parham, P. Conserved and variable residues within the Bw4 motif of HLA-B make separable contributions to recognition by the NKB1 killer cell-inhibitory receptor. J. Immunol. 1997, 158, 5237–5241. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, G.M.; Guinan, K.J.; Cunningham, R.T.; Middleton, D.; Parham, P.; Gardiner, C.M. Functional polymorphism of the KIR3DL1/S1 receptor on human NK cells. J. Immunol. 2007, 178, 235–241. [Google Scholar] [CrossRef]
- Pando, M.J.; Gardiner, C.M.; Gleimer, M.; McQueen, K.L.; Parham, P. The protein made from a common allele of KIR3DL1 (3DL1*004) is poorly expressed at cell surfaces due to substitution at positions 86 in Ig domain 0 and 182 in Ig domain 1. J. Immunol. 2003, 171, 6640–6649. [Google Scholar] [CrossRef]
- Taner, S.B.; Pando, M.J.; Roberts, A.; Schellekens, J.; Marsh, S.G.E.; Malmberg, K.-J.; Parham, P.; Brodsky, F.M. Interactions of NK cell receptor KIR3DL1*004 with chaperones and conformation-specific antibody reveal a functional folded state as well as predominant intracellular retention. J. Immunol. 2011, 186, 62–72. [Google Scholar] [CrossRef]
- Forlenza, C.J.; Boudreau, J.E.; Zheng, J.; Le Luduec, J.-B.; Chamberlain, E.; Heller, G.; Cheung, N.-K.V.; Hsu, K.C. KIR3DL1 Allelic Polymorphism and HLA-B Epitopes Modulate Response to Anti-GD2 Monoclonal Antibody in Patients With Neuroblastoma. J. Clin. Oncol. 2016, 34, 2443–2451. [Google Scholar] [CrossRef]
- Martin, M.P.; Qi, Y.; Gao, X.; Yamada, E.; Martin, J.N.; Pereyra, F.; Colombo, S.; Brown, E.E.; Shupert, W.L.; Phair, J.; et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat. Genet. 2007, 39, 733–740. [Google Scholar] [CrossRef]
- Yawata, M.; Yawata, N.; Draghi, M.; Little, A.-M.; Partheniou, F.; Parham, P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J. Exp. Med. 2006, 203, 633–645. [Google Scholar] [CrossRef]
- Tao, S.D.; He, Y.M.; Ying, Y.L.; He, J.; Zhu, F.M.; Lv, H.J. KIR3DL1 genetic diversity and phenotypic variation in the Chinese Han population. Genes Immun. 2014, 15, 8–15. [Google Scholar] [CrossRef]
- Gagne, K.; Busson, M.; Bignon, J.-D.; Balère-Appert, M.-L.; Loiseau, P.; Dormoy, A.; Dubois, V.; Perrier, P.; Jollet, I.; Bois, M.; et al. Donor KIR3DL1/3DS1 gene and recipient Bw4 KIR ligand as prognostic markers for outcome in unrelated hematopoietic stem cell transplantation. Biol. Blood Marrow Transplant. 2009, 15, 1366–1375. [Google Scholar] [CrossRef] [PubMed]
- Rettman, P.; Willem, C.; Volteau, C.; Legrand, N.; Chevallier, P.; Lodé, L.; Esbelin, J.; Cesbron, A.; Bonneville, M.; Moreau, P.; et al. Impact of Graft-Versus-Graft Natural Killer Cell Alloreactivity on Single Unit Dominance After Double Umbilical Cord Blood Transplantation. Transplantation 2017, 101, 2092–2101. [Google Scholar] [CrossRef] [PubMed]
- Gagne, K.; Willem, C.; Legrand, N.; Djaoud, Z.; David, G.; Rettman, P.; Bressollette-Bodin, C.; Senitzer, D.; Esbelin, J.; Cesbron-Gautier, A.; et al. Both the nature of KIR3DL1 alleles and the KIR3DL1/S1 allele combination affect the KIR3DL1 NK-cell repertoire in the French population. Eur. J. Immunol. 2013, 43, 1085–1098. [Google Scholar] [CrossRef] [PubMed]
- Morvan, M.; Willem, C.; Gagne, K.; Kerdudou, N.; David, G.; Sébille, V.; Folléa, G.; Bignon, J.-D.; Retière, C. Phenotypic and functional analyses of KIR3DL1+ and KIR3DS1+ NK cell subsets demonstrate differential regulation by Bw4 molecules and induced KIR3DS1 expression on stimulated NK cells. J. Immunol. 2009, 182, 6727–6735. [Google Scholar] [CrossRef]
- Norman, P.J.; Abi-Rached, L.; Gendzekhadze, K.; Korbel, D.; Gleimer, M.; Rowley, D.; Bruno, D.; Carrington, C.V.F.; Chandanayingyong, D.; Chang, Y.-H.; et al. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat. Genet. 2007, 39, 1092–1099. [Google Scholar] [CrossRef]
- Maniangou, B.; Legrand, N.; Alizadeh, M.; Guyet, U.; Willem, C.; David, G.; Charpentier, E.; Walencik, A.; Retière, C.; Gagne, K. Killer Immunoglobulin-Like Receptor Allele Determination Using Next-Generation Sequencing Technology. Front. Immunol. 2017, 8, 547. [Google Scholar] [CrossRef]
- Le Bourgeois, A.; Labopin, M.; Leclerc, M.; de Latour, R.P.; Bourhis, J.-H.; Ceballos, P.; Orvain, C.; Wallet, H.L.; Bilger, K.; Blaise, D.; et al. Clofarabine/busulfan-based reduced intensity conditioning regimens provides very good survivals in acute myeloid leukemia patients in complete remission at transplant: A retrospective study on behalf of the SFGM-TC. Oncotarget 2018, 9, 36603–36612. [Google Scholar] [CrossRef]
- Duléry, R.; Ménard, A.-L.; Chantepie, S.; El-Cheikh, J.; François, S.; Delage, J.; Giannotti, F.; Ruggeri, A.; Brissot, E.; Battipaglia, G.; et al. Sequential Conditioning with Thiotepa in T Cell- Replete Hematopoietic Stem Cell Transplantation for the Treatment of Refractory Hematologic Malignancies: Comparison with Matched Related, Haplo-Mismatched, and Unrelated Donors. Biol. Blood Marrow Transplant. 2018, 24, 1013–1021. [Google Scholar] [CrossRef]
- Chevallier, P.; Peterlin, P.; Garnier, A.; Le Bourgeois, A.; Mahé, B.; Dubruille, V.; Blin, N.; Touzeau, C.; Gastinne, T.; Lok, A.; et al. Clofarabine-based reduced intensity conditioning regimen with peripheral blood stem cell graft and post-transplant cyclophosphamide in adults with myeloid malignancies. Oncotarget 2018, 9, 33528–33535. [Google Scholar] [CrossRef]
- Luznik, L.; O’Donnell, P.V.; Symons, H.J.; Chen, A.R.; Leffell, M.S.; Zahurak, M.; Gooley, T.A.; Piantadosi, S.; Kaup, M.; Ambinder, R.F.; et al. HLA-Haploidentical Bone Marrow Transplantation for Hematologic Malignancies Using Nonmyeloablative Conditioning and High-Dose, Posttransplantation Cyclophosphamide. Biol. Blood Marrow Transplant. J. Am. Soc. Blood Marrow Transplant. 2008, 14, 641–650. [Google Scholar] [CrossRef]
- Le Bourgeois, A.; Labopin, M.; Marçais, A.; de Latour, R.P.; Blaise, D.; Chantepie, S.; N’Guyen, S.; Maillard, N.; Forcade, E.; Yakoub-Agha, I.; et al. Sequential allogeneic hematopoietic stem cell transplantation for active refractory/relapsed myeloid malignancies: Results of a reduced-intensity conditioning preceded by clofarabine and cytosine arabinoside, a retrospective study on behalf of the SFGM-TC. Ann. Hematol. 2020, 99, 1855–1862. [Google Scholar] [CrossRef] [PubMed]
- Willem, C.; Makanga, D.R.; Guillaume, T.; Maniangou, B.; Legrand, N.; Gagne, K.; Peterlin, P.; Garnier, A.; Béné, M.C.; Cesbron, A.; et al. Impact of KIR/HLA Incompatibilities on NK Cell Reconstitution and Clinical Outcome after T Cell-Replete Haploidentical Hematopoietic Stem Cell Transplantation with Posttransplant Cyclophosphamide. J. Immunol. 2019, 202, 2141–2152. [Google Scholar] [CrossRef] [PubMed]
- Dubreuil, L.; Maniangou, B.; Chevallier, P.; Quéméner, A.; Legrand, N.; Béné, M.C.; Willem, C.; David, G.; Alizadeh, M.; Makanga, D.R.; et al. Centromeric KIR AA Individuals Harbor Particular KIR Alleles Conferring Beneficial NK Cell Features with Implications in Haplo-Identical Hematopoietic Stem Cell Transplantation. Cancers 2020, 12, 3595. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.Y.; Gaidulis, L.; Miller, M.M.; Goto, R.M.; Rodriguez, R.; Forman, S.J.; Senitzer, D. Development of a multiplex PCR-SSP method for Killer-cell immunoglobulin-like receptor genotyping. Tissue Antigens 2004, 64, 462–468. [Google Scholar] [CrossRef] [PubMed]
- Hsu, K.C.; Chida, S.; Geraghty, D.E.; Dupont, B. The killer cell immunoglobulin-like receptor (KIR) genomic region: Gene-order, haplotypes and allelic polymorphism. Immunol. Rev. 2002, 190, 40–52. [Google Scholar] [CrossRef]
- Williams, F.; Maxwell, L.D.; Halfpenny, I.A.; Meenagh, A.; Sleator, C.; Curran, M.D.; Middleton, D. Multiple copies of KIR 3DL/S1 and KIR 2DL4 genes identified in a number of individuals. Hum. Immunol. 2003, 64, 729–732. [Google Scholar] [CrossRef]
- Misra, M.K.; Augusto, D.G.; Martin, G.M.; Nemat-Gorgani, N.; Sauter, J.; Hofmann, J.A.; Traherne, J.A.; González-Quezada, B.; Gorodezky, C.; Bultitude, W.P.; et al. Report from the Killer-cell Immunoglobulin-like Receptors (KIR) component of the 17th International HLA and Immunogenetics Workshop. Hum. Immunol. 2018, 79, 825–833. [Google Scholar] [CrossRef]
- Amorim, L.M.; Augusto, D.G.; Nemat-Gorgani, N.; Montero-Martin, G.; Marin, W.M.; Shams, H.; Dandekar, R.; Caillier, S.; Parham, P.; Fernández-Viña, M.A.; et al. High-Resolution Characterization of KIR Genes in a Large North American Cohort Reveals Novel Details of Structural and Sequence Diversity. Front. Immunol. 2021, 12, 674778. [Google Scholar] [CrossRef]
- Gonzalez-Galarza, F.F.; McCabe, A.; Santos, E.J.M.D.; Jones, J.; Takeshita, L.; Ortega-Rivera, N.D.; Cid-Pavon, G.M.D.; Ramsbottom, K.; Ghattaoraya, G.; Alfirevic, A.; et al. Allele frequency net database (AFND) 2020 update: Gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res. 2020, 48, D783–D788. [Google Scholar] [CrossRef] [PubMed]
- Harrison, G.F.; Leaton, L.A.; Harrison, E.A.; Kichula, K.M.; Viken, M.K.; Shortt, J.; Gignoux, C.R.; Lie, B.A.; Vukcevic, D.; Leslie, S.; et al. Allele imputation for the killer cell immunoglobulin-like receptor KIR3DL1/S1. PLoS Comput. Biol. 2022, 18, e1009059. [Google Scholar] [CrossRef] [PubMed]
- Alicata, C.; Ashouri, E.; Nemat-Gorgani, N.; Guethlein, L.A.; Marin, W.M.; Tao, S.; Moretta, L.; Hollenbach, J.A.; Trowsdale, J.; Traherne, J.A.; et al. KIR Variation in Iranians Combines High Haplotype and Allotype Diversity With an Abundance of Functional Inhibitory Receptors. Front. Immunol. 2020, 11, 556. [Google Scholar] [CrossRef] [PubMed]
- Dubreuil, L.; Chevallier, P.; Retière, C.; Gagne, K. Relevance of Polymorphic KIR and HLA Class I Genes in NK-Cell-Based Immunotherapies for Adult Leukemic Patients. Cancers 2021, 13, 3767. [Google Scholar] [CrossRef] [PubMed]
- Solomon, S.R.; Aubrey, M.T.; Zhang, X.; Jackson, K.C.; Roark, C.L.; Freed, B.M.; Morris, L.E.; Holland, H.K.; Solh, M.M.; Bashey, A. Lineage-Specific Relapse Prediction After Haploidentical Transplantation With Post-Transplant Cyclophosphamide Based on Recipient HLA-B-Leader Genotype and HLA-C-Group KIR Ligand. Transplant. Cell. Ther. 2022, 28, 601.e1–601.e8. [Google Scholar] [CrossRef]
- van der Ploeg, K.; Le Luduec, J.-B.; Stevenson, P.A.; Park, S.; Gooley, T.A.; Petersdorf, E.W.; Shaffer, B.C.; Hsu, K.C. HLA-A Alleles Influencing NK Cell Function Impact AML Relapse Following Allogeneic Hematopoietic Cell Transplantation. Blood Adv. 2020, 4, 4955–4964. [Google Scholar] [CrossRef]
- Solomon, S.R.; Aubrey, M.T.; Zhang, X.; Piluso, A.; Freed, B.M.; Brown, S.; Jackson, K.C.; Morris, L.E.; Holland, H.K.; Solh, M.M.; et al. Selecting the Best Donor for Haploidentical Transplant: Impact of HLA, Killer Cell Immunoglobulin-Like Receptor Genotyping, and Other Clinical Variables. Biol. Blood Marrow Transplant. J. Am. Soc. Blood Marrow Transplant. 2018, 24, 789–798. [Google Scholar] [CrossRef]
- Shimoni, A.; Labopin, M.; Finke, J.; Ciceri, F.; Deconinck, E.; Kröger, N.; Gramatzki, M.; Stelljes, M.; Blaise, D.; Stoelzel, F.; et al. Donor Selection for a Second Allogeneic Stem Cell Transplantation in AML Patients Relapsing after a First Transplant: A Study of the Acute Leukemia Working Party of EBMT. Blood Cancer J. 2019, 9, 88. [Google Scholar] [CrossRef]
- Cieri, N.; Greco, R.; Crucitti, L.; Morelli, M.; Giglio, F.; Levati, G.; Assanelli, A.; Carrabba, M.G.; Bellio, L.; Milani, R.; et al. Post-Transplantation Cyclophosphamide and Sirolimus after Haploidentical Hematopoietic Stem Cell Transplantation Using a Treosulfan-Based Myeloablative Conditioning and Peripheral Blood Stem Cells. Biol. Blood Marrow Transplant. 2015, 21, 1506–1514. [Google Scholar] [CrossRef]
- Solomon, S.R.; Solh, M.; Morris, L.E.; Holland, H.K.; Bashey, A. Myeloablative Conditioning with PBSC Grafts for T Cell-Replete Haploidentical Donor Transplantation Using Posttransplant Cyclophosphamide. Adv. Hematol. 2016, 2016, 9736564. [Google Scholar] [CrossRef]
- Makanga, D.R.; Da Rin de Lorenzo, F.; David, G.; Willem, C.; Dubreuil, L.; Legrand, N.; Guillaume, T.; Peterlin, P.; Lebourgeois, A.; Béné, M.C.; et al. Genetic and Molecular Basis of Heterogeneous NK Cell Responses against Acute Leukemia. Cancers 2020, 12, 1927. [Google Scholar] [CrossRef] [PubMed]
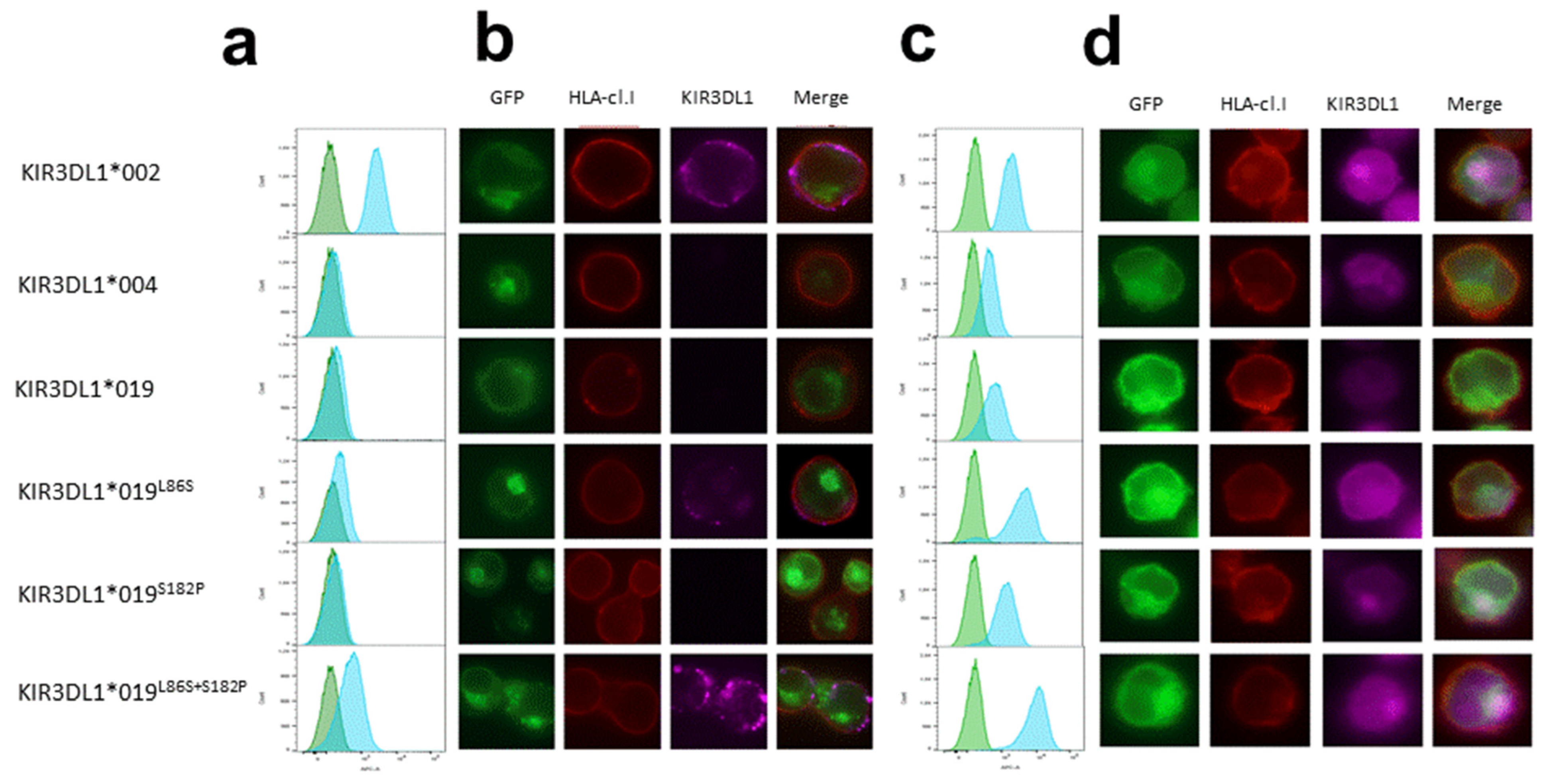

| DOMAIN | Lea. | D0 | D1 | D2 | Stem | Trm | Cytop. | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| POSITION | −20 | −9 | 2 | 30 | 31 | 44 | 47 | 54 | 86 | 182 | 238 | 283 | 320 | 343 | 373 |
| KIR3DL1*002 | L | F | V | Y | R | R | V | L | S | P | R | W | I | C | E |
| KIR3DL1*004 | S | L | M | Y | H | G | I | I | L | S | G | L | V | Y | Q |
| KIR3DL1*019 | S | L | M | C | H | G | I | I | L | S | G | L | V | Y | Q |
| KIR3DL1*019L86S | S | L | M | C | H | G | I | I | S | S | G | L | V | Y | Q |
| KIR3DL1*019S182P | S | L | M | C | H | G | I | I | L | P | G | L | V | Y | Q |
| KIR3DL1*019L86S+S182P | S | L | M | C | H | G | I | I | S | P | G | L | V | Y | Q |
| Univariate Analysis | Multivariate Analysis | |||||
|---|---|---|---|---|---|---|
| Variable | n (%) ** | HR (95% CI) | p-Value | HR (95% CI) | p-Value | |
| Age | 105 (100%) | 0.96 (0.94–0.98) | 0.001 | 0.97 (0.94–1.00) | 0.02 | |
| Diseases | AML | 77 (73.3%) | Reference | Reference | ||
| Other myeloid | 28 (26.7%) | 0.34 (0.14–0.82) | 0.02 | 0.24 (0.06–0.95) | 0.04 | |
| DRI | Low–intermediate | 57 (54.3%) | Reference | Reference | ||
| High–very high | 48 (45.7%) | 4.79 (2.31–9.96) | <0.001 | 2.60 (0.91–7.44) | 0.08 | |
| Status | CR = 1 | 48 (45.7%) | Reference | Reference | ||
| CR > 1 | 10 (9.5%) | 3.09 (1.12–8.53) | 0.03 | 1.35 (0.35–5.16) | 0.67 | |
| No CR | 47 (44.8%) | 3.42 (1.51–7.73) | 0.003 | 4.58 (1.04–20.20) | 0.04 | |
| Conditioning | RIC ** | 83 (79%) | Reference | Reference | ||
| Sequential | 22 (21%) | 3.90 (2.02–7.52) | <0.001 | 0.42 (0.11–1.70) | 0.23 | |
| KIR3DL1 allotype | Expressed | 91 (86.7%) | Reference | Reference | ||
| Null | 14 (13.3%) | 2.49 (1.24–4.96) | 0.01 | 2.10 (1.11–3.98) | 0.02 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Legrand, N.; Salameh, P.; Jullien, M.; Chevallier, P.; Ferron, E.; David, G.; Devilder, M.-C.; Willem, C.; Gendzekhadze, K.; Parham, P.; et al. Non-Expressed Donor KIR3DL1 Alleles May Represent a Risk Factor for Relapse after T-Replete Haploidentical Hematopoietic Stem Cell Transplantation. Cancers 2023, 15, 2754. https://doi.org/10.3390/cancers15102754
Legrand N, Salameh P, Jullien M, Chevallier P, Ferron E, David G, Devilder M-C, Willem C, Gendzekhadze K, Parham P, et al. Non-Expressed Donor KIR3DL1 Alleles May Represent a Risk Factor for Relapse after T-Replete Haploidentical Hematopoietic Stem Cell Transplantation. Cancers. 2023; 15(10):2754. https://doi.org/10.3390/cancers15102754
Chicago/Turabian StyleLegrand, Nolwenn, Perla Salameh, Maxime Jullien, Patrice Chevallier, Enora Ferron, Gaelle David, Marie-Claire Devilder, Catherine Willem, Ketevan Gendzekhadze, Peter Parham, and et al. 2023. "Non-Expressed Donor KIR3DL1 Alleles May Represent a Risk Factor for Relapse after T-Replete Haploidentical Hematopoietic Stem Cell Transplantation" Cancers 15, no. 10: 2754. https://doi.org/10.3390/cancers15102754
APA StyleLegrand, N., Salameh, P., Jullien, M., Chevallier, P., Ferron, E., David, G., Devilder, M.-C., Willem, C., Gendzekhadze, K., Parham, P., Retière, C., & Gagne, K. (2023). Non-Expressed Donor KIR3DL1 Alleles May Represent a Risk Factor for Relapse after T-Replete Haploidentical Hematopoietic Stem Cell Transplantation. Cancers, 15(10), 2754. https://doi.org/10.3390/cancers15102754






