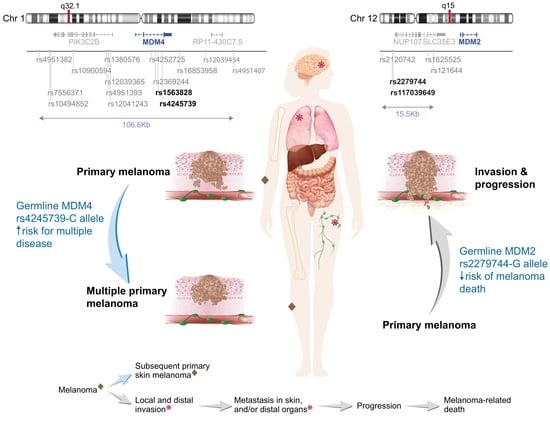
Sex-Specific Associations of MDM2 and MDM4 Variants with Risk of Multiple Primary Melanomas and Melanoma Survival in Non-Hispanic Whites
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Biospecimens and Genotyping
2.3. Statistical Analyses
2.4. Assessment of Other Credible Risk Variants as Potential Candidates for the Study of Melanoma Risk and Outcomes
3. Results
3.1. Study Population Characteristics
3.2. Effect of MDM2 and MDM4 Variants on Risk of Multiple Melanoma
3.3. Associations between MDM2 and MDM4 Variants and Survival
3.4. Other MDM2 and MDM4 Candidate Loci for the Study of Melanoma Risk and Outcomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Olsen, C.M.; Thompson, J.F.; Pandeya, N.; Whiteman, D.C. Evaluation of Sex-Specific Incidence of Melanoma. JAMA Dermatol. 2020, 156, 553–560. [Google Scholar] [CrossRef]
- Ferlay, J.E.M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Today; International Agency for Research on Cancer: Lyon, France, 2020. [Google Scholar]
- Kocarnik, J.M.; Park, S.L.; Han, J.; Dumitrescu, L.; Cheng, I.; Wilkens, L.R.; Schumacher, F.R.; Kolonel, L.; Carlson, C.S.; Crawford, D.C.; et al. Pleiotropic and Sex-Specific Effects of Cancer GWAS SNPs on Melanoma Risk in the Population Architecture Using Genomics and Epidemiology (PAGE) Study. PLoS ONE 2015, 10, e0120491. [Google Scholar] [CrossRef] [PubMed]
- Hernando, B.; Ibarrola-Villava, M.; Fernandez, L.P.; Peña-Chilet, M.; Llorca-Cardeñosa, M.; Oltra, S.S.; Alonso, S.; Boyano, M.D.; Martinez-Cadenas, C.; Ribas, G. Sex-specific genetic effects associated with pigmentation, sensitivity to sunlight, and melanoma in a population of Spanish origin. Biol. Sex. Differ. 2016, 7, 17. [Google Scholar] [CrossRef] [PubMed]
- Sangalli, A.; Orlandi, E.; Poli, A.; Maurichi, A.; Santinami, M.; Nicolis, M.; Ferronato, S.; Malerba, G.; Rodolfo, M.; Gomez Lira, M. Sex-specific effect of RNASEL rs486907 and miR-146a rs2910164 polymorphisms’ interaction as a susceptibility factor for melanoma skin cancer. Melanoma Res. 2017, 27, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Lira, F.E.; Podlipnik, S.; Potrony, M.; Tell-Marti, G.; Calbet-Llopart, N.; Barreiro, A.; Carrera, C.; Malvehy, J.; Puig, S. Inherited MC1R variants in patients with melanoma are associated with better survival in women. Br. J. Dermatol. 2020, 182, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Visconti, A.; Ribero, S.; Sanna, M.; Spector, T.D.; Bataille, V.; Falchi, M. Body site-specific genetic effects influence naevus count distribution in women. Pigment. Cell. Melanoma Res. 2020, 33, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Benna, C.; Rajendran, S.; Spiro, G.; Menin, C.; Dall’Olmo, L.; Rossi, C.R.; Mocellin, S. Gender-specific associations between polymorphisms of the circadian gene RORA and cutaneous melanoma susceptibility. J. Transl. Med. 2021, 19, 57. [Google Scholar] [CrossRef]
- Nan, H.; Qureshi, A.A.; Hunter, D.J.; Han, J. A functional SNP in the MDM2 promoter, pigmentary phenotypes, and risk of skin cancer. Cancer Causes Control 2009, 20, 171–179. [Google Scholar] [CrossRef]
- Firoz, E.F.; Warycha, M.; Zakrzewski, J.; Pollens, D.; Wang, G.; Shapiro, R.; Berman, R.; Pavlick, A.; Manga, P.; Ostrer, H.; et al. Association of MDM2 SNP309, age of onset, and gender in cutaneous melanoma. Clin. Cancer Res. 2009, 15, 2573–2580. [Google Scholar] [CrossRef]
- Gluck, I.; Simon, A.J.; Catane, R.; Pfeffer, R.; Schachter, J.; Rechavi, G.; Bar, J. Germline analysis of thymidine/guanidine polymorphism at position 309 of the Mdm2 promoter in malignant melanoma patients. Melanoma Res. 2009, 19, 199–202. [Google Scholar] [CrossRef]
- Capasso, M.; Ayala, F.; Avvisati, R.A.; Russo, R.; Gambale, A.; Mozzillo, N.; Ascierto, P.A.; Iolascon, A. MDM2 SNP309 and p53 Arg72Pro in cutaneous melanoma: Association between SNP309 GG genotype and tumor Breslow thickness. J. Hum. Genet. 2010, 55, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Cotignola, J.; Chou, J.F.; Roy, P.; Mitra, N.; Busam, K.; Halpern, A.C.; Orlow, I. Investigation of the effect of MDM2 SNP309 and TP53 Arg72Pro polymorphisms on the age of onset of cutaneous melanoma. J. Investig. Dermatol. 2012, 132, 1471–1478. [Google Scholar] [CrossRef] [PubMed]
- Kinyamu, H.K.; Archer, T.K. Estrogen receptor-dependent proteasomal degradation of the glucocorticoid receptor is coupled to an increase in mdm2 protein expression. Mol. Cell. Biol. 2003, 23, 5867–5881. [Google Scholar] [CrossRef] [PubMed]
- Bond, G.L.; Hu, W.; Bond, E.E.; Robins, H.; Lutzker, S.G.; Arva, N.C.; Bargonetti, J.; Bartel, F.; Taubert, H.; Wuerl, P.; et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell 2004, 119, 591–602. [Google Scholar] [CrossRef]
- Thunell, L.K.; Bivik, C.; Waster, P.; Fredrikson, M.; Stjernstrom, A.; Synnerstad, I.; Rosdahl, I.; Enerback, C. MDM2 SNP309 promoter polymorphism confers risk for hereditary melanoma. Melanoma Res. 2014, 24, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Knappskog, S.; Bjørnslett, M.; Myklebust, L.M.; Huijts, P.E.; Vreeswijk, M.P.; Edvardsen, H.; Guo, Y.; Zhang, X.; Yang, M.; Ylisaukko-Oja, S.K.; et al. The MDM2 promoter SNP285C/309G haplotype diminishes Sp1 transcription factor binding and reduces risk for breast and ovarian cancer in Caucasians. Cancer Cell. 2011, 19, 273–282. [Google Scholar] [CrossRef]
- Karni-Schmidt, O.; Lokshin, M.; Prives, C. The Roles of MDM2 and MDMX in Cancer. Annu. Rev. Pathol. 2016, 11, 617–644. [Google Scholar] [CrossRef]
- Toledo, F.; Wahl, G.M. MDM2 and MDM4: p53 regulators as targets in anticancer therapy. Int. J. Biochem. Cell. Biol. 2007, 39, 1476–1482. [Google Scholar] [CrossRef]
- Wynendaele, J.; Böhnke, A.; Leucci, E.; Nielsen, S.J.; Lambertz, I.; Hammer, S.; Sbrzesny, N.; Kubitza, D.; Wolf, A.; Gradhand, E.; et al. An illegitimate microRNA target site within the 3′ UTR of MDM4 affects ovarian cancer progression and chemosensitivity. Cancer Res. 2010, 70, 9641–9649. [Google Scholar] [CrossRef]
- Stegeman, S.; Moya, L.; Selth, L.A.; Spurdle, A.B.; Clements, J.A.; Batra, J. A genetic variant of MDM4 influences regulation by multiple microRNAs in prostate cancer. Endocr. Relat. Cancer 2015, 22, 265–276. [Google Scholar] [CrossRef]
- Begg, C.B.; Orlow, I.; Hummer, A.J.; Armstrong, B.K.; Kricker, A.; Marrett, L.D.; Millikan, R.C.; Gruber, S.B.; Anton-Culver, H.; Zanetti, R.; et al. Lifetime risk of melanoma in CDKN2A mutation carriers in a population-based sample. J. Natl. Cancer Inst. 2005, 97, 1507–1515. [Google Scholar] [CrossRef] [PubMed]
- Begg, C.B.; Hummer, A.J.; Mujumdar, U.; Armstrong, B.K.; Kricker, A.; Marrett, L.D.; Millikan, R.C.; Gruber, S.B.; Culver, H.A.; Zanetti, R.; et al. A design for cancer case-control studies using only incident cases: Experience with the GEM study of melanoma. Int. J. Epidemiol. 2006, 35, 756–764. [Google Scholar] [CrossRef]
- Orlow, I.; Roy, P.; Reiner, A.S.; Yoo, S.; Patel, H.; Paine, S.; Armstrong, B.K.; Kricker, A.; Marrett, L.D.; Millikan, R.C.; et al. Vitamin D receptor polymorphisms in patients with cutaneous melanoma. Int. J. Cancer 2012, 130, 405–418. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef] [PubMed]
- Stephens, M.; Smith, N.J.; Donnelly, P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001, 68, 978–989. [Google Scholar] [CrossRef]
- Stephens, M.; Donnelly, P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 2003, 73, 1162–1169. [Google Scholar] [CrossRef]
- Selvin, S. The Analysis of Contingency Table Data: Logistic Model I; Oxford University Press: New York, NY, USA, 1996; pp. 213–214. [Google Scholar]
- Marshall, S.W. Power for tests of interaction: Effect of raising the Type I error rate. Epidemiol. Perspect. Innov. 2007, 4, 4. [Google Scholar] [CrossRef]
- Durand, C.P. Does raising type 1 error rate improve power to detect interactions in linear regression models? A simulation study. PLoS ONE 2013, 8, e71079. [Google Scholar] [CrossRef]
- Team, T.R.D.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2008. [Google Scholar]
- Damsky, W.E.; Rosenbaum, L.E.; Bosenberg, M. Decoding melanoma metastasis. Cancers 2010, 3, 126–163. [Google Scholar] [CrossRef]
- Pluta, J.; Pyle, L.C.; Nead, K.T.; Wilf, R.; Li, M.; Mitra, N.; Weathers, B.; D’Andrea, K.; Almstrup, K.; Anson-Cartwright, L.; et al. Identification of 22 susceptibility loci associated with testicular germ cell tumors. Nat. Commun. 2021, 12, 4487. [Google Scholar] [CrossRef]
- Machiela, M. National Cancer Institute: Division of Cancer Epidemiology & Genetics. Available online: https://web.archive.org/web/20230221161649/https://ldlink.nci.nih.gov/ (accessed on 15 October 2021).
- Machiela, M.J.; Chanock, S.J. LDlink: A web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics 2015, 31, 3555–3557. [Google Scholar] [CrossRef] [PubMed]
- RegulomeDB. Available online: https://web.archive.org/web/20230221161914/https://regulomedb.org/regulome-search/ (accessed on 15 October 2021).
- Boyle, A.P.; Hong, E.L.; Hariharan, M.; Cheng, Y.; Schaub, M.A.; Kasowski, M.; Karczewski, K.J.; Park, J.; Hitz, B.C.; Weng, S.; et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012, 22, 1790–1797. [Google Scholar] [CrossRef] [PubMed]
- GTEx Portal. Available online: https://web.archive.org/web/20230220193714/https://gtexportal.org/home/index.html (accessed on 15 October 2021).
- Polsky, D.; Melzer, K.; Hazan, C.; Panageas, K.S.; Busam, K.; Drobnjak, M.; Kamino, H.; Spira, J.G.; Kopf, A.W.; Houghton, A.; et al. HDM2 protein overexpression and prognosis in primary malignant melanoma. J. Natl. Cancer Inst. 2002, 94, 1803–1806. [Google Scholar] [CrossRef] [PubMed]
- Nelson, R.A.; Levine, A.M.; Bernstein, L. Reproductive factors and risk of intermediate- or high-grade B-Cell non-Hodgkin’s lymphoma in women. J. Clin. Oncol. 2001, 19, 1381–1387. [Google Scholar] [CrossRef]
- Moussa, R.S.; Kovacevic, Z.; Richardson, D.R. Differential targeting of the cyclin-dependent kinase inhibitor, p21CIP1/WAF1, by chelators with anti-proliferative activity in a range of tumor cell-types. Oncotarget 2015, 6, 29694–29711. [Google Scholar] [CrossRef]
- Knappskog, S.; Trovik, J.; Marcickiewicz, J.; Tingulstad, S.; Staff, A.C.; MoMaTEC study group; Romundstad, P.; Hveem, K.; Vatten, L.; Salvesen, H.B.; et al. SNP285C modulates oestrogen receptor/Sp1 binding to the MDM2 promoter and reduces the risk of endometrial but not prostatic cancer. Eur. J. Cancer 2012, 48, 1988–1996. [Google Scholar] [CrossRef]
- Cancer Genome Atlas, N. Genomic Classification of Cutaneous Melanoma. Cell 2015, 161, 1681–1696. [Google Scholar] [CrossRef]
- Box, N.F.; Terzian, T. The role of p53 in pigmentation, tanning and melanoma. Pigment. Cell. Melanoma Res. 2008, 21, 525–533. [Google Scholar] [CrossRef]
- Leroy, B.; Anderson, M.; Soussi, T. TP53 mutations in human cancer: Database reassessment and prospects for the next decade. Hum. Mutat. 2014, 35, 672–688. [Google Scholar] [CrossRef]
- Duffy, M.J.; Synnott, N.C.; O’Grady, S.; Crown, J. Targeting p53 for the treatment of cancer. Semin. Cancer Biol. 2020, 79, 58–67. [Google Scholar] [CrossRef]
- de Polo, A.; Luo, Z.; Gerarduzzi, C.; Chen, X.; Little, J.B.; Yuan, Z.M. AXL receptor signalling suppresses p53 in melanoma through stabilization of the MDMX-MDM2 complex. J. Mol. Cell. Biol. 2017, 9, 154–165. [Google Scholar] [CrossRef] [PubMed]
- Jacinta-Fernandes, A.; Xavier, J.M.; Magno, R.; Lage, J.G.; Maia, A.T. Allele-specific miRNA-binding analysis identifies candidate target genes for breast cancer risk. NPJ Genom. Med. 2020, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Di Leva, G.; Piovan, C.; Gasparini, P.; Ngankeu, A.; Taccioli, C.; Briskin, D.; Cheung, D.G.; Bolon, B.; Anderlucci, L.; Alder, H.; et al. Estrogen mediated-activation of miR-191/425 cluster modulates tumorigenicity of breast cancer cells depending on estrogen receptor status. PLoS Genet. 2013, 9, e1003311. [Google Scholar] [CrossRef]
- Nagpal, N.; Ahmad, H.M.; Molparia, B.; Kulshreshtha, R. MicroRNA-191, an estrogen-responsive microRNA, functions as an oncogenic regulator in human breast cancer. Carcinogenesis 2013, 34, 1889–1899. [Google Scholar] [CrossRef]
- Zhou, L.; Zhang, X.; Li, Z.; Zhou, C.; Li, M.; Tang, X.; Lu, C.; Li, H.; Yuan, Q.; Yang, M. Association of a genetic variation in a miR-191 binding site in MDM4 with risk of esophageal squamous cell carcinoma. PLoS ONE 2013, 8, e64331. [Google Scholar] [CrossRef]
- Garcia-Closas, M.; Couch, F.J.; Lindstrom, S.; Michailidou, K.; Schmidt, M.K.; Brook, M.N.; Orr, N.; Rhie, S.K.; Riboli, E.; Feigelson, H.S.; et al. Genome-wide association studies identify four ER negative-specific breast cancer risk loci. Nat. Genet. 2013, 45, 392–398. [Google Scholar] [CrossRef]
- Xu, C.; Zhu, J.; Fu, W.; Liang, Z.; Song, S.; Zhao, Y.; Lyu, L.; Zhang, A.; He, J.; Duan, P. MDM4 rs4245739 A > C polymorphism correlates with reduced overall cancer risk in a meta-analysis of 69477 subjects. Oncotarget 2016, 7, 71718–71726. [Google Scholar] [CrossRef]
- Zhou, R.; Li, Y.; Wang, N.; Niu, C.; Huang, X.; Cao, S.; Huo, X. MDM4 polymorphisms associated with the risk but not the prognosis of esophageal cancer in Cixian high-incidence region from northern China. Asia Pac. J. Clin. Oncol. 2022, 18, e435–e441. [Google Scholar] [CrossRef]
- Gansmo, L.B.; Romundstad, P.; Birkeland, E.; Hveem, K.; Vatten, L.; Knappskog, S.; Lonning, P.E. MDM4 SNP34091 (rs4245739) and its effect on breast-, colon-, lung-, and prostate cancer risk. Cancer Med. 2015, 4, 1901–1907. [Google Scholar] [CrossRef]
- Gansmo, L.B.; Bjornslett, M.; Halle, M.K.; Salvesen, H.B.; Dorum, A.; Birkeland, E.; Hveem, K.; Romundstad, P.; Vatten, L.; Lonning, P.E.; et al. The MDM4 SNP34091 (rs4245739) C-allele is associated with increased risk of ovarian-but not endometrial cancer. Tumour Biol. 2016, 37, 10697–10702. [Google Scholar] [CrossRef]
- Dika, E.; Patrizi, A.; Lambertini, M.; Manuelpillai, N.; Fiorentino, M.; Altimari, A.; Ferracin, M.; Lauriola, M.; Fabbri, E.; Campione, E.; et al. Estrogen Receptors and Melanoma: A Review. Cells 2019, 8, 1463. [Google Scholar] [CrossRef] [PubMed]
- Weinberg-Shukron, A.; Renbaum, P.; Kalifa, R.; Zeligson, S.; Ben-Neriah, Z.; Dreifuss, A.; Abu-Rayyan, A.; Maatuk, N.; Fardian, N.; Rekler, D.; et al. A mutation in the nucleoporin-107 gene causes XX gonadal dysgenesis. J. Clin. Investig. 2015, 125, 4295–4304. [Google Scholar] [CrossRef] [PubMed]
- Alanee, S.; Delfino, K.; Wilber, A.; Robinson, K.; Brard, L.; Semaan, A. Single nucleotide variant in Nucleoporin 107 may be predictive of sensitivity to chemotherapy in patients with ovarian cancer. Pharm. Genom. 2017, 27, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Shi, R.; Xu, L.; Huang, L.; Cheng, J.X. Nucleoporin 107 Promotes the Survival of Tumor Cells in Cervical Cancers. Gynecol. Obstet. Investig. 2020, 85, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Jabs, V.; Edlund, K.; Konig, H.; Grinberg, M.; Madjar, K.; Rahnenfuhrer, J.; Ekman, S.; Bergkvist, M.; Holmberg, L.; Ickstadt, K.; et al. Integrative analysis of genome-wide gene copy number changes and gene expression in non-small cell lung cancer. PLoS ONE 2017, 12, e0187246. [Google Scholar] [CrossRef]
- Huang, L.; Wang, T.; Wang, F.; Hu, X.; Zhan, G.; Jin, X.; Zhang, L.; Li, Y. NUP37 silencing induces inhibition of cell proliferation, G1 phase cell cycle arrest and apoptosis in non-small cell lung cancer cells. Pathol. Res. Pract. 2020, 216, 152836. [Google Scholar] [CrossRef]
- Song, J.; Xu, Q.; Zhang, H.; Yin, X.; Zhu, C.; Zhao, K.; Zhu, J. Five key lncRNAs considered as prognostic targets for predicting pancreatic ductal adenocarcinoma. J. Cell. Biochem. 2018, 119, 4559–4569. [Google Scholar] [CrossRef]
| Characteristic a | Total N (%) | N Females (%) | N Males (%) |
|---|---|---|---|
| Melanoma participants | 3663 | 1595 (43.5) | 2068 (56.5) |
| Age at diagnosis in years, median (IQR) | |||
| 53.0 (26.0) | 53.0 (26.0) | 64.0 (21.0) | |
| <40 | 348 (21.8) | 348 (21.8) | 161 (7.8) |
| 40–49 | 328 (20.6) | 328 (20.6) | 244 (11.8) |
| 50–59 | 326 (20.4) | 326 (20.4) | 430 (20.8) |
| 60–69 | 245 (15.4) | 245 (15.4) | 504 (24.4) |
| 70–79 | 263 (16.5) | 263 (16.5) | 555 (26.8) |
| >80 | 85 (5.3) | 85 (5.3) | 174 (8.4) |
| Multiple vs. single primary melanoma status | |||
| Single primary | 1184 (74.2) | 1184 (74.2) | 1274 (61.6) |
| Multiple primaries | 411 (25.8) | 411 (25.8) | 794 (38.4) |
| Breslow thickness in mm, median (IQR) | |||
| 0.60 (0.65) | 0.60 (0.65) | 0.65 (0.90) | |
| In situ | 114 (7.2) | 114 (7.2) | 187 (9.0) |
| 0.01–1.00 | 1056 (66.2) | 1056 (66.2) | 1202 (58.1) |
| 1.01–2.00 | 235 (14.7) | 235 (14.7) | 366 (17.7) |
| 2.01–4.00 | 114 (7.2) | 114 (7.2) | 165 (8.0) |
| >4.00 | 43 (2.7) | 43 (2.7) | 103 (5.0) |
| Missing | 33 (2.1) | 33 (2.1) | 45 (2.2) |
| Anatomic site | |||
| Head/neck | 212 (13.3) | 212 (13.3) | 460 (22.2) |
| Trunk | 476 (29.8) | 476 (29.8) | 1110 (53.7) |
| Upper extremities | 365 (22.9) | 365 (22.9) | 303 (14.7) |
| Lower extremities | 542 (34.0) | 542 (34.0) | 195 (9.4) |
| All Cases (N = 3616) | Females (N = 1583) | Males (N = 2033) | ||||
|---|---|---|---|---|---|---|
| Gene Variants, Major/Minor Alleles | OR (95% CI) a | Ptrend a | OR (95% CI) c | Ptrend c | OR (95% CI) c | Ptrend c |
| MDM2 | ||||||
| rs117039649 (SNP285), G/C | 0.99 (0.75–1.30) | 0.92 | 1.21 (0.79–1.85) | 0.37 | 0.87 (0.60–1.25) | 0.44 |
| rs2279744 (SNP309), T/G | 1.04 (0.93–1.16) | 0.52 | 1.00 (0.84–1.18) | 0.99 | 1.06 (0.92–1.23) | 0.40 |
| Pinteraction = 0.24 (rs117039649) and 0.61 (rs2279744) b | ||||||
| MDM4 | ||||||
| rs1563828, C/T | 0.99 (0.88–1.11) | 0.81 | 1.13 (0.94–1.36) | 0.19 | 0.90 (0.77–1.04) | 0.16 |
| rs4245739, A/C | 1.03 (0.91–1.16) | 0.69 | 1.25 (1.03–1.51) | 0.03 | 0.90 (0.77–1.05) | 0.18 |
| Pinteraction = 0.06 (rs1563828) and 0.01 (rs4245739) b | ||||||
| All (N = 3663) | Females (N = 1595) | Males (N = 2068) | |||||
|---|---|---|---|---|---|---|---|
| Haplotype | Frequency | OR (95% CI) a | Pglobal a | OR (95% CI) c | Pglobal c | OR (95% CI) c | Pglobal c |
| MDM2 | |||||||
| rs117039649-rs2279744 (SNP285-SNP309) | |||||||
| G–T | 0.64 | 1 (ref) | 1 (ref) | 1 (ref) | |||
| G–G | 0.32 | 1.02 (0.91–1.14) | 0.985 (0.83–1.18) | 1.034 (0.89–1.20) | |||
| C–T | 0.01 | 0.50 (0.21–1.20) | 1.717 (0.52–5.72) | 0.152 (0.03–0.68) | |||
| C–G | 0.03 | 1.12 (0.82–1.54) | 0.42 | 1.12 (0.69–1.82) | 0.75 | 1.156 (0.77–1.75) | 0.09 |
| Pinteraction = 0.08 b | |||||||
| MDM4 | |||||||
| rs1563828-rs4245739 | |||||||
| C–A | 0.68 | 1 (ref) | 1 (ref) | 1 (ref) | |||
| T–C | 0.26 | 1.03 (0.91–1.17) | 0.20 | 1.24 (1.02–1.50) | 0.09 | 0.91 (0.78–1.08) | 0.31 |
| C–C | 0.01 | 0.76 (0.44–0.31) | 0.72(0.20–2.56) | 0.73 (0.40–1.34) | |||
| T–A | 0.05 | 0.79 (0.61–1.02) | 0.80 (0.53–1.19) | 0.78 (0.55–1.09) | |||
| Pinteraction = 0.15 b | |||||||
| All (N = 3521) | Females (N = 1552) | Males (N = 1969) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene Variants, Major/Minor Alleles | N Total/N Deaths a | HR (95% CI) b | Ptrend b | N Total/N Deaths a | HR (95% CI) d | Ptrend d | N Total/N Deaths a | HR (95% CI) d | Ptrend d |
| MDM2 | |||||||||
| rs117039649 (SNP285), G/C | 3521/248 | 0.65 (0.38–1.12) | 0.12 | 1552/70 | 0.68 (0.27–1.69) | 0.41 | 1969/178 | 0.61 (0.31–1.21) | 0.16 |
| rs2279744 (SNP309), T/G | 3367/236 | 0.84 (0.69–1.02) | 0.07 | 1484/67 | 0.63 (0.42–0.95) | 0.03 | 1883/169 | 0.94 (0.75–1.17) | 0.57 |
| Pinteraction = 0.77 (rs117039649) and 0.13 (rs2279744) c | |||||||||
| MDM4 | |||||||||
| rs1563828, C/T | 3484/245 | 0.92 (0.75–1.12) | 0.39 | 1541/70 | 1.09 (0.77–1.55) | 0.63 | 1943/175 | 0.84 (0.66–1.07) | 0.16 |
| rs4245739, A/C | 3480/250 | 0.89 (0.72–1.1) | 0.28 | 1532/70 | 1.03 (0.71–1.50) | 0.89 | 1948/180 | 0.82 (0.64–1.06) | 0.12 |
| Pinteraction = 0.25 (rs1563828) and 0.33 (rs4245739) c | |||||||||
| All (N = 3521) | Females (N = 1552) | Males (N = 1969) | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Haplotype | Frequency | HR (95% CI) a | Pglobal a | Frequency | HR (95% CI) c | Pglobal c | Frequency | HR (95% CI) c | Pglobal c |
| MDM2 | |||||||||
| rs117039649-rs2279744 (SNP285-SNP309) | |||||||||
| G-T | 0.642 | 1 (ref) | 0.639 | 1 (ref) | 0.644 | 1 (ref) | |||
| G-G | 0.322 | 0.86 (0.71–1.06) | 0.325 | 0.62 (0.40–0.94) | 0.320 | 0.98 (0.78–1.24) | |||
| C-T | 0.006 | 0.88 (0.23–3.32) | 0.006 | 0.17 (0–11.25) | 0.006 | 1.45 (0.35–6.10) | |||
| C-G | 0.030 | 0.55 (0.28–1.08) | 0.18 | 0.030 | 0.72 (0.25–2.05) | 0.12 | 0.030 | 0.45 (0.18–1.10) | 0.38 |
| Pinteraction = 0.21 b | |||||||||
| MDM4 | |||||||||
| rs1563828-rs4245739 | |||||||||
| C-A | 0.679 | 1 (ref) | 0.682 | 1 (ref) | 0.677 | 1 (ref) | |||
| T-C | 0.262 | 0.93 (0.75–1.15) | 0.56 | 0.261 | 1.07 (0.73–1.56) | 0.263 | 0.87 (0.67–1.12) | ||
| T-A | 0.050 | 0.88 (0.57–1.37) | 0.052 | 1.28 (0.62–2.62) | 0.049 | 0.75 (0.42–1.32) | |||
| C-C | 0.009 | 0.02 (0–15.83) | 0.006 | 0.02 (0–31.02) | 0.87 | 0.011 | 0.03 (0–26.12) | 0.34 | |
| Pinteraction = 0.67 b | |||||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ward, S.V.; Autuori, I.; Luo, L.; LaPilla, E.; Yoo, S.; Sharma, A.; Busam, K.J.; Olilla, D.W.; Dwyer, T.; Anton-Culver, H.; et al. Sex-Specific Associations of MDM2 and MDM4 Variants with Risk of Multiple Primary Melanomas and Melanoma Survival in Non-Hispanic Whites. Cancers 2023, 15, 2707. https://doi.org/10.3390/cancers15102707
Ward SV, Autuori I, Luo L, LaPilla E, Yoo S, Sharma A, Busam KJ, Olilla DW, Dwyer T, Anton-Culver H, et al. Sex-Specific Associations of MDM2 and MDM4 Variants with Risk of Multiple Primary Melanomas and Melanoma Survival in Non-Hispanic Whites. Cancers. 2023; 15(10):2707. https://doi.org/10.3390/cancers15102707
Chicago/Turabian StyleWard, Sarah V., Isidora Autuori, Li Luo, Emily LaPilla, Sarah Yoo, Ajay Sharma, Klaus J. Busam, David W. Olilla, Terence Dwyer, Hoda Anton-Culver, and et al. 2023. "Sex-Specific Associations of MDM2 and MDM4 Variants with Risk of Multiple Primary Melanomas and Melanoma Survival in Non-Hispanic Whites" Cancers 15, no. 10: 2707. https://doi.org/10.3390/cancers15102707
APA StyleWard, S. V., Autuori, I., Luo, L., LaPilla, E., Yoo, S., Sharma, A., Busam, K. J., Olilla, D. W., Dwyer, T., Anton-Culver, H., Zanetti, R., Sacchetto, L., Cust, A. E., Gallagher, R. P., Kanetsky, P. A., Rosso, S., Begg, C. B., Berwick, M., Thomas, N. E., & Orlow, I., on behalf of the GEM Study Group. (2023). Sex-Specific Associations of MDM2 and MDM4 Variants with Risk of Multiple Primary Melanomas and Melanoma Survival in Non-Hispanic Whites. Cancers, 15(10), 2707. https://doi.org/10.3390/cancers15102707