Exosome-Derived microRNA: Implications in Melanoma Progression, Diagnosis and Treatment
Abstract
Simple Summary
Abstract
1. Introduction
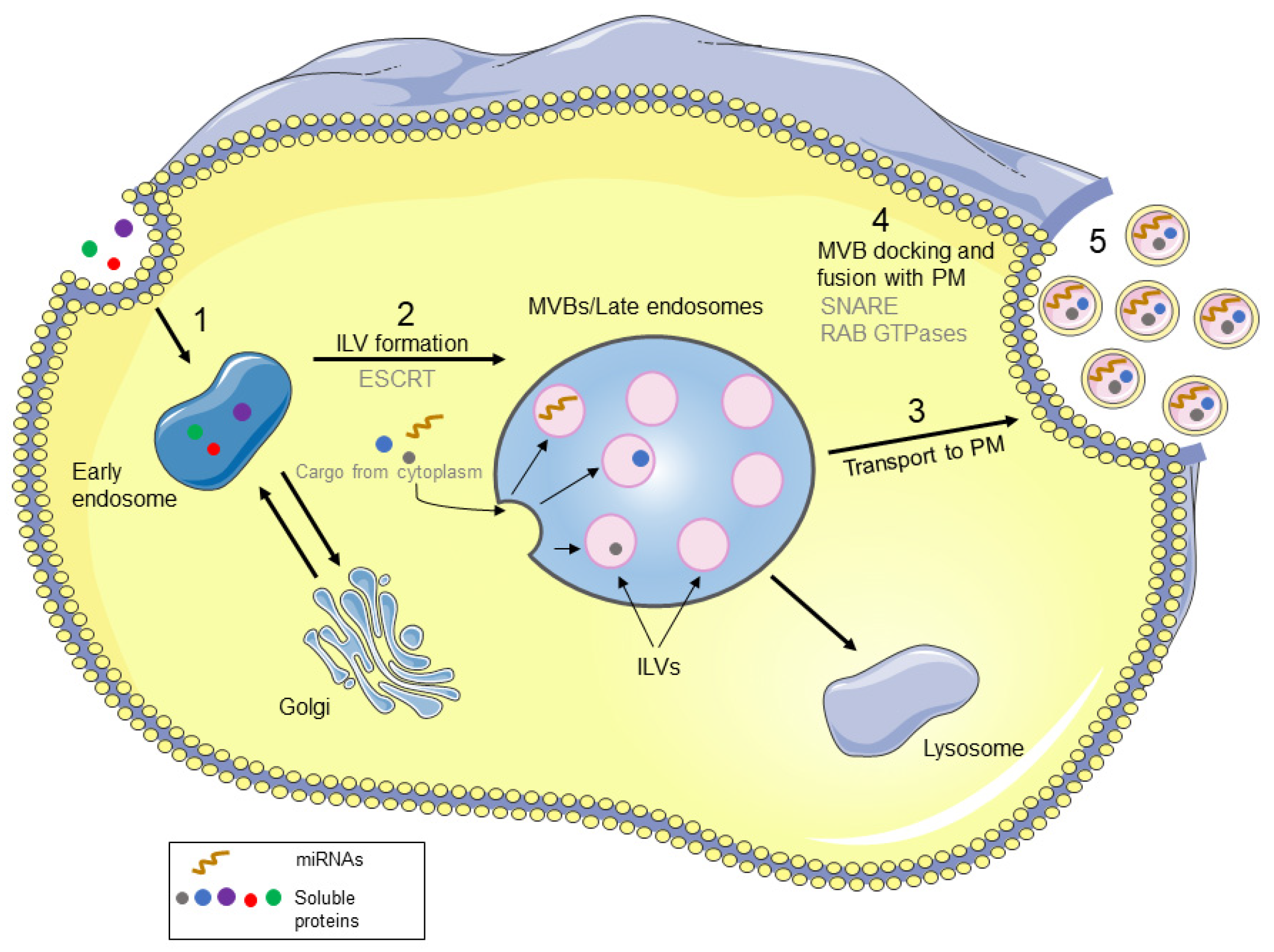
2. Biological Properties and Biogenesis of Exosomes
3. Exosome-Derived microRNAs
3.1. Sorting of miRNAs into Exosomes
3.2. miRNAs in Melanoma-Derived Exosomes
4. Roles of Exosome-Derived miRNAs in Melanoma Progression
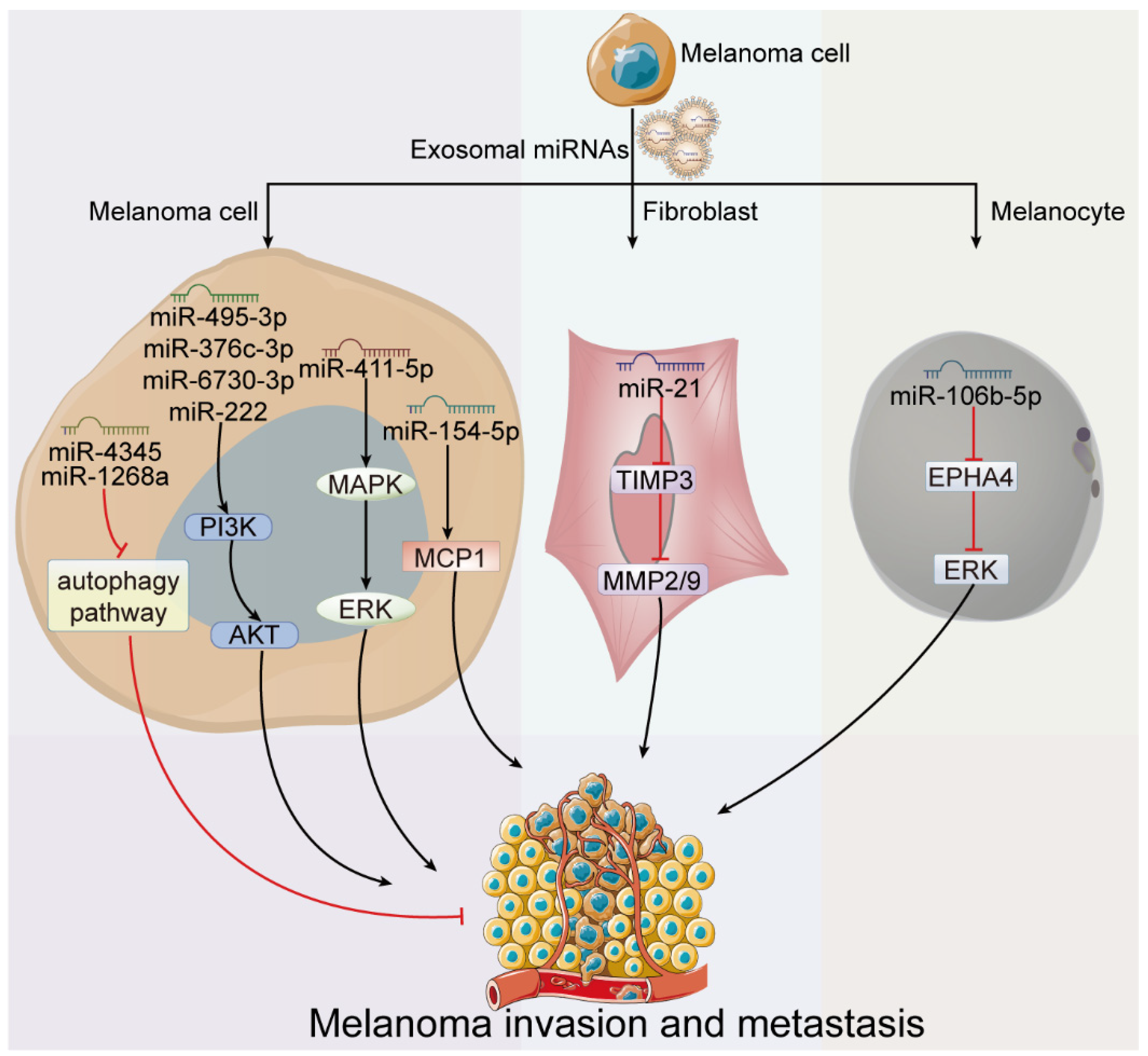
4.1. Exosome-Derived miRNAs in Melanoma Invasion and Metastasis
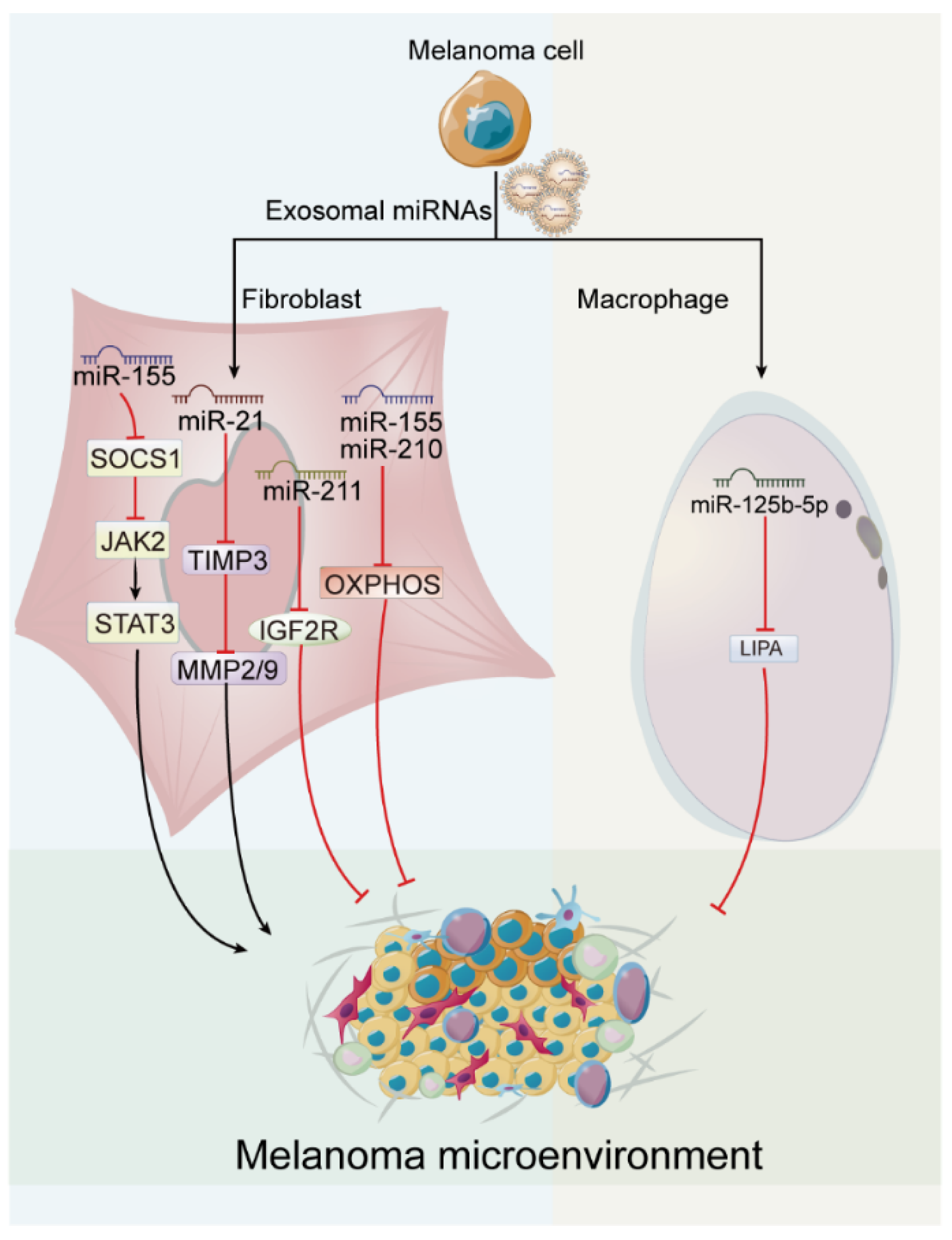
4.2. Exosome-Derived miRNAs in Melanoma Microenvironment
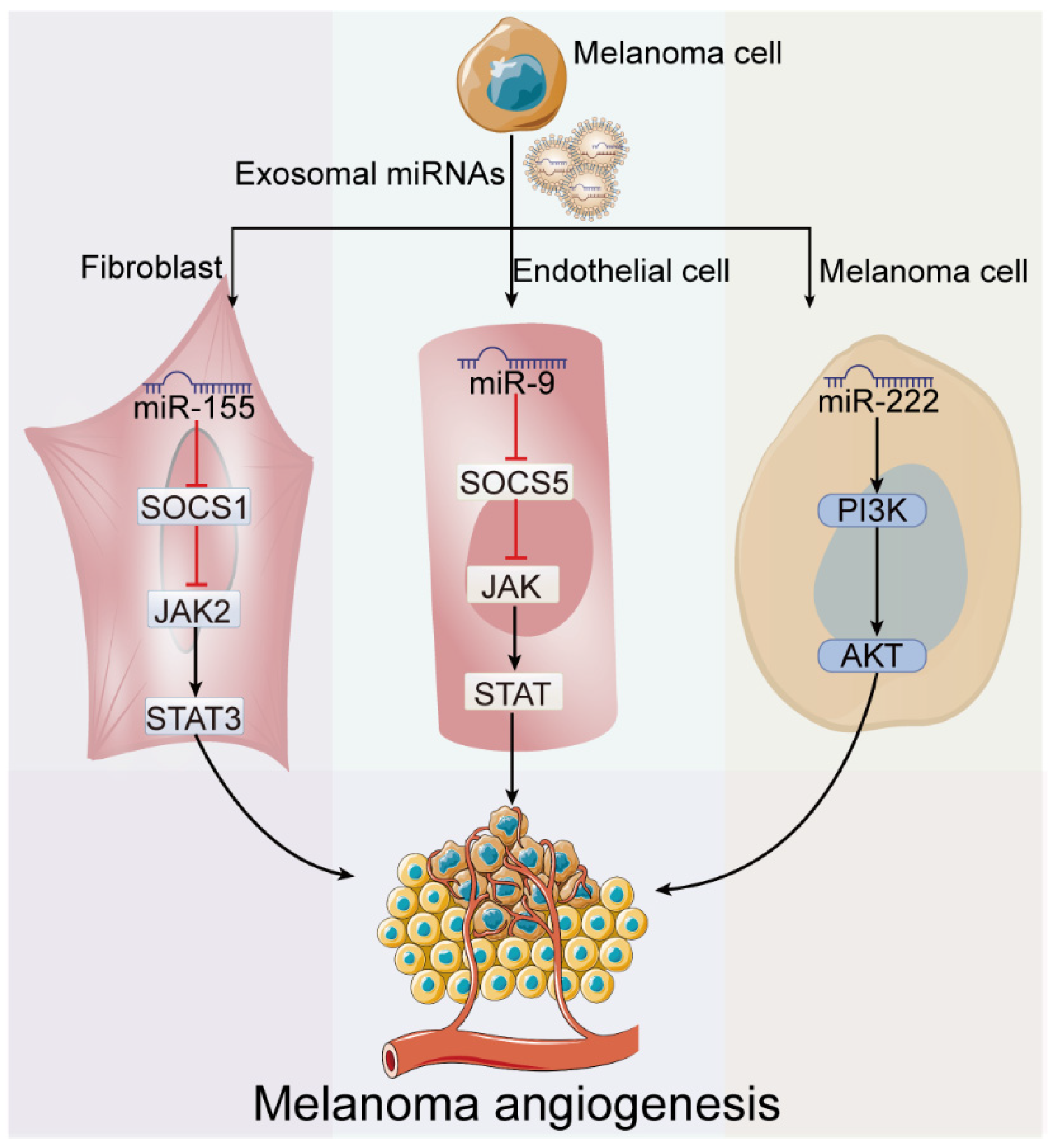
4.3. Exosome-Derived miRNAs in Melanoma Angiogenesis
4.4. Exosome-Derived miRNAs in Melanoma Immune Escape
5. Exosome-Derived miRNAs as Potential Marker for Melanoma Diagnosis
6. miRNAs Based on Exosomes in Melanoma Treatment
7. Future Perspectives and Conclusion Remarks
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Arnold, M.; Singh, D.; Laversanne, M.; Vignat, J.; Vaccarella, S.; Meheus, F.; Cust, A.E.; de Vries, E.; Whiteman, D.C.; Bray, F. Global Burden of Cutaneous Melanoma in 2020 and Projections to 2040. JAMA Dermatol. 2022, 158, 495. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Fong, M.Y.; Min, Y.; Somlo, G.; Liu, L.; Palomares, M.R.; Yu, Y.; Chow, A.; O’Connor, S.T.F.; Chin, A.R.; et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014, 25, 501–515. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Martin, P.; Fogarty, B.; Brown, A.; Schurman, K.; Phipps, R.; Yin, V.P.; Lockman, P.; Bai, S. Exosome Delivered Anticancer Drugs Across the Blood-Brain Barrier for Brain Cancer Therapy in Danio Rerio. Pharm. Res. 2015, 32, 2003–2014. [Google Scholar] [CrossRef]
- Cao, L.-Q.; Yang, X.-W.; Chen, Y.-B.; Zhang, D.-W.; Jiang, X.-F.; Xue, P. Exosomal miR-21 regulates the TETs/PTENp1/PTEN pathway to promote hepatocellular carcinoma growth. Mol. Cancer 2019, 18, 148. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.-H.; Tian, D.; Yang, Z.-C.; Li, J.-L. Exosomal miR-21 promotes proliferation, invasion and therapy resistance of colon adenocarcinoma cells through its target PDCD4. Sci. Rep. 2020, 10, 8271. [Google Scholar] [CrossRef] [PubMed]
- Trams, E.G.; Lauter, C.J.; Salem, N., Jr.; Heine, U. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim. Biophys. Acta 1981, 645, 63–70. [Google Scholar] [CrossRef]
- Beach, A.; Zhang, H.-G.; Ratajczak, M.Z.; Kakar, S.S. Exosomes: An overview of biogenesis, composition and role in ovarian cancer. J. Ovarian Res. 2014, 7, 14. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef]
- Yu, H.; Sun, T.; An, J.; Wen, L.; Liu, F.; Bu, Z.; Cui, Y.; Feng, J. Potential Roles of Exosomes in Parkinson’s Disease: From Pathogenesis, Diagnosis, and Treatment to Prognosis. Front. Cell Dev. Biol. 2020, 8, 86. [Google Scholar] [CrossRef]
- Mantile, F.; Franco, P.; Stoppelli, M.P.; Liguori, G.L. Biological role and clinical relevance of extracellular vesicles as key mediators of cell communication in cancer. In Advances in Biomembranes and Lipid Self-Assembly2021; Academic Press: Cambridge, MA, USA, 2021; pp. 37–117. [Google Scholar]
- Zhang, B.; Yin, Y.; Lai, R.C.; Lim, S.K. Immunotherapeutic Potential of Extracellular Vesicles. Front. Immunol. 2014, 5, 518. [Google Scholar] [CrossRef]
- Guo, M.; Hao, Y.; Feng, Y.; Li, H.; Mao, Y.; Dong, Q.; Cui, M. Microglial Exosomes in Neurodegenerative Disease. Front. Mol. Neurosci. 2021, 14, 630808. [Google Scholar] [CrossRef] [PubMed]
- Henne, W.M.; Buchkovich, N.J.; Emr, S.D. The ESCRT Pathway. Dev. Cell 2011, 21, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Kajimoto, T.; Okada, T.; Miya, S.; Zhang, L.; Nakamura, S.-I. Ongoing activation of sphingosine 1-phosphate receptors mediates maturation of exosomal multivesicular endosomes. Nat. Commun. 2013, 4, 2712. [Google Scholar] [CrossRef]
- Marsh, M.; van Meer, G. CELL BIOLOGY: No ESCRTs for Exosomes. Science 2008, 319, 1191–1192. [Google Scholar] [CrossRef]
- Teng, F.; Fussenegger, M. Shedding Light on Extracellular Vesicle Biogenesis and Bioengineering. Adv. Sci. 2020, 8, 2003505. [Google Scholar] [CrossRef]
- Hutagalung, A.H.; Novick, P.J. Role of Rab GTPases in Membrane Traffic and Cell Physiology. Physiol. Rev. 2011, 91, 119–149. [Google Scholar] [CrossRef] [PubMed]
- Ostrowski, M.; Carmo, N.B.; Krumeich, S.; Fanget, I.; Raposo, G.; Savina, A.; Moita, C.F.; Schauer, K.; Hume, A.N.; Freitas, R.P.; et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat. Cell Biol. 2010, 12, 19–30. [Google Scholar] [CrossRef]
- Lespagnol, A.; Duflaut, D.; Beekman, C.; Blanc, L.; Fiucci, G.; Marine, J.-C.; Vidal, M.; Amson, R.; Telerman, A. Exosome secretion, including the DNA damage-induced p53-dependent secretory pathway, is severely compromised in TSAP6/Steap3-null mice. Cell Death Differ. 2008, 15, 1723–1733. [Google Scholar] [CrossRef]
- Monleón, I.; Martínez-Lorenzo, M.J.; Monteagudo, L.; Lasierra, P.; Taulés, M.; Iturralde, M.; Piñeiro, A.; Larrad, L.; Alava, M.A.; Naval, J.; et al. Differential Secretion of Fas Ligand- or APO2 Ligand/TNF-Related Apoptosis-Inducing Ligand-Carrying Microvesicles During Activation-Induced Death of Human T Cells. J. Immunol. 2001, 167, 6736–6744. [Google Scholar] [CrossRef]
- Pegtel, D.M.; Cosmopoulos, K.; Thorley-Lawson, D.A.; van Eijndhoven, M.A.J.; Hopmans, E.S.; Lindenberg, J.L.; de Gruijl, T.D.; Würdinger, T.; Middeldorp, J.M. Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. USA 2010, 107, 6328–6333. [Google Scholar] [CrossRef]
- Squadrito, M.L.; Baer, C.; Burdet, F.; Maderna, C.; Gilfillan, G.D.; Lyle, R.; Ibberson, M.; De Palma, M. Endogenous RNAs Modulate MicroRNA Sorting to Exosomes and Transfer to Acceptor Cells. Cell Rep. 2014, 8, 1432–1446. [Google Scholar] [CrossRef] [PubMed]
- Ratajczak, J.; Miękus, K.; Kucia, M.; Zhang, J.; Reca, R.; Dvorak, P.; Ratajczak, M.Z. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: Evidence for horizontal transfer of mRNA and protein delivery. Leukemia 2006, 20, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Skog, J.; Würdinger, T.; Van Rijn, S.; Meijer, D.H.; Gainche, L.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Palma, J.; Yaddanapudi, S.C.; Pigati, L.; Havens, M.A.; Jeong, S.; Weiner, G.A.; Weimer, K.M.E.; Stern, B.; Hastings, M.L.; Duelli, D.M. MicroRNAs are exported from malignant cells in customized particles. Nucleic Acids Res. 2012, 40, 9125–9138. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhang, S.; Weber, J.; Baxter, D.; Galas, D.J. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010, 38, 7248–7259. [Google Scholar] [CrossRef]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and Exosomal MicroRNA: Trafficking, Sorting, and Function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Iguchi, H.; Hagiwara, K.; Yoshioka, Y.; Takeshita, F.; Ochiya, T. Neutral Sphingomyelinase 2 (nSMase2)-dependent Exosomal Transfer of Angiogenic MicroRNAs Regulate Cancer Cell Metastasis. J. Biol. Chem. 2013, 288, 10849–10859. [Google Scholar] [CrossRef]
- Garcia-Martin, R.; Wang, G.; Brandão, B.B.; Zanotto, T.M.; Shah, S.; Patel, S.K.; Schilling, B.; Kahn, C.R. MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature 2021, 601, 446–451. [Google Scholar] [CrossRef]
- Villarroya-Beltri, C.; Gutierrez-Vazquez, C.; Sanchez-Cabo, F.; Pérez-Hernández, D.; Vázquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sánchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef]
- Koppers-Lalic, D.; Hackenberg, M.; Bijnsdorp, I.V.; van Eijndhoven, M.A.; Sadek, P.; Sie, D.; Zini, N.; Middeldorp, J.M.; Ylstra, B.; de Menezes, R.X.; et al. Nontemplated Nucleotide Additions Distinguish the Small RNA Composition in Cells from Exosomes. Cell Rep. 2014, 8, 1649–1658. [Google Scholar] [CrossRef]
- Frank, F.; Sonenberg, N.; Nagar, B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 2010, 465, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Guduric-Fuchs, J.; O’Connor, A.; Camp, B.; O’Neill, C.L.; Medina, R.J.; A Simpson, D. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genom. 2012, 13, 357. [Google Scholar] [CrossRef] [PubMed]
- Gibbings, D.J.; Ciaudo, C.; Erhardt, M.; Voinnet, O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 2009, 11, 1143–1149. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Pressman, S.; Andress, A.P.; Kim, K.; White, J.L.; Cassidy, J.J.; Li, X.; Lubell, K.; Lim, D.-H.; Cho, I.S.; et al. Silencing by small RNAs is linked to endosomal trafficking. Nature 2009, 11, 1150–1156. [Google Scholar] [CrossRef]
- Vignard, V.; Labbé, M.; Marec, N.; André-Grégoire, G.; Jouand, N.; Fonteneau, J.-F.; Labarrière, N.; Fradin, D. MicroRNAs in Tumor Exosomes Drive Immune Escape in Melanoma. Cancer Immunol. Res. 2020, 8, 255–267. [Google Scholar] [CrossRef] [PubMed]
- Xiao, D.; Ohlendorf, J.; Chen, Y.; Taylor, D.D.; Rai, S.N.; Waigel, S.; Zacharias, W.; Hao, H.; McMasters, K.M. Identifying mRNA, MicroRNA and Protein Profiles of Melanoma Exosomes. PLOS ONE 2012, 7, e46874. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Yan, T.; Huang, C.; Xu, Z.; Wang, L.; Jiang, E.; Wang, H.; Chen, Y.; Liu, K.; Shao, Z.; et al. Melanoma cell-secreted exosomal miR-155-5p induce proangiogenic switch of cancer-associated fibroblasts via SOCS1/JAK2/STAT3 signaling pathway. J. Exp. Clin. Cancer Res. 2018, 37, 242. [Google Scholar] [CrossRef]
- Gerloff, D.; Lützkendorf, J.; Moritz, R.K.; Wersig, T.; Mäder, K.; Müller, L.P.; Sunderkötter, C. Melanoma-Derived Exosomal miR-125b-5p Educates Tumor Associated Macrophages (TAMs) by Targeting Lysosomal Acid Lipase A (LIPA). Cancers 2020, 12, 464. [Google Scholar] [CrossRef]
- Dika, E.; Broseghini, E.; Porcellini, E.; Lambertini, M.; Riefolo, M.; Durante, G.; Loher, P.; Roncarati, R.; Bassi, C.; Misciali, C.; et al. Unraveling the role of microRNA/isomiR network in multiple primary melanoma pathogenesis. Cell Death Dis. 2021, 12, 473. [Google Scholar] [CrossRef]
- Felicetti, F.; De Feo, A.; Coscia, C.; Puglisi, R.; Pedini, F.; Pasquini, L.; Bellenghi, M.; Errico, M.C.; Pagani, E.; Carè, A. Exosome-mediated transfer of miR-222 is sufficient to increase tumor malignancy in melanoma. J. Transl. Med. 2016, 14, 56. [Google Scholar] [CrossRef]
- Liu, D.; Li, X.; Zeng, B.; Zhao, Q.; Chen, H.; Zhang, Y.; Chen, Y.; Wang, J.; Xing, H.R. Exosomal microRNA-4535 of Melanoma Stem Cells Promotes Metastasis by Inhibiting Autophagy Pathway. Stem Cell Rev. Rep. 2022, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, D.; Chen, H.; Bin Zeng, B.; Zhao, Q.; Zhang, Y.; Chen, Y.; Wang, J.; Xing, H.R. Melanoma stem cells promote metastasis via exosomal miR-1268a inactivation of autophagy. Biol. Res. 2022, 55, 29. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Zeng, B.; Li, X.; Zhao, Q.; Liu, D.; Chen, Y.; Zhang, Y.; Wang, J.; Xing, H.R. High-Metastatic Melanoma Cells Promote the Metastatic Capability of Low-Metastatic Melanoma Cells via Exosomal Transfer of miR-411-5p. Front. Oncol. 2022, 12, 895164. [Google Scholar] [CrossRef] [PubMed]
- Pegoraro, A.; De Marchi, E.; Ferracin, M.; Orioli, E.; Zanoni, M.; Bassi, C.; Tesei, A.; Capece, M.; Dika, E.; Negrini, M.; et al. P2X7 promotes metastatic spreading and triggers release of miRNA-containing exosomes and microvesicles from melanoma cells. Cell Death Dis. 2021, 12, 1088. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Jo, H.; Kwon, Y.; Jeong, M.S.; Jung, H.S.; Kim, Y.; Jeoung, D. MiR-154-5p-MCP1 Axis Regulates Allergic Inflammation by Mediating Cellular Interactions. Front. Immunol. 2021, 12, 663726. [Google Scholar] [CrossRef]
- Bae, I.-S.; Kim, S.H. Milk Exosome-Derived MicroRNA-2478 Suppresses Melanogenesis through the Akt-GSK3β Pathway. Cells 2021, 10, 2848. [Google Scholar] [CrossRef]
- Wang, C.; Wang, Y.; Chang, X.; Ba, X.; Hu, N.; Liu, Q.; Fang, L.; Wang, Z. Melanoma-Derived Exosomes Endow Fibroblasts with an Invasive Potential via miR-21 Target Signaling Pathway. Cancer Manag. Res. 2020, 12, 12965–12974. [Google Scholar] [CrossRef]
- Luan, W.; Ding, Y.; Xi, H.; Ruan, H.; Lu, F.; Ma, S.; Wang, J. Exosomal miR-106b-5p derived from melanoma cell promotes primary melanocytes epithelial-mesenchymal transition through targeting EphA4. J. Exp. Clin. Cancer Res. 2021, 40, 107. [Google Scholar] [CrossRef]
- Xiao, D.; Barry, S.; Kmetz, D.; Egger, M.; Pan, J.; Rai, S.N.; Qu, J.; McMasters, K.M.; Hao, H. Melanoma cell–derived exosomes promote epithelial–mesenchymal transition in primary melanocytes through paracrine/autocrine signaling in the tumor microenvironment. Cancer Lett. 2016, 376, 318–327. [Google Scholar] [CrossRef]
- Li, J.; Chen, J.; Wang, S.; Li, P.; Zheng, C.; Zhou, X.; Tao, Y.; Chen, X.; Sun, L.; Wang, A.; et al. Blockage of transferred exosome-shuttled miR-494 inhibits melanoma growth and metastasis. J. Cell. Physiol. 2019, 234, 15763–15774. [Google Scholar] [CrossRef]
- Dror, S.; Sander, L.; Schwartz, H.; Sheinboim, D.; Barzilai, A.; Dishon, Y.; Apcher, S.; Golan, T.; Greenberger, S.; Barshack, I.; et al. Melanoma miRNA trafficking controls tumour primary niche formation. Nature 2016, 18, 1006–1017. [Google Scholar] [CrossRef] [PubMed]
- La Shu, S.; Yang, Y.; Allen, C.L.; Maguire, O.; Minderman, H.; Sen, A.; Ciesielski, M.J.; Collins, K.A.; Bush, P.J.; Singh, P.; et al. Metabolic reprogramming of stromal fibroblasts by melanoma exosome microRNA favours a pre-metastatic microenvironment. Sci. Rep. 2018, 8, 12905. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, G.; Wu, X.; Jiang, Z.; Kasman, I.; Yao, J.; Guan, Y.; Oeh, J.; Modrusan, Z.; Bais, C.; Sampath, D.; et al. Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J. 2012, 31, 3513–3523. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Karisma, V.W.; Liu, H.; Zhong, L. MicroRNA-300: A Transcellular Mediator in Exosome Regulates Melanoma Progression. Front. Oncol. 2019, 9, 1005. [Google Scholar] [CrossRef]
- Lee, J.-H.; Dindorf, J.; Eberhardt, M.; Lai, X.; Ostalecki, C.; Koliha, N.; Gross, S.; Blume, K.; Bruns, H.; Wild, S.; et al. Innate extracellular vesicles from melanoma patients suppress β-catenin in tumor cells by miRNA-34a. Life Sci. Alliance 2019, 2, e201800205. [Google Scholar] [CrossRef]
- Guo, D.; Lui, G.Y.; Lai, S.L.; Wilmott, J.S.; Tikoo, S.; Jackett, L.A.; Quek, C.; Brown, D.L.; Sharp, D.M.; Kwan, R.Y.; et al. RAB27A promotes melanoma cell invasion and metastasis via regulation of pro-invasive exosomes. Int. J. Cancer 2018, 144, 3070–3085. [Google Scholar] [CrossRef]
- Parmiani, G. Melanoma Cancer Stem Cells: Markers and Functions. Cancers 2016, 8, 34. [Google Scholar] [CrossRef]
- Xia, Y.; Zhou, Y.; Han, H.; Li, P.; Wei, W.; Lin, N. lncRNA NEAT1 facilitates melanoma cell proliferation, migration, and invasion via regulating miR-495-3p and E2F3. J. Cell. Physiol. 2019, 234, 19592–19601. [Google Scholar] [CrossRef]
- Bilke, S.; Schwentner, R.; Yang, F.; Kauer, M.; Jug, G.; Walker, R.L.; Davis, S.; Zhu, Y.J.; Pineda, M.; Meltzer, P.S.; et al. Oncogenic ETS fusions deregulate E2F3 target genes in Ewing sarcoma and prostate cancer. Genome Res. 2013, 23, 1797–1809. [Google Scholar] [CrossRef]
- Liu, L.; Qiu, M.; Tan, G.; Liang, Z.; Qin, Y.; Chen, L.; Chen, H.; Liu, J. miR-200c Inhibits invasion, migration and proliferation of bladder cancer cells through down-regulation of BMI-1 and E2F3. J. Transl. Med. 2014, 12, 305. [Google Scholar] [CrossRef]
- Segura, M.F.; Hanniford, D.; Menendez, S.; Reavie, L.; Zou, X.; Alvarez-Diaz, S.; Zakrzewski, J.; Blochin, E.; Rose, A.; Bogunovic, D.; et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc. Natl. Acad. Sci. USA 2009, 106, 1814–1819. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Ji, J.; Xu, Y.; Liu, Y.; Shi, L.; Liu, Y.; Lu, X.; Zhao, Y.; Luo, F.; Wang, B.; et al. MicroRNA-191, by promoting the EMT and increasing CSC-like properties, is involved in neoplastic and metastatic properties of transformed human bronchial epithelial cells. Mol. Carcinog. 2014, 54, E148–E161. [Google Scholar] [CrossRef] [PubMed]
- Truffi, M.; Sorrentino, L.; Corsi, F. Fibroblasts in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1234, 15–29. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Sahoo, A.; Sawada, J.; Marchica, J.; Sahoo, S.; Layng, F.I.A.L.; Finlay, D.; Mazar, J.; Joshi, P.; Komatsu, M.; et al. Exo-some-mediated MIR211 modulates tumor microenvironment via the DUSP6-ERK5 axis and contributes to BRAFV600E in-hibitor resistance in melanoma. bioRxiv 2019, 42, 548818. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Gholipour, M.; Taheri, M. MicroRNA Signature in Melanoma: Biomarkers and Therapeutic Targets. Front. Oncol. 2021, 11, 608987. [Google Scholar] [CrossRef] [PubMed]
- Munir, M.; Kay, M.; Kang, M.; Rahman, M.; Al-Harrasi, A.; Choudhury, M.; Moustaid-Moussa, N.; Hussain, F.; Rahman, S. Tumor-Associated Macrophages as Multifaceted Regulators of Breast Tumor Growth. Int. J. Mol. Sci. 2021, 22, 6526. [Google Scholar] [CrossRef]
- Fu, L.-Q.; Du, W.-L.; Cai, M.-H.; Yao, J.-Y.; Zhao, Y.-Y.; Mou, X.-Z. The roles of tumor-associated macrophages in tumor angiogenesis and metastasis. Cell. Immunol. 2020, 353, 104119. [Google Scholar] [CrossRef]
- Pan, Y.; Yu, Y.; Wang, X.; Zhang, T. Tumor-Associated Macrophages in Tumor Immunity. Front. Immunol. 2020, 11, 583084. [Google Scholar] [CrossRef]
- Wang, T.; Ge, Y.; Xiao, M.; Lopez-Coral, A.; Azuma, R.; Somasundaram, R.; Zhang, G.; Wei, Z.; Xu, X.; Rauscher, F.J., 3rd; et al. Melanoma-derived conditioned media efficiently induce the differentiation of monocytes to macrophages that display a highly invasive gene signature. Pigment. Cell Melanoma Res. 2012, 25, 493–505. [Google Scholar] [CrossRef]
- Bardi, G.T.; Smith, M.A.; Hood, J.L. Melanoma exosomes promote mixed M1 and M2 macrophage polarization. Cytokine 2018, 105, 63–72. [Google Scholar] [CrossRef]
- Huang, S.C.-C.; Everts, B.; Ivanova, Y.; O’Sullivan, D.; Nascimento, M.; Smith, A.M.; Beatty, W.; Love-Gregory, L.; Lam, W.Y.; O’Neill, C.M.; et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat. Immunol. 2014, 15, 846–855. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.; Yin, H.; Li, X.; Zhu, G.; He, W.; Gou, X. Bladder cancer cell-secreted exosomal miR-21 activates the PI3K/AKT pathway in macrophages to promote cancer progression. Int. J. Oncol. 2019, 56, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Weng, Y.-S.; Tseng, H.-Y.; Chen, Y.-A.; Shen, P.-C.; Al Haq, A.T.; Chen, L.-M.; Tung, Y.-C.; Hsu, H.-L. MCT-1/miR-34a/IL-6/IL-6R signaling axis promotes EMT progression, cancer stemness and M2 macrophage polarization in triple-negative breast cancer. Mol. Cancer 2019, 18, 42. [Google Scholar] [CrossRef] [PubMed]
- Ragusa, M.; Barbagallo, C.; Statello, L.; Caltabiano, R.; Russo, A.; Puzzo, L.; Avitabile, T.; Longo, A.; Toro, M.D.; Barbagallo, D.; et al. miRNA profiling in vitreous humor, vitreal exosomes and serum from uveal melanoma patients: Pathological and diagnostic implications. Cancer Biol. Ther. 2015, 16, 1387–1396. [Google Scholar] [CrossRef]
- Hood, J.L. Melanoma exosome induction of endothelial cell GM-CSF in pre-metastatic lymph nodes may result in different M1 and M2 macrophage mediated angiogenic processes. Med Hypotheses 2016, 94, 118–122. [Google Scholar] [CrossRef]
- Sun, L.; Li, W.; Lei, F.; Li, X. The regulatory role of micro RNA s in angiogenesis-related diseases. J. Cell. Mol. Med. 2018, 22, 4568–4587. [Google Scholar] [CrossRef]
- Rheinländer, A.; Schraven, B.; Bommhardt, U. CD45 in human physiology and clinical medicine. Immunol. Lett. 2018, 196, 22–32. [Google Scholar] [CrossRef]
- Ju, S.; Zhu, Y.; Liu, L.; Dai, S.; Li, C.; Chen, E.; He, Y.; Zhang, X.; Lu, B. Gadd45b and Gadd45g are important for anti-tumor immune responses. Eur. J. Immunol. 2009, 39, 3010–3018. [Google Scholar] [CrossRef]
- Mosmann, T.R.; Cherwinski, H.; Bond, M.W.; Giedlin, M.A.; Coffman, R.L. Two types of murine helper T cell clone. I. Defini-tion according to profiles of lymphokine activities and secreted proteins. J. Immunol. 1986, 136, 2348–2357. [Google Scholar]
- Qureshi, O.S.; Zheng, Y.; Nakamura, K.; Attridge, K.; Manzotti, C.; Schmidt, E.M.; Baker, J.; Jeffery, L.E.; Kaur, S.; Briggs, Z.; et al. Trans-Endocytosis of CD80 and CD86: A Molecular Basis for the Cell-Extrinsic Function of CTLA-4. Science 2011, 332, 600–603. [Google Scholar] [CrossRef]
- Spranger, S.; Bao, R.; Gajewski, T.F. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature 2015, 523, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, N.; Aharonov, R.; Meiri, E.; Rosenwald, S.; Spector, Y.; Zepeniuk, M.; Benjamin, H.; Shabes, N.; Tabak, S.; Levy, A.; et al. MicroRNAs accurately identify cancer tissue origin. Nat. Biotechnol. 2008, 26, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Stark, M.S.; Klein, K.; Weide, B.; Haydu, L.E.; Pflugfelder, A.; Tang, Y.H.; Palmer, J.M.; Whiteman, D.C.; Scolyer, R.A.; Mann, G.J.; et al. The Prognostic and Predictive Value of Melanoma-related MicroRNAs Using Tissue and Serum: A MicroRNA Expression Analysis. eBioMedicine 2015, 2, 671–680. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.-Y.; Li, P.; He, Q.-Y.; Luo, C.-Q. Circulating miR-221 Expression Level and Prognosis of Cutaneous Malignant Melanoma. J. Pharmacol. Exp. Ther. 2014, 20, 2472–2477. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Wang, H.; Yang, Y.; Guo, S.; Zhang, W.; Liu, Y.; Chunying, L.; Ma, J.; Zhao, T.; Liu, L.; et al. Down-regulated miR-23a Contributes to the Metastasis of Cutaneous Melanoma by Promoting Autophagy. Theranostics 2017, 7, 2231–2249. [Google Scholar] [CrossRef] [PubMed]
- Tembe, V.; Schramm, S.-J.; Stark, M.S.; Patrick, E.; Jayaswal, V.; Tang, Y.H.; Barbour, A.; Hayward, N.K.; Thompson, J.F.; Scolyer, R.A.; et al. MicroRNA and mRNA expression profiling in metastatic melanoma reveal associations with BRAF mutation and patient prognosis. Pigment Cell Melanoma Res. 2015, 28, 254–266. [Google Scholar] [CrossRef]
- Tian, R.; Liu, T.; Qiao, L.; Gao, M.; Li, J. Decreased serum microRNA-206 level predicts unfavorable prognosis in patients with melanoma. Int. J. Clin. Exp. Pathol. 2015, 8, 3097–3103. [Google Scholar]
- Meng, F.; Zhang, Y.; Li, X.; Wang, J.; Wang, Z. Clinical significance of miR-138 in patients with malignant melanoma through targeting of PDK1 in the PI3K/AKT autophagy signaling pathway. Oncol. Rep. 2017, 38, 1655–1662. [Google Scholar] [CrossRef]
- Svedman, F.C.; Lohcharoenkal, W.; Bottai, M.; Brage, S.E.; Sonkoly, E.; Hansson, J.; Pivarcsi, A.; Eriksson, H. Extracellular microvesicle microRNAs as predictive biomarkers for targeted therapy in metastastic cutaneous malignant melanoma. PLOS ONE 2018, 13, e0206942. [Google Scholar] [CrossRef]
- Huber, V.; Vallacchi, V.; Fleming, V.; Hu, X.; Cova, A.; Dugo, M.; Shahaj, E.; Sulsenti, R.; Vergani, E.; Filipazzi, P.; et al. Tumor-derived microRNAs induce myeloid suppressor cells and predict immunotherapy resistance in melanoma. J. Clin. Investig. 2018, 128, 5505–5516. [Google Scholar] [CrossRef]
- Pfeffer, S.R.; Grossmann, K.F.; Cassidy, P.B.; Yang, C.H.; Fan, M.; Kopelovich, L.; Leachman, S.A.; Pfeffer, L.M. Detection of Exosomal miRNAs in the Plasma of Melanoma Patients. J. Clin. Med. 2015, 4, 2012–2027. [Google Scholar] [CrossRef]
- Tengda, L.; Shuping, L.; Mingli, G.; Jie, G.; Yun, L.; Weiwei, Z.; Anmei, D. Serum exosomal microRNAs as potent circulating biomarkers for melanoma. Melanoma Res. 2018, 28, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Alegre, E.; Sanmamed, M.F.; Rodriguez, C.; Carranza, O.; Martín-Algarra, S.; González, A. Study of Circulating MicroRNA-125b Levels in Serum Exosomes in Advanced Melanoma. Arch. Pathol. Lab. Med. 2014, 138, 828–832. [Google Scholar] [CrossRef] [PubMed]
- Wozniak, M.; Peczek, L.; Czernek, L.; Düchler, M. Analysis of the miRNA Profiles of Melanoma Exosomes Derived Under Normoxic and Hypoxic Culture Conditions. Anticancer Res. 2017, 37, 6779–6789. [Google Scholar] [CrossRef]
- Guo, Y.; Zhang, X.; Wang, L.; Li, M.; Shen, M.; Zhou, Z.; Zhu, S.; Li, K.; Fang, Z.; Yan, B.; et al. The plasma exosomal miR-1180-3p serves as a novel potential diagnostic marker for cutaneous melanoma. Cancer Cell Int. 2021, 21, 487. [Google Scholar] [CrossRef] [PubMed]
- Bai, M.; Zhang, H.; Si, L.; Yu, N.; Zeng, A.; Zhao, R. Upregulation of Serum miR-10b Is Associated with Poor Prognosis in Patients with Melanoma. J. Cancer 2017, 8, 2487–2491. [Google Scholar] [CrossRef]
- Fogli, S.; Polini, B.; Carpi, S.; Pardini, B.; Naccarati, A.; Dubbini, N.; Lanza, M.; Breschi, M.C.; Romanini, A.; Nieri, P. Identification of plasma microRNAs as new potential biomarkers with high diagnostic power in human cutaneous melanoma. Tumor Biol. 2017, 39, 1010428317701646. [Google Scholar] [CrossRef]
- Armand-Labit, V.; Meyer, N.; Casanova, A.; Bonnabau, H.; Platzer, V.; Tournier, E.; Sansas, B.; Verdun, S.; Thouvenot, B.; Hilselberger, B.; et al. Identification of a Circulating MicroRNA Profile as a Biomarker of Metastatic Cutaneous Melanoma. Acta Derm. Venereol. 2016, 96, 29–34. [Google Scholar] [CrossRef]
- Shiiyama, R.; Fukushima, S.; Jinnin, M.; Yamashita, J.; Miyashita, A.; Nakahara, S.; Kogi, A.; Aoi, J.; Masuguchi, S.; Inoue, Y.; et al. Sensitive detection of melanoma metastasis using circulating microRNA expression profiles. Melanoma Res. 2013, 23, 366–372. [Google Scholar] [CrossRef]
- Dehghan, F.; Boozarpour, S.; Torabizadeh, Z.; Alijanpour, S. miR-21: A promising biomarker for the early detection of colon cancer. OncoTargets Ther. 2019, 12, 5601–5607. [Google Scholar] [CrossRef]
- Qu, J.; Yang, J.; Chen, M.; Cui, L.; Wang, T.; Gao, W.; Tian, J.; Wei, R. MicroRNA-21 as a diagnostic marker for hepatocellular carcinoma: A systematic review and meta-analysis. Pak. J. Med Sci. 2019, 35, 1466–1471. [Google Scholar] [CrossRef] [PubMed]
- Dioguardi, M.; Caloro, G.A.; Laino, L.; Alovisi, M.; Sovereto, D.; Crincoli, V.; Aiuto, R.; Coccia, E.; Troiano, G.; Muzio, L.L. Circulating miR-21 as a Potential Biomarker for the Diagnosis of Oral Cancer: A Systematic Review with Meta-Analysis. Cancers 2020, 12, 936. [Google Scholar] [CrossRef] [PubMed]
- Badr, F.M. Potential Role of miR-21 in Breast Cancer Diagnosis and Therapy. JSM Biotechnol. Biomed. Eng. 2016, 3, 1068–1075. [Google Scholar]
- Shiao, M.-S.; Chang, J.-M.; Lertkhachonsuk, A.-A.; Rermluk, N.; Jinawath, N. Circulating Exosomal miRNAs as Biomarkers in Epithelial Ovarian Cancer. Biomedicines 2021, 9, 1433. [Google Scholar] [CrossRef]
- Sala, M.; Fuster, J.; Llovet, J.M.; Navasa, M.; Solé, M.; Varela, M.; Pons, F.; Rimola, A.; García-Valdecasas, J.C.; Brú, C.; et al. High pathological risk of recurrence after surgical resection for hepatocellular carcinoma: An indication for salvage liver transplantation. Liver Transplant. 2004, 10, 1294–1300. [Google Scholar] [CrossRef]
- Joly, F.; Ahmed-Lecheheb, D.; Thiery-Vuillemin, A.; Orillard, E.; Coquan, E. Effets secondaires de la chimiothérapie des cancers testiculaires et suivi de l’après cancer. Bull. Cancer 2019, 106, 805–811. [Google Scholar] [CrossRef]
- Michaelidesová, A.; Konířová, J.; Bartůněk, P.; Zíková, M. Effects of Radiation Therapy on Neural Stem Cells. Genes 2019, 10, 640. [Google Scholar] [CrossRef]
- Kim, A.; Cohen, M.S. The discovery of vemurafenib for the treatment of BRAF-mutated metastatic melanoma. Expert Opin. Drug Discov. 2016, 11, 907–916. [Google Scholar] [CrossRef]
- Shi, H.; Hugo, W.; Kong, X.; Hong, A.; Koya, R.C.; Moriceau, G.; Chodon, T.; Guo, R.; Johnson, D.B.; Dahlman, K.B.; et al. Acquired Resistance and Clonal Evolution in Melanoma during BRAF Inhibitor Therapy. Cancer Discov. 2014, 4, 80–93. [Google Scholar] [CrossRef]
- Gibney, G.T.; Zager, J.S. Clinical development of dabrafenib in BRAF mutant melanoma and other malignancies. Expert Opin. Drug Metab. Toxicol. 2013, 9, 893–899. [Google Scholar] [CrossRef]
- Ge, L.; Wu, Y.; Wan, M.; You, Y.; Zhai, Z.; Song, Z. Metformin Increases Sensitivity of Melanoma Cells to Cisplatin by Blocking Exosomal-Mediated miR-34a Secretion. J. Oncol. 2021, 2021, 552523. [Google Scholar] [CrossRef] [PubMed]
- Kumar, P.; El-Sagheer, A.H.; Truong, L.; Brown, T. Locked nucleic acid (LNA) enhances binding affinity of triazole-linked DNA towards RNA. Chem. Commun. 2017, 53, 8910–8913. [Google Scholar] [CrossRef] [PubMed]
- Javanmard, S.H.; Vaseghi, G.; Ghasemi, A.; Rafiee, L.; Ferns, G.A.; Esfahani, H.N.; Nedaeinia, R. Therapeutic inhibition of microRNA-21 (miR-21) using locked-nucleic acid (LNA)-anti-miR and its effects on the biological behaviors of melanoma cancer cells in preclinical studies. Cancer Cell Int. 2020, 20, 384. [Google Scholar] [CrossRef] [PubMed]
- Pencheva, N.; Tran, H.; Buss, C.; Huh, D.; Drobnjak, M.; Busam, K.; Tavazoie, S.F. Convergent Multi-miRNA Targeting of ApoE Drives LRP1/LRP8-Dependent Melanoma Metastasis and Angiogenesis. Cell 2012, 151, 1068–1082. [Google Scholar] [CrossRef] [PubMed]
- Caporali, S.; Amaro, A.; Levati, L.; Alvino, E.; Lacal, P.M.; Mastroeni, S.; Ruffini, F.; Bonmassar, L.; Cappellini, G.C.A.; Felli, N.; et al. miR-126-3p down-regulation contributes to dabrafenib acquired resistance in melanoma by up-regulating ADAM9 and VEGF-A. J. Exp. Clin. Cancer Res. 2019, 38, 272. [Google Scholar] [CrossRef]
- Zheng, X.; Tang, H.; Zhao, X.; Sun, Y.; Jiang, Y.; Liu, Y. Long non-coding RNA FTH1P3 facilitates uveal melanoma cell growth and invasion through miR-224-5p. PLoS ONE 2017, 12, e0184746. [Google Scholar] [CrossRef]
- Hong, D.S.; Kang, Y.K.; Borad, M.; Sachdev, J.; Ejadi, S.; Lim, H.Y.; Brenner, A.J.; Park, K.; Lee, J.L.; Kim, T.Y.; et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer 2020, 122, 1630–1637. [Google Scholar] [CrossRef]
- Lelli, D.; Pedone, C.; Sahebkar, A. Curcumin and treatment of melanoma: The potential role of microRNAs. Biomed. Pharmacother. 2017, 88, 832–834. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Y.; Lv, Y.; Wang, J. Effect of curcumin on the proliferation, apoptosis, migration, and invasion of human melanoma A375 cells. Genet. Mol. Res. 2015, 14, 1056–1067. [Google Scholar] [CrossRef]
- Dahmke, I.N.; Backes, C.; Rudzitis-Auth, J.; Laschke, M.W.; Leidinger, P.; Menger, M.D.; Meese, E.; Mahlknecht, U. Curcumin Intake Affects miRNA Signature in Murine Melanoma with mmu-miR-205-5p Most Significantly Altered. PLOS ONE 2013, 8, e81122. [Google Scholar] [CrossRef]
- Yang, C.H.; Yue, J.; Sims, M.; Pfeffer, L.M. The Curcumin Analog EF24 Targets NF-κB and miRNA-21, and Has Potent Anticancer Activity In Vitro and In Vivo. PLoS ONE 2013, 8, e71130. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, D. Tanshinol inhibits growth of malignant melanoma cells via regulating miR-1207-5p/CHPF pathway. Arch Dermatol. Res. 2019, 312, 373–383. [Google Scholar] [CrossRef]
- Sun, Q.; Cong, R.; Yan, H.; Gu, H.; Zeng, Y.; Liu, N.; Chen, J.; Wang, B. Genistein inhibits growth of human uveal melanoma cells and affects microRNA-27a and target gene expression. Oncol. Rep. 2009, 22, 563–567. [Google Scholar] [CrossRef] [PubMed]
- Varrone, F.; Caputo, E. The miRNAs Role in Melanoma and in Its Resistance to Therapy. Int. J. Mol. Sci. 2020, 21, 878. [Google Scholar] [CrossRef] [PubMed]
- Escudier, B.; Dorval, T.; Chaput, N.; André, F.; Caby, M.-P.; Novault, S.; Flament, C.; Leboulaire, C.; Borg, C.; Amigorena, S.; et al. Vaccination of metastatic melanoma patients with autologous dendritic cell (DC) derived-exosomes: Results of thefirst phase I clinical trial. J. Transl. Med. 2005, 3, 10. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Feng, T.; Liu, B.; Qiu, F.; Xu, Y.; Zhao, Y.; Zheng, Y. Engineered exosomes: Desirable target-tracking characteristics for cerebrovascular and neurodegenerative disease therapies. Theranostics 2021, 11, 8926–8944. [Google Scholar] [CrossRef] [PubMed]
- Lai, J.J.; Chau, Z.L.; Chen, S.; Hill, J.J.; Korpany, K.V.; Liang, N.; Lin, L.; Lin, Y.; Liu, J.K.; Liu, Y.; et al. Exosome Processing and Characterization Approaches for Research and Technology Development. Adv. Sci. 2022, 9, e2103222. [Google Scholar] [CrossRef] [PubMed]
- Bovy, N.; Blomme, B.; Frères, P.; Dederen, S.; Nivelles, O.; Lion, M.; Carnet, O.; Martial, J.A.; Noël, A.; Thiry, M.; et al. Endothelial exosomes contribute to the antitumor response during breast cancer neoadjuvant chemotherapy via microRNA transfer. Oncotarget 2015, 6, 10253–10266. [Google Scholar] [CrossRef]
- O’Brien, K.; Lowry, M.C.; Corcoran, C.; Martinez, V.G.; Daly, M.; Rani, S.; Gallagher, W.M.; Radomski, M.W.; MacLeod, R.A.; O’Driscoll, L. miR-134 in extracellular vesicles reduces triple-negative breast cancer aggression and increases drug sensitivity. Oncotarget 2015, 6, 32774–32789. [Google Scholar] [CrossRef]
- Liang, G.; Kan, S.; Zhu, Y.; Feng, S.; Feng, W.; Gao, S. Engineered exosome-mediated delivery of functionally active miR-26a and its enhanced suppression effect in HepG2 cells. Int. J. Nanomed. 2018, 13, 585–599. [Google Scholar] [CrossRef]
- Ding, Y.; Cao, F.; Sun, H.; Wang, Y.; Liu, S.; Wu, Y.; Cui, Q.; Mei, W.; Li, F. Exosomes derived from human umbilical cord mesenchymal stromal cells deliver exogenous miR-145-5p to inhibit pancreatic ductal adenocarcinoma progression. Cancer Lett. 2018, 442, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Q.; Yi, K.; Qi, H.; Li, S.; Li, X.; Wang, Q.; Wang, Y.; Liu, C.; Qiu, M.; Yuan, X.; et al. Engineering blood exosomes for tumor-targeting efficient gene/chemo combination therapy. Theranostics 2020, 10, 7889–7905. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Mo, H.; He, Z.; Chen, A.; Cheng, P. Extracellular vesicles as an emerging drug delivery system for cancer treatment: Current strategies and recent advances. Biomed. Pharmacother. 2022, 153, 113480. [Google Scholar] [CrossRef] [PubMed]
- Hood, J.L.; Scott, M.J.; Wickline, S.A. Maximizing exosome colloidal stability following electroporation. Anal. Biochem. 2014, 448, 41–49. [Google Scholar] [CrossRef]
- Liang, G.; Zhu, Y.; Ali, D.J.; Tian, T.; Xu, H.; Si, K.; Sun, B.; Chen, B.; Xiao, Z. Engineered exosomes for targeted co-delivery of miR-21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer. J. Nanobiotechnology 2020, 18, 10. [Google Scholar] [CrossRef]
- Hung, M.E.; Leonard, J.N. Stabilization of Exosome-targeting Peptides via Engineered Glycosylation. J. Biol. Chem. 2015, 290, 8166–8172. [Google Scholar] [CrossRef]
- Cui, M.; Wang, H.; Yao, X.; Zhang, D.; Xie, Y.; Cui, R.; Zhang, X. Circulating MicroRNAs in Cancer: Potential and Challenge. Front. Genet. 2019, 10, 626. [Google Scholar] [CrossRef]

| Foundation | miRNA | EP | Target Cell | Target Gene | Mechanism | Ref |
|---|---|---|---|---|---|---|
| Invasion and metastasis | miR-4345 | - | Low metastatic melanoma cell | - | autophagy pathway | [42] |
| miR-1268a | - | Low metastatic melanoma cell | - | autophagy pathway | [43] | |
| miR-411-5p | - | Low metastatic melanoma cell | - | MAPK/ERK pathway | [44] | |
| miR-222 | - | Melanoma cell | p27Kip1/CDKN1B, c-KIT receptor and c-FOS | PI3K/AKT pathway | [41] | |
| miR-495-3p miR-376c-3p miR-6730-3p | - | Melanoma cell | - | PI3K/AKT pathway | [45] | |
| miR-154-5p | - | Melanoma cell | MCP1 | - | [46] | |
| miR-2478 | - | Melanoma cell | RAP1A | Akt/GSK3β pathway | [47] | |
| miR-21 | ↑ | Fibroblast | TIMP3 and MMP | - | [48] | |
| miR-106b-5p | ↑ | Melanocyte | EPHA4 | EPHA4/ERK pathway | [49] | |
| miR-191 let-7i let-7a | ↑ | Primary melanocyte | LIN28B and HMGA2 | EMT | [50] | |
| miR-494 | ↓ | - | Bcl-2 | - | [51] | |
| Tumor microeniron-ment | miR-21 | ↑ | Fibroblast | TIMP3 and MMP | - | [48] |
| miR-155 | ↑ | CAF | SOCS1 | JAK2/STAT3 pathway | [38] | |
| miR-211 | ↑ | Fibroblast | IGF2R | MAPK pathway | [52] | |
| miR-155 miR-210 | ↑ | Stromal fibroblast | - | OXPHOS | [53] | |
| miR-125b-5p | ↑ | Macrophage | LIPA | switch of TAM phenotype | [39] | |
| Angiogenes-is | miR-155 | ↑ | Fibroblast | SOCS1 | JAK2/STAT3 pathway | [38] |
| miR-9 | ↑ | Endothelial cell | SOCS5 | JAK/STAT pathway | [54] | |
| miR-222 | - | Melanoma cell | - | PI3K/AKT pathway | [41] | |
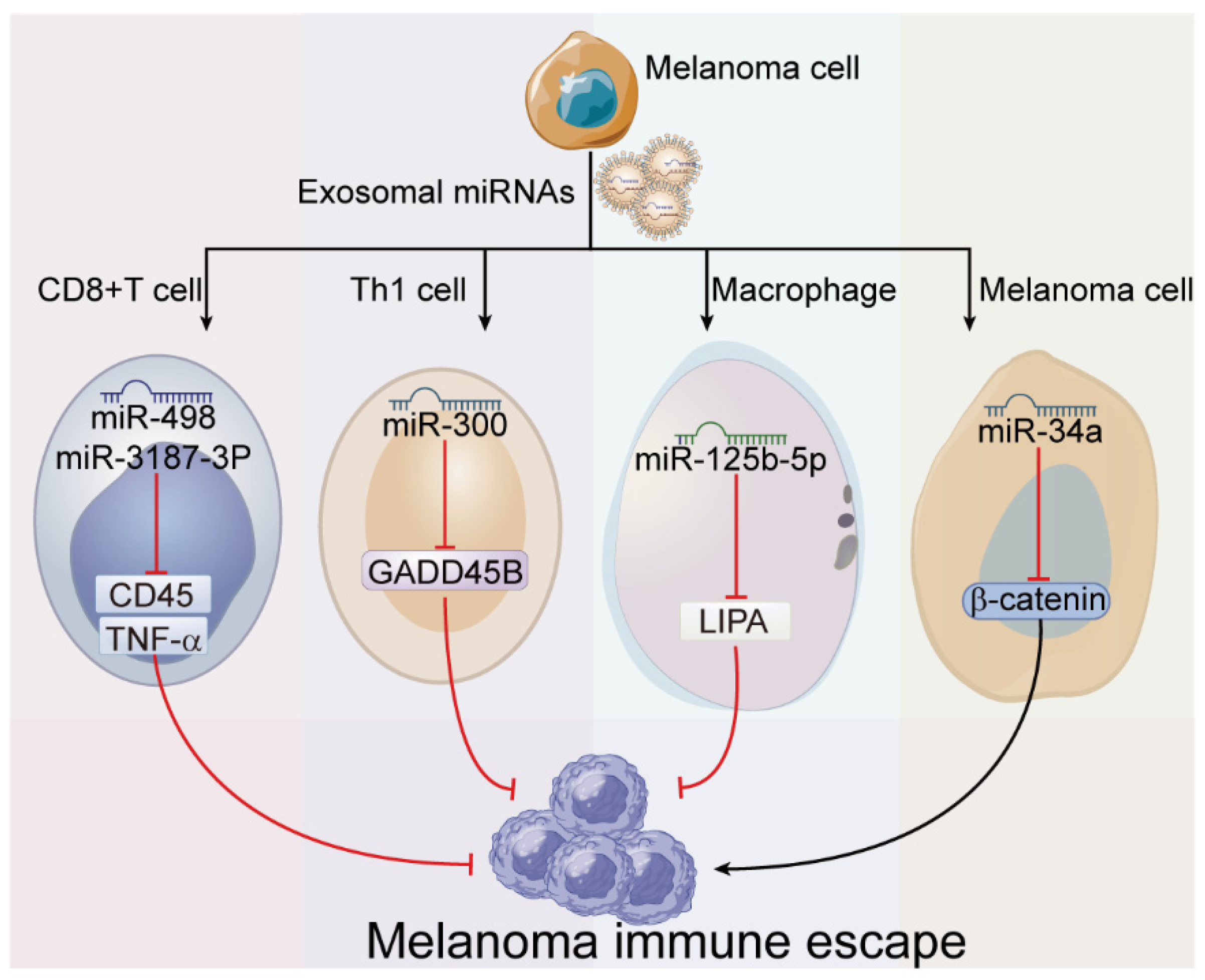
| Immune escape | miR-122 miR-149 miR-498 miR-181 miR-3187-3p | - | CD8+ T cell | TNF-α PTPRC | - | [36] |
| miR-300 | ↓ | Th 1 cell | GADD45B | - | [55] | |
| miR-125b-5p | ↑ | Macrophage | LIPA | switch of TAM phenotype | [39] | |
| miR-34a | - | Melanoma cell | β-catenin | - | [56] |
| miRNA | EP | Samples | Target | Effect | Refs |
|---|---|---|---|---|---|
| miR-211-5p | ↑ | Serum | - | Associated with disease stage and survival | [84] |
| miR-16 | ↑ | Serum | - | Associated with disease stage | [84] |
| miR-4487 | ↓ | Serum | - | Associated with diseases stage and survival | [84] |
| miR-221 | ↑ | Serum | - | Associated with patient survival and advanced clinical stage | [85] |
| miR-23a | ↓ | Serum | ATG12 | Associated with patient survival and tumor thickness | [86] |
| miR-150-5p | ↓ | Serum | - | Associated with patient survival | [87] |
| miR-142-3p | ↓ | Serum | - | Distinguish between stage III and stage IV | [87] |
| miR-206 | ↓ | Serum | - | Associated with poor prognosis and clinical stage | [88] |
| miR-106b-5p | ↑ | Serum exosome | EPHA4 | Activates the ERK pathway | [49] |
| miR-138 | ↓ | Whole blood | - | Associated with survival | [89] |
| let-7g-5p | ↓ | Plasma exosome | MAPK | High expression levels associated with better disease control | [90] |
| miR-34a | ↑ | Plasma exosome | β-catenin | Prevents tumor relapse and suppresses tumor cell proliferation | [56] |
| miR-146a miR-155 miR-125b miR-100 miR-125a miR-146b miR-99b | ↑ | Melanoma exosome | CTLA4 PD-1 | Convert myeloid cells into myeloid-derived suppressor cells | [91] |
| miR-495-3p miR-376c-3p miR-6730-3p | ↑ | Melanoma exosome | - | Promote melanoma growth and metastasis | [45] |
| miR-191 let-7a | ↑ | Serum exosome | - | Regulate the EMT process | [50] |
| miR-122 miR-149 miR-498 miR-3187-3p | ↑ | Melanoma exosome | TNF-α PTPRC | Promote melanoma immune escape | [36] |
| miR-17 miR-19a miR-21 miR-126 miR-149 | ↑ | Plasma exosome | - | Associated with melanoma development and metastasis | [92] |
| miR-532-5p miR-106b | ↑ | Serum exosome | - | Associated with metastasis and disease stage | [93] |
| miR-125b | ↓ | Serum exosome | - | Associated with advanced melanoma | [94] |
| miR-494-5p miR-4497 miR-513a-5p | ↑ | Melanoma exosome | - | May be related to melanoma growth and progression | [95] |
| miR-125b-5P miR-3934-5p | ↓ | Melanoma exosome | - | May be related to melanoma growth and progression | [95] |
| miR-1180-3p | ↓ | Plasma exosome | - | Promote melanoma growth | [96] |
| miR-10b | ↑ | Serum | - | High levels associated with short disease-free survival and overall survival | [97] |
| miR-149-3p miR-150-5p miR-193a-3p | ↑ ↑ ↓ | Plasma | - | Triple classifier is suitable for early diagnosis of melanoma | [98] |
| miR-1246 miR-185 | ↑ | Plasma | Distinguish metastatic melanoma | [99] | |
| miR-9 | ↑ | Serum | Distinguish metastatic melanoma | [100] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, Q.; Li, Z.; Li, Y.; Li, Y.; Zhang, Y.; Gui, R.; Cui, Y.; Zhang, Q.; Qian, L.; Xiong, Y.; et al. Exosome-Derived microRNA: Implications in Melanoma Progression, Diagnosis and Treatment. Cancers 2023, 15, 80. https://doi.org/10.3390/cancers15010080
Ye Q, Li Z, Li Y, Li Y, Zhang Y, Gui R, Cui Y, Zhang Q, Qian L, Xiong Y, et al. Exosome-Derived microRNA: Implications in Melanoma Progression, Diagnosis and Treatment. Cancers. 2023; 15(1):80. https://doi.org/10.3390/cancers15010080
Chicago/Turabian StyleYe, Qiang, Zi Li, Yang Li, Yirong Li, Yan Zhang, Runlin Gui, Yue Cui, Qi Zhang, Lu Qian, Yuyan Xiong, and et al. 2023. "Exosome-Derived microRNA: Implications in Melanoma Progression, Diagnosis and Treatment" Cancers 15, no. 1: 80. https://doi.org/10.3390/cancers15010080
APA StyleYe, Q., Li, Z., Li, Y., Li, Y., Zhang, Y., Gui, R., Cui, Y., Zhang, Q., Qian, L., Xiong, Y., & Yu, Y. (2023). Exosome-Derived microRNA: Implications in Melanoma Progression, Diagnosis and Treatment. Cancers, 15(1), 80. https://doi.org/10.3390/cancers15010080






