Non-Coding RNAs in Airway Diseases: A Brief Overview of Recent Data
Abstract
Simple Summary
Abstract
1. Lung Diseases
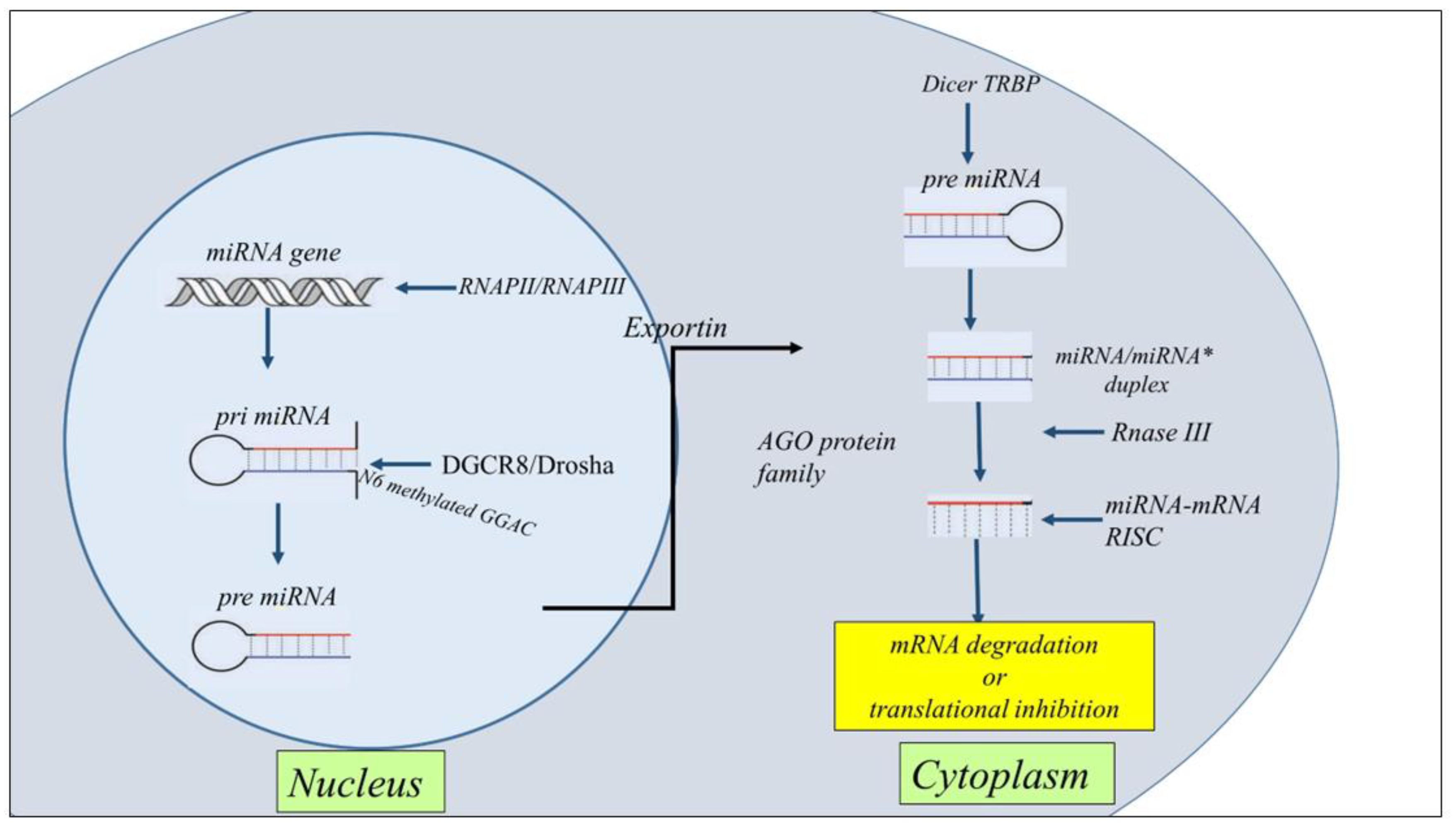
2. Nc-RNA Biogenesis
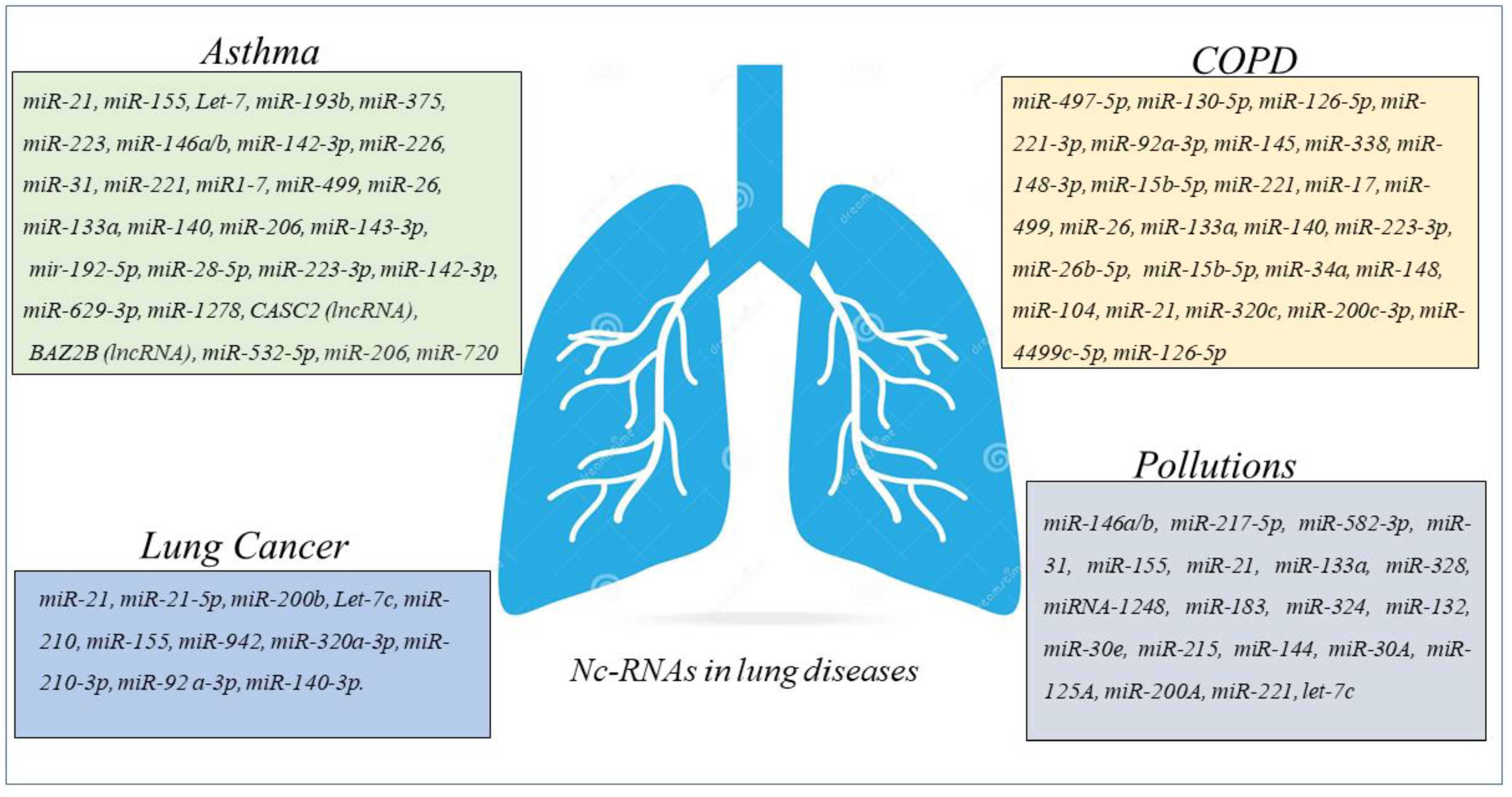
3. Nc-RNAs in Asthma
4. ncRNAs in COPD
5. ncRNAs in Lung Cancer
6. Environmental Pollution and ncRNAs in the Airways
7. ncRNAs as Therapeutic Approach in Lung Diseases
8. Extracellular Vesicles and nc-RNA in Airway Diseases
9. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dey, S.; Eapen, M.S.; Chia, C.; Gaikwad, A.V.; Wark, P.A.B.; Sohal, S.S. Pathogenesis, Clinical Features of Asthma COPD Overlap, and Therapeutic Modalities. Am. J. Physiol. Lung Cell. Mol. Physiol. 2022, 322, L64–L83. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.J.; Brightling, C. Pathogenesis of Asthma: Implications for Precision Medicine. Clin. Sci. 2017, 131, 1723–1735. [Google Scholar] [CrossRef] [PubMed]
- Zahran, H.S.; Bailey, C.M.; Damon, S.A.; Garbe, P.L.; Breysse, P.N. Vital Signs: Asthma in Children—United States, 2001–2016. MMWR. Morb. Mortal. Wkly. Rep. 2018, 67, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Kwah, J.H.; Peters, A.T. Asthma in Adults: Principles of Treatment. Allergy Asthma Proc. 2019, 40, 396–402. [Google Scholar] [CrossRef]
- Barnes, P.J. Inflammatory Mechanisms in Patients with Chronic Obstructive Pulmonary Disease. J. Allergy Clin. Immunol. 2016, 138, 16–27. [Google Scholar] [CrossRef]
- Barnes, P.J.; Burney, P.G.J.; Silverman, E.K.; Celli, B.R.; Vestbo, J.; Wedzicha, J.A.; Wouters, E.F.M. Chronic Obstructive Pulmonary Disease. Nat. Rev. Dis. Prim. 2015, 1, 15076. [Google Scholar] [CrossRef]
- Negewo, N.A.; Gibson, P.G.; McDonald, V.M. COPD and Its Comorbidities: Impact, Measurement and Mechanisms. Respirology 2015, 20, 1160–1171. [Google Scholar] [CrossRef]
- Mathioudakis, A.G.; Vanfleteren, L.E.G.W.; Lahousse, L.; Higham, A.; Allinson, J.P.; Gotera, C.; Visca, D.; Singh, D.; Spanevello, A. Current Developments and Future Directions in COPD. Eur. Respir. Rev. Off. J. Eur. Respir. Soc. 2020, 29, 200289. [Google Scholar] [CrossRef]
- Hillas, G.; Papaporfyriou, A.; Dimakou, K.; Papaioannou, A.I. Pharmacological Treatment of Stable COPD: Need for a Simplified Approach. Postgrad. Med. 2020, 132, 126–131. [Google Scholar] [CrossRef]
- Alshabanat, A.; Zafari, Z.; Albanyan, O.; Dairi, M.; FitzGerald, J.M. Asthma and COPD Overlap Syndrome (ACOS): A Systematic Review and Meta Analysis. PLoS ONE 2015, 10, e0136065. [Google Scholar] [CrossRef]
- Barnes, P.J.; Adcock, I.M. Chronic Obstructive Pulmonary Disease and Lung Cancer: A Lethal Association. Am. J. Respir. Crit. Care Med. 2011, 184, 866–867. [Google Scholar] [CrossRef] [PubMed]
- Durham, A.L.; Adcock, I.M. The Relationship between COPD and Lung Cancer. Lung Cancer 2015, 90, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.; Anzueto, A.; Miravitlles, M.; Arvis, P.; Haverstock, D.; Trajanovic, M.; Sethi, S. Prognostic Factors for Clinical Failure of Exacerbations in Elderly Outpatients with Moderate-to-Severe COPD. Int. J. Chron. Obstruct. Pulmon. Dis. 2015, 10, 985–993. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Reck, M.; Heigener, D.F.; Mok, T.; Soria, J.-C.; Rabe, K.F. Management of Non-Small-Cell Lung Cancer: Recent Developments. Lancet 2013, 382, 709–719. [Google Scholar] [CrossRef] [PubMed]
- Comer, B.S.; Ba, M.; Singer, C.A.; Gerthoffer, W.T. Epigenetic Targets for Novel Therapies of Lung Diseases. Pharmacol. Ther. 2015, 147, 91–110. [Google Scholar] [CrossRef]
- Tubita, V.; Callejas-Díaz, B.; Roca-Ferrer, J.; Marin, C.; Liu, Z.; Wang, D.Y.; Mullol, J. Role of MicroRNAs in Inflammatory Upper Airway Diseases. Allergy 2021, 76, 1967–1980. [Google Scholar] [CrossRef]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and Their Integrated Networks. J. Integr. Bioinform. 2019, 16. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene Regulation by Long Non-Coding RNAs and Its Biological Functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Dong, H.; Zhou, J.; Cheng, Y.; Wang, M.; Wang, S.; Xu, H. Biogenesis, Functions, and Role of CircRNAs in Lung Cancer. Cancer Manag. Res. 2021, 13, 6651–6671. [Google Scholar] [CrossRef]
- Fatica, A.; Bozzoni, I. Long Non-Coding RNAs: New Players in Cell Differentiation and Development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef]
- Maoz, R.; Garfinkel, B.P.; Soreq, H. Alzheimer’s Disease and NcRNAs. Adv. Exp. Med. Biol. 2017, 978, 337–361. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Wu, Y.; Wang, W.; Su, W.; Liu, Y.; Wang, Y.; Fan, C.; Li, X.; Li, G.; Li, Y.; et al. Circular RNAs (CircRNAs) in Cancer. Cancer Lett. 2018, 425, 134–142. [Google Scholar] [CrossRef]
- Zhang, S.-J.; Chen, X.; Li, C.-P.; Li, X.-M.; Liu, C.; Liu, B.-H.; Shan, K.; Jiang, Q.; Zhao, C.; Yan, B. Identification and Characterization of Circular RNAs as a New Class of Putative Biomarkers in Diabetes Retinopathy. Investig. Ophthalmol. Vis. Sci. 2017, 58, 6500–6509. [Google Scholar] [CrossRef] [PubMed]
- Ullah, S.; John, P.; Bhatti, A. MicroRNAs with a Role in Gene Regulation and in Human Diseases. Mol. Biol. Rep. 2014, 41, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Tüfekci, K.U.; Oner, M.G.; Meuwissen, R.L.J.; Genç, S. The Role of MicroRNAs in Human Diseases. Methods Mol. Biol. 2014, 1107, 33–50. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Lu, H.; Song, G. MiR-221-3p and MiR-92a-3p Enhances Smoking-Induced Inflammation in COPD. J. Clin. Lab. Anal. 2021, 35, e23857. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, R.; Lu, Y.; Zeng, Y. The MicroRNA-1278/SHP-1/STAT3 Pathway Is Involved in Airway Smooth Muscle Cell Proliferation in a Model of Severe Asthma Both Intracellularly and Extracellularly. Mol. Cell. Biochem. 2022, 477, 1439–1451. [Google Scholar] [CrossRef] [PubMed]
- Soni, D.K.; Biswas, R. Role of Non-Coding RNAs in Post-Transcriptional Regulation of Lung Diseases. Front. Genet. 2021, 12, 767348. [Google Scholar] [CrossRef]
- Tanzer, A.; Stadler, P.F. Molecular Evolution of a MicroRNA Cluster. J. Mol. Biol. 2004, 339, 327–335. [Google Scholar] [CrossRef]
- de Rie, D.; Abugessaisa, I.; Alam, T.; Arner, E.; Arner, P.; Ashoor, H.; Åström, G.; Babina, M.; Bertin, N.; Burroughs, A.M.; et al. An Integrated Expression Atlas of MiRNAs and Their Promoters in Human and Mouse. Nat. Biotechnol. 2017, 35, 872–878. [Google Scholar] [CrossRef] [PubMed]
- Alsop, E.; Meechoovet, B.; Kitchen, R.; Sweeney, T.; Beach, T.G.; Serrano, G.E.; Hutchins, E.; Ghiran, I.; Reiman, R.; Syring, M.; et al. A Novel Tissue Atlas and Online Tool for the Interrogation of Small RNA Expression in Human Tissues and Biofluids. Front. Cell Dev. Biol. 2022, 10, 804164. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of MicroRNA Biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Makarova, J.A.; Shkurnikov, M.U.; Wicklein, D.; Lange, T.; Samatov, T.R.; Turchinovich, A.A.; Tonevitsky, A.G. Intracellular and Extracellular MicroRNA: An Update on Localization and Biological Role. Prog. Histochem. Cytochem. 2016, 51, 33–49. [Google Scholar] [CrossRef]
- Munjas, J.; Sopić, M.; Stefanović, A.; Košir, R.; Ninić, A.; Joksić, I.; Antonić, T.; Spasojević-Kalimanovska, V.; Prosenc Zmrzljak, U. Non-Coding RNAs in Preeclampsia-Molecular Mechanisms and Diagnostic Potential. Int. J. Mol. Sci. 2021, 22, 10652. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Fan, R.; Xiao, C.; Wan, X.; Cha, W.; Miao, Y.; Zhou, Y.; Qin, C.; Cui, T.; Su, F.; Shan, X. Small Molecules with Big Roles in MicroRNA Chemical Biology and MicroRNA-Targeted Therapeutics. RNA Biol. 2019, 16, 707–718. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Huang, P.; Qiu, C. Progresses in Epigenetic Studies of Asthma from the Perspective of High-Throughput Analysis Technologies: A Narrative Review. Ann. Transl. Med. 2022, 10, 493. [Google Scholar] [CrossRef]
- Gomez, J.L. Epigenetics in Asthma. Curr. Allergy Asthma Rep. 2019, 19, 56. [Google Scholar] [CrossRef]
- Heffler, E.; Allegra, A.; Pioggia, G.; Picardi, G.; Musolino, C.; Gangemi, S. MicroRNA Profiling in Asthma: Potential Biomarkers and Therapeutic Targets. Am. J. Respir. Cell Mol. Biol. 2017, 57, 642–650. [Google Scholar] [CrossRef]
- Farmanzadeh, A.; Qujeq, D.; Yousefi, T. The Interaction Network of MicroRNAs with Cytokines and Signaling Pathways in Allergic Asthma. MicroRNA 2022, 11, 104–117. [Google Scholar] [CrossRef]
- Wang, X.; Chen, H.; Liu, J.; Gai, L.; Yan, X.; Guo, Z.; Liu, F. Emerging Advances of Non-Coding RNAs and Competitive Endogenous RNA Regulatory Networks in Asthma. Bioengineered 2021, 12, 7820–7836. [Google Scholar] [CrossRef] [PubMed]
- ElKashef, S.M.M.A.E.; Ahmad, S.E.-A.; Soliman, Y.M.A.; Mostafa, M.S. Role of MicroRNA-21 and MicroRNA-155 as Biomarkers for Bronchial Asthma. Innate Immun. 2021, 27, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Specjalski, K.; Niedoszytko, M. MicroRNAs: Future Biomarkers and Targets of Therapy in Asthma? Curr. Opin. Pulm. Med. 2020, 26, 285–292. [Google Scholar] [CrossRef] [PubMed]
- Specjalski, K.; Jassem, E. MicroRNAs: Potential Biomarkers and Targets of Therapy in Allergic Diseases? Arch. Immunol. Ther. Exp. (Warsz). 2019, 67, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Polikepahad, S.; Knight, J.M.; Naghavi, A.O.; Oplt, T.; Creighton, C.J.; Shaw, C.; Benham, A.L.; Kim, J.; Soibam, B.; Harris, R.A.; et al. Proinflammatory Role for Let-7 MicroRNAS in Experimental Asthma. J. Biol. Chem. 2010, 285, 30139–30149. [Google Scholar] [CrossRef]
- Lu, T.X.; Hartner, J.; Lim, E.-J.; Fabry, V.; Mingler, M.K.; Cole, E.T.; Orkin, S.H.; Aronow, B.J.; Rothenberg, M.E. MicroRNA-21 Limits in Vivo Immune Response-Mediated Activation of the IL-12/IFN-Gamma Pathway, Th1 Polarization, and the Severity of Delayed-Type Hypersensitivity. J. Immunol. 2011, 187, 3362–3373. [Google Scholar] [CrossRef]
- Dong, J.; Sun, D.; Lu, F. Association of Two Polymorphisms of MiRNA-146a Rs2910164 (G > C) and MiRNA-499 Rs3746444 (T > C) with Asthma: A Meta-Analysis. J. Asthma Off. J. Assoc. Care Asthma 2021, 58, 995–1002. [Google Scholar] [CrossRef]
- Shi, Z.-G.; Sun, Y.; Wang, K.-S.; Jia, J.-D.; Yang, J.; Li, Y.-N. Effects of MiR-26a/MiR-146a/MiR-31 on Airway Inflammation of Asthma Mice and Asthma Children. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 5432–5440. [Google Scholar] [CrossRef]
- Gagliardo, R.; Ferrante, G.; Fasola, S.; Di Vincenzo, S.; Pace, E.; La Grutta, S. Resolvin D1 and MiR-146a Are Independent Distinctive Parameters in Children with Moderate and Severe Asthma. Clin. Exp. Allergy J. Br. Soc. Allergy Clin. Immunol. 2021, 51, 350–353. [Google Scholar] [CrossRef]
- Marques-Rocha, J.L.; Samblas, M.; Milagro, F.I.; Bressan, J.; Martínez, J.A.; Marti, A. Noncoding RNAs, Cytokines, and Inflammation-Related Diseases. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2015, 29, 3595–3611. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Nunez, R.T.; Louafi, F.; Sanchez-Elsner, T. The Interleukin 13 (IL-13) Pathway in Human Macrophages Is Modulated by MicroRNA-155 via Direct Targeting of Interleukin 13 Receptor Alpha1 (IL13Ralpha1). J. Biol. Chem. 2011, 286, 1786–1794. [Google Scholar] [CrossRef] [PubMed]
- Malmhäll, C.; Alawieh, S.; Lu, Y.; Sjöstrand, M.; Bossios, A.; Eldh, M.; Rådinger, M. MicroRNA-155 Is Essential for T(H)2-Mediated Allergen-Induced Eosinophilic Inflammation in the Lung. J. Allergy Clin. Immunol. 2014, 133, 1429–1438.e7. [Google Scholar] [CrossRef] [PubMed]
- Mayoral, R.J.; Deho, L.; Rusca, N.; Bartonicek, N.; Saini, H.K.; Enright, A.J.; Monticelli, S. MiR-221 Influences Effector Functions and Actin Cytoskeleton in Mast Cells. PLoS ONE 2011, 6, e26133. [Google Scholar] [CrossRef]
- Yao, Y.; Chen, C.-L.; Yu, D.; Liu, Z. Roles of Follicular Helper and Regulatory T Cells in Allergic Diseases and Allergen Immunotherapy. Allergy 2021, 76, 456–470. [Google Scholar] [CrossRef]
- Yang, H.-Y.; Barbi, J.; Wu, C.-Y.; Zheng, Y.; Vignali, P.D.A.; Wu, X.; Tao, J.-H.; Park, B.V.; Bandara, S.; Novack, L.; et al. MiR-17 Resulted in Enhanced Suppressive Activity. Ectopic of Genes Encoding Effector Cytokines. Thus, MiR-17 Provides a Potent Layer of Treg Cell Control Expression of MiR-17 Imparted Effector-T-Cell-like Characteristics to Treg Cells via the de-repression. Immunity 2016, 45, 83–93. [Google Scholar] [CrossRef]
- Johansson, K.; Malmhäll, C.; Ramos-Ramírez, P.; Rådinger, M. MicroRNA-155 Is a Critical Regulator of Type 2 Innate Lymphoid Cells and IL-33 Signaling in Experimental Models of Allergic Airway Inflammation. J. Allergy Clin. Immunol. 2017, 139, 1007–1016.e9. [Google Scholar] [CrossRef]
- Cheng, W.; Yan, K.; Xie, L.Y.; Chen, F.; Yu, H.C.; Huang, Y.X.; Dang, C.X. MiR-143-3p Controls TGF-Β1-Induced Cell Proliferation and Extracellular Matrix Production in Airway Smooth Muscle via Negative Regulation of the Nuclear Factor of Activated T Cells 1. Mol. Immunol. 2016, 78, 133–139. [Google Scholar] [CrossRef]
- Lou, L.; Tian, M.; Chang, J.; Li, F.; Zhang, G. MiRNA-192-5p Attenuates Airway Remodeling and Autophagy in Asthma by Targeting MMP-16 and ATG7. Biomed. Pharmacother. 2020, 122, 109692. [Google Scholar] [CrossRef]
- Maneechotesuwan, K. Role of MicroRNA in Severe Asthma. Respir. Investig. 2019, 57, 9–19. [Google Scholar] [CrossRef]
- Perry, M.M.; Baker, J.E.; Gibeon, D.S.; Adcock, I.M.; Chung, K.F. Airway Smooth Muscle Hyperproliferation Is Regulated by MicroRNA-221 in Severe Asthma. Am. J. Respir. Cell Mol. Biol. 2014, 50, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Tsitsiou, E.; Williams, A.E.; Moschos, S.A.; Patel, K.; Rossios, C.; Jiang, X.; Adams, O.-D.; Macedo, P.; Booton, R.; Gibeon, D.; et al. Transcriptome Analysis Shows Activation of Circulating CD8+ T Cells in Patients with Severe Asthma. J. Allergy Clin. Immunol. 2012, 129, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Maes, T.; Cobos, F.A.; Schleich, F.; Sorbello, V.; Henket, M.; De Preter, K.; Bracke, K.R.; Conickx, G.; Mesnil, C.; Vandesompele, J.; et al. Asthma Inflammatory Phenotypes Show Differential MicroRNA Expression in Sputum. J. Allergy Clin. Immunol. 2016, 137, 1433–1446. [Google Scholar] [CrossRef] [PubMed]
- Kyyaly, M.A.; Sanchez-Elsner, T.; He, P.; Sones, C.L.; Arshad, S.H.; Kurukulaaratchy, R.J. Circulating MiRNAs-A Potential Tool to Identify Severe Asthma Risk? Clin. Transl. Allergy 2021, 11, e12040. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Sun, Z.; Ren, T.; Lei, W. Differential Expression of LncRNA CASC2 in the Serum of Childhood Asthma and Its Role in Airway Smooth Muscle Cells Proliferation and Migration. J. Asthma Allergy 2022, 15, 197–207. [Google Scholar] [CrossRef]
- Xia, L.; Wang, X.; Liu, L.; Fu, J.; Xiao, W.; Liang, Q.; Han, X.; Huang, S.; Sun, L.; Gao, Y.; et al. Lnc-BAZ2B Promotes M2 Macrophage Activation and Inflammation in Children with Asthma through Stabilizing BAZ2B Pre-MRNA. J. Allergy Clin. Immunol. 2021, 147, 921–932.e9. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Panganiban, R.; Kho, A.T.; McGeachie, M.J.; Farnam, L.; Chase, R.P.; Weiss, S.T.; Lu, Q.; Tantisira, K.G. Circulating MicroRNAs and Treatment Response in Childhood Asthma. Am. J. Respir. Crit. Care Med. 2020, 202, 65–72. [Google Scholar] [CrossRef]
- Karam, R.A.; Abd Elrahman, D.M. Differential Expression of MiR-155 and Let-7a in the Plasma of Childhood Asthma: Potential Biomarkers for Diagnosis and Severity. Clin. Biochem. 2019, 68, 30–36. [Google Scholar] [CrossRef]
- Kho, A.T.; McGeachie, M.J.; Moore, K.G.; Sylvia, J.M.; Weiss, S.T.; Tantisira, K.G. Circulating MicroRNAs and Prediction of Asthma Exacerbation in Childhood Asthma. Respir. Res. 2018, 19, 128. [Google Scholar] [CrossRef]
- Zhuang, Y.; Hobbs, B.D.; Hersh, C.P.; Kechris, K. Identifying MiRNA-MRNA Networks Associated With COPD Phenotypes. Front. Genet. 2021, 12, 748356. [Google Scholar] [CrossRef]
- Barreiro, E. The Role of MicroRNAs in COPD Muscle Dysfunction and Mass Loss: Implications on the Clinic. Expert Rev. Respir. Med. 2016, 10, 1011–1022. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Ye, M.; Wang, J.; Ye, L.; Jin, M. Construction of Potential MiRNA-MRNA Regulatory Network in COPD Plasma by Bioinformatics Analysis. Int. J. Chron. Obstruct. Pulmon. Dis. 2020, 15, 2135–2145. [Google Scholar] [CrossRef] [PubMed]
- Lacedonia, D.; Palladino, G.P.; Foschino-Barbaro, M.P.; Scioscia, G.; Carpagnano, G.E. Expression Profiling of MiRNA-145 and MiRNA-338 in Serum and Sputum of Patients with COPD, Asthma, and Asthma-COPD Overlap Syndrome Phenotype. Int. J. Chronic Obstr. Pulm. Dis. 2017, 12, 1811. [Google Scholar] [CrossRef] [PubMed]
- Hirai, K.; Shirai, T.; Shimoshikiryo, T.; Ueda, M.; Gon, Y.; Maruoka, S.; Itoh, K. Circulating MicroRNA-15b-5p as a Biomarker for Asthma-COPD Overlap. Allergy 2021, 76, 766–774. [Google Scholar] [CrossRef] [PubMed]
- Barnes, P.J.; Baker, J.; Donnelly, L.E. Cellular Senescence as a Mechanism and Target in Chronic Lung Diseases. Am. J. Respir. Crit. Care Med. 2019, 200, 556–564. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, H.; Raman, I.; Yan, M.; Chen, Q.; Li, Q.-Z. Peripheral Blood Mononuclear Cell Gene Expression in Chronic Obstructive Pulmonary Disease: MiRNA and MRNA Regulation. J. Inflamm. Res. 2022, 15, 2167–2180. [Google Scholar] [CrossRef]
- Kim, R.Y.; Sunkara, K.P.; Bracke, K.R.; Jarnicki, A.G.; Donovan, C.; Hsu, A.C.; Ieni, A.; Beckett, E.L.; Galvão, I.; Wijnant, S.; et al. A MicroRNA-21-Mediated SATB1/S100A9/NF-ΚB Axis Promotes Chronic Obstructive Pulmonary Disease Pathogenesis. Sci. Transl. Med. 2021, 13, eaav7223. [Google Scholar] [CrossRef]
- Ferraro, M.; Di Vincenzo, S.; Sangiorgi, C.; Leto Barone, S.; Gangemi, S.; Lanata, L.; Pace, E. Carbocysteine Modifies Circulating MiR-21, IL-8, SRAGE, and FAGEs Levels in Mild Acute Exacerbated COPD Patients: A Pilot Study. Pharmaceuticals 2022, 15, 218. [Google Scholar] [CrossRef]
- Cerón-Pisa, N.; Iglesias, A.; Shafiek, H.; Martín-Medina, A.; Esteva-Socias, M.; Muncunill, J.; Fleischer, A.; Verdú, J.; Cosío, B.G.; Sauleda, J. Hsa-Mir-320c, Hsa-Mir-200c-3p, and Hsa-Mir-449c-5p as Potential Specific MiRNA Biomarkers of COPD: A Pilot Study. Pathophysiol. Off. J. Int. Soc. Pathophysiol. 2022, 29, 143–156. [Google Scholar] [CrossRef]
- Liu, P.; Wang, Y.; Zhang, N.; Zhao, X.; Li, R.; Wang, Y.; Chen, C.; Wang, D.; Zhang, X.; Chen, L.; et al. Comprehensive Identification of RNA Transcripts and Construction of RNA Network in Chronic Obstructive Pulmonary Disease. Respir. Res. 2022, 23, 154. [Google Scholar] [CrossRef]
- Qiao, X.; Hou, G.; He, Y.-L.; Song, D.-F.; An, Y.; Altawil, A.; Zhou, X.-M.; Wang, Q.-Y.; Kang, J.; Yin, Y. The Novel Regulatory Role of the LncRNA-MiRNA-MRNA Axis in Chronic Inflammatory Airway Diseases. Front. Mol. Biosci. 2022, 9, 927549. [Google Scholar] [CrossRef]
- Paul, S.; Ruiz-Manriquez, L.M.; Ambriz-Gonzalez, H.; Medina-Gomez, D.; Valenzuela-Coronado, E.; Moreno-Gomez, P.; Pathak, S.; Chakraborty, S.; Srivastava, A. Impact of Smoking-Induced Dysregulated Human MiRNAs in Chronic Disease Development and Their Potential Use in Prognostic and Therapeutic Purposes. J. Biochem. Mol. Toxicol. 2022, 36, e23134. [Google Scholar] [CrossRef] [PubMed]
- Mateu-Jimenez, M.; Curull, V.; Rodríguez-Fuster, A.; Aguiló, R.; Sánchez-Font, A.; Pijuan, L.; Gea, J.; Barreiro, E. Profile of Epigenetic Mechanisms in Lung Tumors of Patients with Underlying Chronic Respiratory Conditions. Clin. Epigenetics 2018, 10, 7. [Google Scholar] [CrossRef] [PubMed]
- Rhim, J.; Baek, W.; Seo, Y.; Kim, J.H. From Molecular Mechanisms to Therapeutics: Understanding MicroRNA-21 in Cancer. Cells 2022, 11, 2791. [Google Scholar] [CrossRef]
- Shen, J.; Xia, W.; Khotskaya, Y.B.; Huo, L.; Nakanishi, K.; Lim, S.-O.; Du, Y.; Wang, Y.; Chang, W.-C.; Chen, C.-H.; et al. EGFR Modulates MicroRNA Maturation in Response to Hypoxia through Phosphorylation of AGO2. Nature 2013, 497, 383–387. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Guo, D.; Zhang, Y. Association of MicroRNA-21 with P53 at Mutant Sites R175H and R248Q, Clinicopathological Features, and Prognosis of NSCLC. Mol. Ther. Oncolytics 2020, 19, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Zheng, X.; Shi, P. Electrochemical Sensor Propelled by Exonuclease III for Highly Efficient MicroRNA-155 Detection. Analyst 2022, 147, 4824–4828. [Google Scholar] [CrossRef]
- Yadegar, N.; Dadashi, Z.; Shams, K.; Mohammadi, M.; Abyar, M.; Rafat, M. The Prominent Role of MiR-942 in Carcinogenesis of Tumors. Adv. Biomed. Res. 2022, 11, 63. [Google Scholar] [CrossRef]
- Vykoukal, J.; Fahrmann, J.F.; Patel, N.; Shimizu, M.; Ostrin, E.J.; Dennison, J.B.; Ivan, C.; Goodman, G.E.; Thornquist, M.D.; Barnett, M.J.; et al. Contributions of Circulating MicroRNAs for Early Detection of Lung Cancer. Cancers 2022, 14, 4221. [Google Scholar] [CrossRef]
- Guo, C.; Lv, S.; Liu, Y.; Li, Y. Biomarkers for the Adverse Effects on Respiratory System Health Associated with Atmospheric Particulate Matter Exposure. J. Hazard. Mater. 2022, 421, 126760. [Google Scholar] [CrossRef]
- Liu, G.; Li, Y.; Zhou, J.; Xu, J.; Yang, B. PM2.5 Deregulated MicroRNA and Inflammatory Microenvironment in Lung Injury. Environ. Toxicol. Pharmacol. 2022, 91, 103832. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.; Chen, Q.; Ma, Y. Elevated Expression of MiR-146 Involved in Regulating Mice Pulmonary Dysfunction after Exposure to PM2.5. J. Toxicol. Sci. 2021, 46, 437–443. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.; Li, S.; Ma, X.; Li, R.; Zhang, H.; Li, J.; Yan, X. MiR-217-5p Inhibits Smog (PM2.5)-Induced Inflammation and Oxidative Stress Response of Mouse Lung Tissues and Macrophages through Targeting STAT1. Aging 2022, 14, 6796–6808. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Ju, L.; Li, C.; Cheng, H.; Li, N.; Zhang, Q.; Sun, S.; Ding, L.; Sui, X.; Zhang, C.; et al. MiR-582-3p Participates in the Regulation of Biological Behaviors of A549 Cells by Ambient PM(2.5) Exposure. Environ. Sci. Pollut. Res. Int. 2022, 29, 13624–13634. [Google Scholar] [CrossRef]
- Wang, C.; Wang, J.; Zheng, X.; Zhang, J.; Zhang, J.; Qiao, G.; Liu, H.; Zhao, H.; Bai, J.; Zhang, H.; et al. Epigenetic Regulation Is Involved in Traffic-Related PM(2.5) Aggravating Allergic Airway Inflammation in Rats. Clin. Immunol. 2022, 234, 108914. [Google Scholar] [CrossRef]
- Weidner, J.; Bartel, S.; Kılıç, A.; Zissler, U.M.; Renz, H.; Schwarze, J.; Schmidt-Weber, C.B.; Maes, T.; Rebane, A.; Krauss-Etschmann, S.; et al. Spotlight on MicroRNAs in Allergy and Asthma. Allergy 2021, 76, 1661–1678. [Google Scholar] [CrossRef]
- Makrinioti, H.; Camargo, C.A.; Zhu, Z.; Freishtat, R.J.; Hasegawa, K. Air Pollution, Bronchiolitis, and Asthma: The Role of Nasal MicroRNAs. Lancet. Respir. Med. 2022, 10, 733–734. [Google Scholar] [CrossRef]
- Rider, C.F.; Yamamoto, M.; Günther, O.P.; Hirota, J.A.; Singh, A.; Tebbutt, S.J.; Carlsten, C. Controlled Diesel Exhaust and Allergen Coexposure Modulates MicroRNA and Gene Expression in Humans: Effects on Inflammatory Lung Markers. J. Allergy Clin. Immunol. 2016, 138, 1690–1700. [Google Scholar] [CrossRef]
- Yamamoto, M.; Singh, A.; Sava, F.; Pui, M.; Tebbutt, S.J.; Carlsten, C. MicroRNA Expression in Response to Controlled Exposure to Diesel Exhaust: Attenuation by the Antioxidant N-Acetylcysteine in a Randomized Crossover Study. Environ. Health Perspect. 2013, 121, 670–675. [Google Scholar] [CrossRef]
- Pan, J.; Xue, Y.; Li, S.; Wang, L.; Mei, J.; Ni, D.; Jiang, J.; Zhang, M.; Yi, S.; Zhang, R.; et al. PM(2.5) Induces the Distant Metastasis of Lung Adenocarcinoma via Promoting the Stem Cell Properties of Cancer Cells. Environ. Pollut. 2022, 296, 118718. [Google Scholar] [CrossRef]
- Szymczak, I.; Wieczfinska, J.; Pawliczak, R. Molecular Background of MiRNA Role in Asthma and COPD: An Updated Insight. Biomed Res. Int. 2016, 2016, 7802521. [Google Scholar] [CrossRef]
- Ramelli, S.C.; Gerthoffer, W.T. MicroRNA Targets for Asthma Therapy. Adv. Exp. Med. Biol. 2021, 1303, 89–105. [Google Scholar] [CrossRef]
- Davis, J.S.; Sun, M.; Kho, A.T.; Moore, K.G.; Sylvia, J.M.; Weiss, S.T.; Lu, Q.; Tantisira, K.G. Circulating MicroRNAs and Association with Methacholine PC20 in the Childhood Asthma Management Program (CAMP) Cohort. PLoS ONE 2017, 12, e0180329. [Google Scholar] [CrossRef]
- Wang, Z.; Song, Y.; Jiang, J.; Piao, Y.; Li, L.; Bai, Q.; Xu, C.; Liu, H.; Li, L.; Piao, H.; et al. MicroRNA-182-5p Attenuates Asthmatic Airway Inflammation by Targeting NOX4. Front. Immunol. 2022, 13, 853848. [Google Scholar] [CrossRef]
- Rodrigo-Muñoz, J.M.; Gil-Martínez, M.; Lorente-Sorolla, C.; García-Latorre, R.; Valverde-Monge, M.; Quirce, S.; Sastre, J.; Del Pozo, V. MiR-144-3p Is a Biomarker Related to Severe Corticosteroid-Dependent Asthma. Front. Immunol. 2022, 13, 858722. [Google Scholar] [CrossRef]
- Palumbo, M.L.; Prochnik, A.; Wald, M.R.; Genaro, A.M. Chronic Stress and Glucocorticoid Receptor Resistance in Asthma. Clin. Ther. 2020, 42, 993–1006. [Google Scholar] [CrossRef]
- Kim, R.Y.; Horvat, J.C.; Pinkerton, J.W.; Starkey, M.R.; Essilfie, A.T.; Mayall, J.R.; Nair, P.M.; Hansbro, N.G.; Jones, B.; Haw, T.J.; et al. MicroRNA-21 Drives Severe, Steroid-Insensitive Experimental Asthma by Amplifying Phosphoinositide 3-Kinase-Mediated Suppression of Histone Deacetylase 2. J. Allergy Clin. Immunol. 2017, 139, 519–532. [Google Scholar] [CrossRef]
- Liang, J.; Liu, X.-H.; Chen, X.-M.; Song, X.-L.; Li, W.; Huang, Y. Emerging Roles of Non-Coding RNAs in Childhood Asthma. Front. Pharmacol. 2022, 13, 856104. [Google Scholar] [CrossRef]
- Mei, D.; Tan, W.S.D.; Tay, Y.; Mukhopadhyay, A.; Wong, W.S.F. Therapeutic RNA Strategies for Chronic Obstructive Pulmonary Disease. Trends Pharmacol. Sci. 2020, 41, 475–486. [Google Scholar] [CrossRef]
- Wang, D.; He, S.; Liu, B.; Liu, C. MiR-27-3p Regulates TLR2/4-Dependent Mouse Alveolar Macrophage Activation by Targetting PPARγ. Clin. Sci. 2018, 132, 943–958. [Google Scholar] [CrossRef]
- Togo, S.; Holz, O.; Liu, X.; Sugiura, H.; Kamio, K.; Wang, X.; Kawasaki, S.; Ahn, Y.; Fredriksson, K.; Skold, C.M.; et al. Lung Fibroblast Repair Functions in Patients with Chronic Obstructive Pulmonary Disease Are Altered by Multiple Mechanisms. Am. J. Respir. Crit. Care Med. 2008, 178, 248–260. [Google Scholar] [CrossRef]
- Ikari, J.; Smith, L.M.; Nelson, A.J.; Iwasawa, S.; Gunji, Y.; Farid, M.; Wang, X.; Basma, H.; Feghali-Bostwick, C.; Liu, X.; et al. Effect of Culture Conditions on MicroRNA Expression in Primary Adult Control and COPD Lung Fibroblasts in Vitro. In Vitro Cell. Dev. Biol. Anim. 2015, 51, 390–399. [Google Scholar] [CrossRef]
- Ikari, J.; Nelson, A.J.; Obaid, J.; Giron-Martinez, A.; Ikari, K.; Makino, F.; Iwasawa, S.; Gunji, Y.; Farid, M.; Wang, X.; et al. Reduced MicroRNA-503 Expression Augments Lung Fibroblast VEGF Production in Chronic Obstructive Pulmonary Disease. PLoS ONE 2017, 12, e0184039. [Google Scholar] [CrossRef]
- Zhang, L.; Valizadeh, H.; Alipourfard, I.; Bidares, R.; Aghebati-Maleki, L.; Ahmadi, M. Epigenetic Modifications and Therapy in Chronic Obstructive Pulmonary Disease (COPD): An Update Review. COPD 2020, 17, 333–342. [Google Scholar] [CrossRef]
- Rüger, J.; Ioannou, S.; Castanotto, D.; Stein, C.A. Oligonucleotides to the (Gene) Rescue: FDA Approvals 2017–2019. Trends Pharmacol. Sci. 2020, 41, 27–41. [Google Scholar] [CrossRef]
- Ma, P.; Han, W.; Meng, C.; Tan, X.; Liu, P.; Dong, L. LINC02389/MiR-7-5p Regulated Cisplatin Resistance of Non-Small-Cell Lung Cancer via Promoting Oxidative Stress. Anal. Cell. Pathol. 2022, 2022, 6100176. [Google Scholar] [CrossRef]
- Fariha, A.; Hami, I.; Tonmoy, M.I.Q.; Akter, S.; Al Reza, H.; Bahadur, N.M.; Rahaman, M.M.; Hossain, M.S. Cell Cycle Associated MiRNAs as Target and Therapeutics in Lung Cancer Treatment. Heliyon 2022, 8, e11081. [Google Scholar] [CrossRef]
- Murugan, D.; Rangasamy, L. A Perspective to Weaponize MicroRNAs against Lung Cancer. Non-Coding RNA Res. 2023, 8, 18–32. [Google Scholar] [CrossRef]
- Mohan, A.; Agarwal, S.; Clauss, M.; Britt, N.S.; Dhillon, N.K. Extracellular Vesicles: Novel Communicators in Lung Diseases. Respir. Res. 2020, 21, 175. [Google Scholar] [CrossRef]
- Li, Y.; Yin, Z.; Fan, J.; Zhang, S.; Yang, W. The Roles of Exosomal MiRNAs and LncRNAs in Lung Diseases. Signal Transduct. Target. Ther. 2019, 4, 47. [Google Scholar] [CrossRef]
- Kaur, G.; Maremanda, K.P.; Campos, M.; Chand, H.S.; Li, F.; Hirani, N.; Haseeb, M.A.; Li, D.; Rahman, I. Distinct Exosomal MiRNA Profiles from BALF and Lung Tissue of COPD and IPF Patients. Int. J. Mol. Sci. 2021, 22, 11830. [Google Scholar] [CrossRef]
- Shan, C.; Liang, Y.; Cai, H.; Wang, F.; Chen, X.; Yin, Q.; Wang, K.; Wang, Y. Emerging Function and Clinical Significance of Extracellular Vesicle Noncoding RNAs in Lung Cancer. Mol. Ther. Oncolytics 2022, 24, 814–833. [Google Scholar] [CrossRef]
- Cao, H.; Zhang, P.; Yu, H.; Xi, J. Extracellular Vesicles-Encapsulated MiR-153-3p Potentiate the Survival and Invasion of Lung Adenocarcinoma. Mol. Cells 2022, 45, 376–387. [Google Scholar] [CrossRef]
- Poroyko, V.; Mirzapoiazova, T.; Nam, A.; Mambetsariev, I.; Mambetsariev, B.; Wu, X.; Husain, A.; Vokes, E.E.; Wheeler, D.L.; Salgia, R. Exosomal MiRNAs Species in the Blood of Small Cell and Non-Small Cell Lung Cancer Patients. Oncotarget 2018, 9, 19793–19806. [Google Scholar] [CrossRef]
- Duréndez-Sáez, E.; Torres-Martinez, S.; Calabuig-Fariñas, S.; Meri-Abad, M.; Ferrero-Gimeno, M.; Camps, C. Exosomal MicroRNAs in Non-Small Cell Lung Cancer. Transl. Cancer Res. 2021, 10, 3128–3139. [Google Scholar] [CrossRef]
- Zheng, R.; Du, M.; Tian, M.; Zhu, Z.; Wei, C.; Chu, H.; Gan, C.; Liang, J.; Xue, R.; Gao, F.; et al. Fine Particulate Matter Induces Childhood Asthma Attacks via Extracellular Vesicle-Packaged Let-7i-5p-Mediated Modulation of the MAPK Signaling Pathway. Adv. Sci. 2022, 9, e2102460. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Albano, G.D.; Gagliardo, R.; Montalbano, A.M.; Profita, M. Non-Coding RNAs in Airway Diseases: A Brief Overview of Recent Data. Cancers 2023, 15, 54. https://doi.org/10.3390/cancers15010054
Albano GD, Gagliardo R, Montalbano AM, Profita M. Non-Coding RNAs in Airway Diseases: A Brief Overview of Recent Data. Cancers. 2023; 15(1):54. https://doi.org/10.3390/cancers15010054
Chicago/Turabian StyleAlbano, Giusy Daniela, Rosalia Gagliardo, Angela Marina Montalbano, and Mirella Profita. 2023. "Non-Coding RNAs in Airway Diseases: A Brief Overview of Recent Data" Cancers 15, no. 1: 54. https://doi.org/10.3390/cancers15010054
APA StyleAlbano, G. D., Gagliardo, R., Montalbano, A. M., & Profita, M. (2023). Non-Coding RNAs in Airway Diseases: A Brief Overview of Recent Data. Cancers, 15(1), 54. https://doi.org/10.3390/cancers15010054





