The Emerging Role of Tumor Microenvironmental Stimuli in Regulating Metabolic Rewiring of Liver Cancer Stem Cells
Abstract
Simple Summary
Abstract
1. Introduction
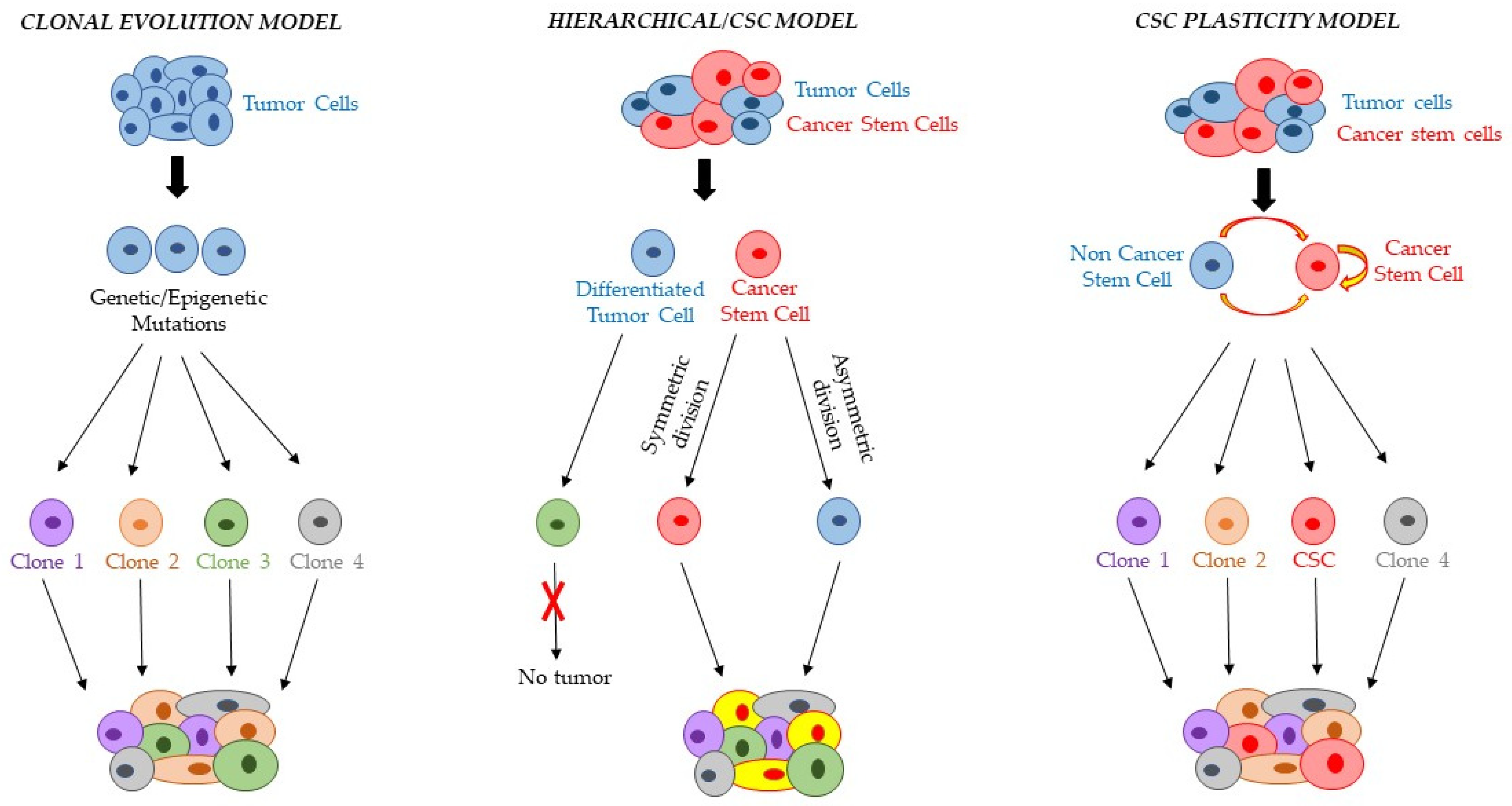
2. CSC and Intratumor Heterogeneity
3. Liver Tumor Metabolic Reprogramming and CSCs
3.1. Glucose Metabolism
3.2. Lipid Metabolism
3.3. Iron Metabolism
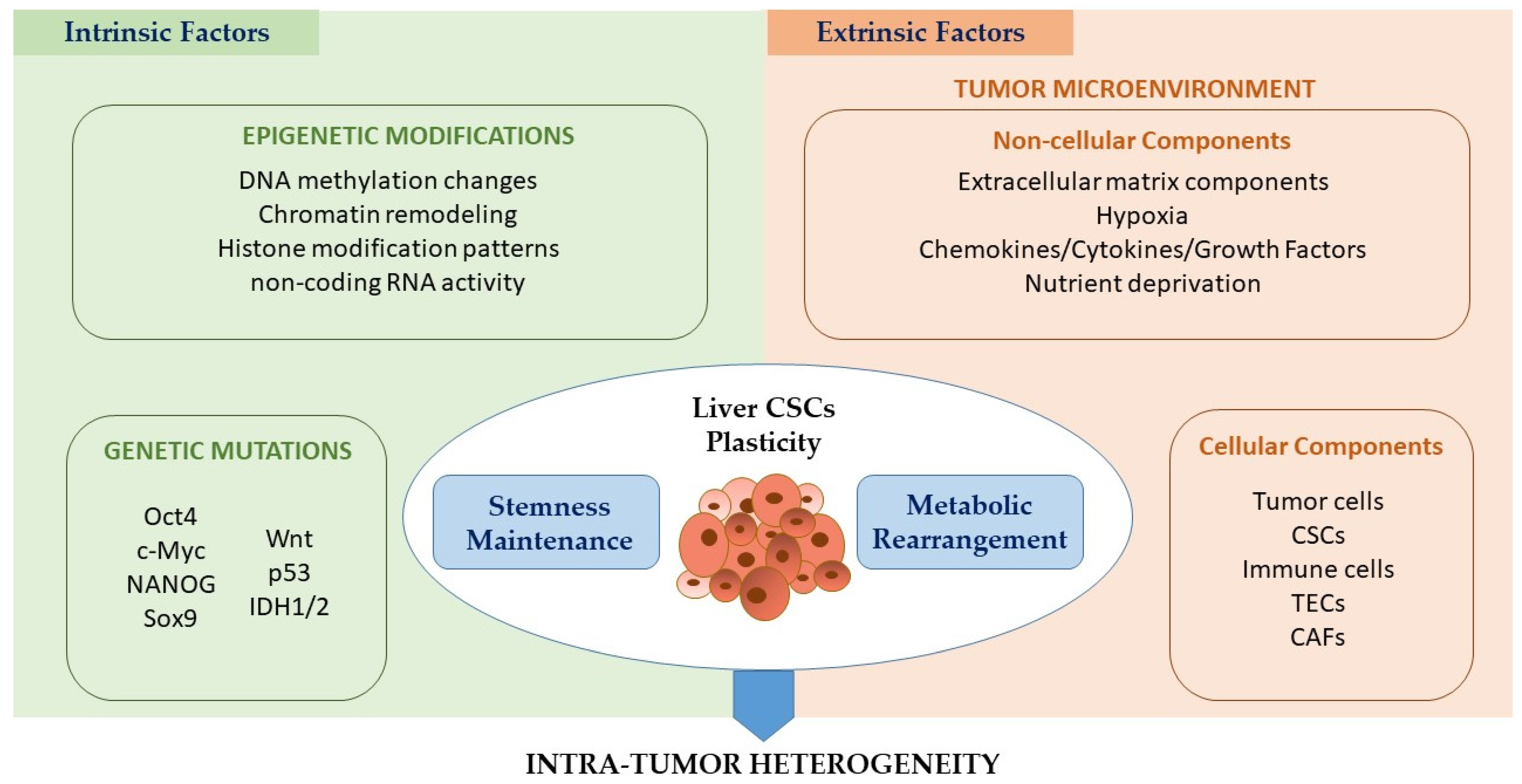
4. Effect of TME on Liver CSC Metabolic Plasticity
4.1. Intrinsic Factors Controlling CSC Plasticity
4.2. Extrinsic Cues Regulating CSC Plasticity: Effect of Non-cellular and Cellular Components of the TME
4.2.1. Role of TME Noncellular Components in Controlling CSC Metabolic Plasticity
4.2.2. Regulatory Effect of TME Cellular Components on Liver CSC Metabolic Features
| Mediators | Secretory Cells | Receptor Cells | Refs |
|---|---|---|---|
| DAMPS | TECs | Immune cells | [125] |
| Angiocrin factors | Liver CSCs | [97,126,127,128] | |
| IL-6, HGF | CAFs | CD24+ HCC CSCs | [134] |
| IL-6, IL-33 | MDSCs | [136] | |
| Lactate, Piruvate | Liver CSCs | [137] | |
| LTB4 | MDSCs | CCA CSCs | [136] |
| TNFα, IL-1β, IFN-γ | Inflammatory Cells | Liver CSCs | [115] |
| IL-6, TGF-β, TNFα | TAMs | HCC CSCs | [113,115,144,145] |
| S100A9 | Liver CSCs | [146] | |
| BMP2, TGF-β2 | TANs | HCC CSCs | [147] |
5. Conclusions
Future Directions: Targeting CSC Metabolism as a Promising Therapeutic Opportunity
Author Contributions
Funding
Conflicts of Interest
References
- Banales, J.M.; Cardinale, V.; Carpino, G.; Marzioni, M.; Andersen, J.B.; Invernizzi, P.; Lind, G.E.; Folseraas, T.; Forbes, S.J.; Fouassier, L.; et al. Expert consensus document: Cholangiocarcinoma: Current knowledge and future perspectives consensus statement from the European Network for the Study of Cholangiocarcinoma (ENS-CCA). Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 261–280. [Google Scholar] [CrossRef] [PubMed]
- El-Serag, H.B. Hepatocellular carcinoma. N. Engl. J. Med. 2011, 365, 1118–1127. [Google Scholar] [CrossRef]
- Oikawa, T. Cancer Stem cells and their cellular origins in primary liver and biliary tract cancers. Hepatology 2016, 64, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Herreros-Pomares, A. Identification, Culture and Targeting of Cancer Stem Cells. Life 2022, 12, 184. [Google Scholar] [CrossRef] [PubMed]
- Palorini, R.; Votta, G.; Balestrieri, C.; Monestiroli, A.; Olivieri, S.; Vento, R.; Chiaradonna, F. Energy metabolism characterization of a novel cancer stem cell-like line 3AB-OS. J. Cell Biochem. 2014, 115, 368–379. [Google Scholar] [CrossRef]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef]
- Yamashita, T.; Wang, X.W. Cancer stem cells in the development of liver cancer. J. Clin. Investig. 2013, 123, 1911–1918. [Google Scholar] [CrossRef]
- Correnti, M.; Booijink, R.; Di Maira, G.; Raggi, C.; Marra, F. Stemness features in liver cancer. Hepatoma Res. 2018, 4, 69. [Google Scholar] [CrossRef]
- Kaushik, V.; Kulkarni, Y.; Felix, K.; Azad, N.; Iyer, A.K.V.; Yakisich, J.S. Alternative models of cancer stem cells: The stemness phenotype model, 10 years later. World J. Stem Cells 2021, 13, 934–943. [Google Scholar] [CrossRef]
- Borovski, T.; De Sousa E Melo, F.; Vermeulen, L.; Medema, J.P. Cancer stem cell niche: The place to be. Cancer Res. 2011, 71, 634–639. [Google Scholar] [CrossRef] [PubMed]
- Meacham, C.E.; Morrison, S.J. Tumour heterogeneity and cancer cell plasticity. Nature 2013, 501, 328–337. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, C.A.; Pollett, A.; Gallinger, S.; Dick, J.E. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 2007, 445, 106–110. [Google Scholar] [CrossRef] [PubMed]
- Plaks, V.; Kong, N.; Werb, Z. The cancer stem cell niche: How essential is the niche in regulating stemness of tumor cells? Cell Stem Cell 2015, 16, 225–238. [Google Scholar] [CrossRef]
- Prasetyanti, P.R.; Medema, J.P. Intra-tumor heterogeneity from a cancer stem cell perspective. Mol. Cancer 2017, 16, 41. [Google Scholar] [CrossRef]
- Cruz, M.H.; Sidén, A.; Calaf, G.M.; Delwar, Z.M.; Yakisich, J.S. The stemness phenotype model. ISRN Oncol. 2012, 2012, 392647. [Google Scholar] [CrossRef]
- Da Silva-Diz, V.; Lorenzo-Sanz, L.; Bernat-Peguera, A.; Lopez-Cerda, M.; Muñoz, P. Cancer cell plasticity: Impact on tumor progression and therapy response. Semin. Cancer Biol. 2018, 53, 48–58. [Google Scholar] [CrossRef]
- Recalcati, S.; Correnti, M.; Gammella, E.; Raggi, C.; Invernizzi, P.; Cairo, G. Iron Metabolism in Liver Cancer Stem Cells. Front. Oncol. 2019, 9, 149. [Google Scholar] [CrossRef]
- Ma, S.; Chan, K.W.; Hu, L.; Lee, T.K.; Wo, J.Y.; Ng, I.O.; Zheng, B.J.; Guan, X.Y. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology 2007, 132, 2542–2556. [Google Scholar] [CrossRef]
- Marquardt, J.U.; Raggi, C.; Andersen, J.B.; Seo, D.; Avital, I.; Geller, D.; Lee, Y.H.; Kitade, M.; Holczbauer, A.; Gillen, M.C.; et al. Human hepatic cancer stem cells are characterized by common stemness traits and diverse oncogenic pathways. Hepatology 2011, 54, 1031–1042. [Google Scholar] [CrossRef]
- Raggi, C.; Factor, V.M.; Seo, D.; Holczbauer, A.; Gillen, M.C.; Marquardt, J.U.; Andersen, J.B.; Durkin, M.; Thorgeirsson, S.S. Epigenetic reprogramming modulates malignant properties of human liver cancer. Hepatology 2014, 59, 2251–2262. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, T.; Ji, J.; Budhu, A.; Forgues, M.; Yang, W.; Wang, H.Y.; Jia, H.; Ye, Q.; Qin, L.X.; Wauthier, E.; et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology 2009, 136, 1012–1024. [Google Scholar] [CrossRef] [PubMed]
- Kokuryo, T.; Yokoyama, Y.; Nagino, M. Recent advances in cancer stem cell research for cholangiocarcinoma. J. Hepatobiliary Pancreat. Sci. 2012, 19, 606–613. [Google Scholar] [CrossRef] [PubMed]
- Raggi, C.; Correnti, M.; Sica, A.; Andersen, J.B.; Cardinale, V.; Alvaro, D.; Chiorino, G.; Forti, E.; Glaser, S.; Alpini, G.; et al. Cholangiocarcinoma stem-like subset shapes tumor-initiating niche by educating associated macrophages. J. Hepatol. 2016, 66, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Chaffer, C.L.; Brueckmann, I.; Scheel, C.; Kaestli, A.J.; Wiggins, P.A.; Rodrigues, L.O.; Brooks, M.; Reinhardt, F.; Su, Y.; Polyak, K.; et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc. Natl. Acad. Sci. USA 2011, 108, 7950–7955. [Google Scholar] [CrossRef]
- Fekir, K.; Dubois-Pot-Schneider, H.; Désert, R.; Daniel, Y.; Glaise, D.; Rauch, C.; Morel, F.; Fromenty, B.; Musso, O.; Cabillic, F.; et al. Retrodifferentiation of Human Tumor Hepatocytes to Stem Cells Leads to Metabolic Reprogramming and Chemoresistance. Cancer Res. 2019, 79, 1869–1883. [Google Scholar] [CrossRef]
- Rais, Y.; Zviran, A.; Geula, S.; Gafni, O.; Chomsky, E.; Viukov, S.; Mansour, A.A.; Caspi, I.; Krupalnik, V.; Zerbib, M.; et al. Deterministic direct reprogramming of somatic cells to pluripotency. Nature 2013, 502, 65–70. [Google Scholar] [CrossRef]
- Pastore, M.; Lori, G.; Gentilini, A.; Taddei, M.L.; Di Maira, G.; Campani, C.; Recalcati, S.; Invernizzi, P.; Marra, F.; Raggi, C. Multifaceted Aspects of Metabolic Plasticity in Human Cholangiocarcinoma: An Overview of Current Perspectives. Cells 2020, 9, 596. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Chandel, N.S. Fundamentals of cancer metabolism. Sci. Adv. 2016, 2, e1600200. [Google Scholar] [CrossRef]
- Sharma, A.; Boise, L.H.; Shanmugam, M. Cancer Metabolism and the Evasion of Apoptotic Cell Death. Cancers 2019, 11, 1144. [Google Scholar] [CrossRef]
- Chae, Y.C.; Kim, J.H. Cancer stem cell metabolism: Target for cancer therapy. BMB Rep. 2018, 51, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Chen, X.; Wang, L.; Chen, S. The sweet trap in tumors: Aerobic glycolysis and potential targets for therapy. Oncotarget 2016, 7, 38908–38926. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Chen, H.H.; Gao, C.Y.; Zhang, X.X.; Jiang, J.X.; Zhang, Y.; Fang, J.; Zhao, F.; Chen, Z.G. Energy metabolism in cancer stem cells. World J. Stem Cells 2020, 12, 448–461. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J. Energy metabolism of cancer: Glycolysis versus oxidative phosphorylation (Review). Oncol. Lett. 2012, 4, 1151–1157. [Google Scholar] [CrossRef]
- Zhang, H. HIF-1 suppresses lipid catabolism to promote cancer progression. Mol. Cell Oncol. 2015, 2, e980184. [Google Scholar] [CrossRef]
- Xu, S.; Herschman, H.R. A Tumor Agnostic Therapeutic Strategy for Hexokinase 1-Null/Hexokinase 2-Positive Cancers. Cancer Res. 2019, 79, 5907–5914. [Google Scholar] [CrossRef]
- Du, D.; Liu, C.; Qin, M.; Zhang, X.; Xi, T.; Yuan, S.; Hao, H.; Xiong, J. Metabolic dysregulation and emerging therapeutical targets for hepatocellular carcinoma. Acta Pharm. Sin. B 2022, 12, 558–580. [Google Scholar] [CrossRef]
- Kubo, Y.; Aishima, S.; Tanaka, Y.; Shindo, K.; Mizuuchi, Y.; Abe, K.; Shirabe, K.; Maehara, Y.; Honda, H.; Oda, Y. Different expression of glucose transporters in the progression of intrahepatic cholangiocarcinoma. Hum. Pathol. 2014, 45, 1610–1617. [Google Scholar] [CrossRef]
- Suzuki, H.; Komuta, M.; Bolog, A.; Yokobori, T.; Wada, S.; Araki, K.; Kubo, N.; Watanabe, A.; Tsukagoshi, M.; Kuwano, H. Relationship between 18-F-fluoro-deoxy-D-glucose uptake and expression of glucose transporter 1 and pyruvate kinase M2 in intrahepatic cholangiocarcinoma. Dig. Liver Dis. 2015, 47, 590–596. [Google Scholar] [CrossRef]
- Sanmai, S.; Proungvitaya, T.; Limpaiboon, T.; Chua-On, D.; Seubwai, W.; Roytrakul, S.; Wongkham, S.; Wongkham, C.; Somintara, O.; Sangkhamanon, S.; et al. Serum pyruvate dehydrogenase kinase as a prognostic marker for cholangiocarcinoma. Oncol. Lett. 2019, 17, 5275–5282. [Google Scholar] [CrossRef] [PubMed]
- Raggi, C.; Taddei, M.L.; Sacco, E.; Navari, N.; Correnti, M.; Piombanti, B.; Pastore, M.; Campani, C.; Pranzini, E.; Iorio, J.; et al. Mitochondrial oxidative metabolism contributes to a cancer stem cell phenotype in cholangiocarcinoma. J. Hepatol. 2021, 74, 1373–1385. [Google Scholar] [CrossRef] [PubMed]
- Thonsri, U.; Seubwai, W.; Waraasawapati, S.; Sawanyawisuth, K.; Vaeteewoottacharn, K.; Boonmars, T.; Cha’on, U. Overexpression of lactate dehydrogenase A in cholangiocarcinoma is correlated with poor prognosis. Histol. Histopathol. 2017, 32, 503–510. [Google Scholar] [CrossRef]
- Yu, Y.; Liao, M.; Liu, R.; Chen, J.; Feng, H.; Fu, Z. Overexpression of lactate dehydrogenase-A in human intrahepatic cholangiocarcinoma: Its implication for treatment. World J. Surg. Oncol. 2014, 12, 78. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, M.; Yao, X.; Fei, Y.; Lin, Z.; Li, Z.; Cai, K.; Luo, Z. HCAR1/MCT1 Regulates Tumor Ferroptosis through the Lactate-Mediated AMPK-SCD1 Activity and Its Therapeutic Implications. Cell Rep. 2020, 33, 108487. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Sheng, J.; Shen, L.; Su, J.; Xu, Y.; Xie, Q.; Wu, Y.; Zhang, X.; Sun, L. Autophagy inhibitor chloroquine increases sensitivity to cisplatin in QBC939 cholangiocarcinoma cells by mitochondrial ROS. PLoS ONE 2017, 12, e0173712. [Google Scholar] [CrossRef]
- Hong, X.; Song, R.; Song, H.; Zheng, T.; Wang, J.; Liang, Y.; Qi, S.; Lu, Z.; Song, X.; Jiang, H.; et al. PTEN antagonises Tcl1/hnRNPK-mediated G6PD pre-mRNA splicing which contributes to hepatocarcinogenesis. Gut 2014, 63, 1635–1647. [Google Scholar] [CrossRef]
- Qin, Z.; Xiang, C.; Zhong, F.; Liu, Y.; Dong, Q.; Li, K.; Shi, W.; Ding, C.; Qin, L.; He, F. Transketolase (TKT) activity and nuclear localization promote hepatocellular carcinoma in a metabolic and a non-metabolic manner. J. Exp. Clin. Cancer Res. 2019, 38, 154. [Google Scholar] [CrossRef]
- Sancho, P.; Barneda, D.; Heeschen, C. Hallmarks of cancer stem cell metabolism. Br. J. Cancer 2016, 114, 1305–1312. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Wang, W.H.; Che, L.; Lan, Y.; Zhang, L.Y.; Zhan, D.L.; Huang, Z.Y.; Lin, Z.N.; Lin, Y.C. BNIP3L-Dependent Mitophagy Promotes HBx-Induced Cancer Stemness of Hepatocellular Carcinoma Cells via Glycolysis Metabolism Reprogramming. Cancers 2020, 12, 655. [Google Scholar] [CrossRef]
- Kowalik, M.A.; Puliga, E.; Cabras, L.; Sulas, P.; Petrelli, A.; Perra, A.; Ledda-Columbano, G.M.; Morandi, A.; Merlin, S.; Orrù, C.; et al. Thyroid hormone inhibits hepatocellular carcinoma progression via induction of differentiation and metabolic reprogramming. J. Hepatol. 2020, 72, 1159–1169. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; Kwon, H.; Han, C.; Zhang, J.; Dash, S.; Lim, K.; Wu, T. Active glycolytic metabolism in CD133(+) hepatocellular cancer stem cells: Regulation by MIR-122. Oncotarget 2015, 6, 40822–40835. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.W.; Leung, C.O.; Chan, K.K.; Ho, D.W.; Leung, M.S.; Wong, C.M.; Ng, I.O.; Lo, R.C. Deregulated GATA6 modulates stem cell-like properties and metabolic phenotype in hepatocellular carcinoma. Int. J. Cancer 2019, 145, 1860–1873. [Google Scholar] [CrossRef]
- Hur, W.; Ryu, J.Y.; Kim, H.U.; Hong, S.W.; Lee, E.B.; Lee, S.Y.; Yoon, S.K. Systems approach to characterize the metabolism of liver cancer stem cells expressing CD133. Sci. Rep. 2017, 7, 45557. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Tian, R.; Yang, X.; Wang, H.; Shi, Y.; Fan, X.; Zhang, J.; Chen, Y.; Zhang, K.; Chen, Z.; et al. KCNN4 Promotes the Stemness Potentials of Liver Cancer Stem Cells by Enhancing Glucose Metabolism. Int. J. Mol. Sci. 2022, 23, 6958. [Google Scholar] [CrossRef]
- Tamai, K.; Nakamura-Shima, M.; Shibuya-Takahashi, R.; Kanno, S.I.; Yasui, A.; Mochizuki, M.; Iwai, W.; Wakui, Y.; Abue, M.; Yamamoto, K.; et al. BEX2 suppresses mitochondrial activity and is required for dormant cancer stem cell maintenance in intrahepatic cholangiocarcinoma. Sci. Rep. 2020, 10, 21592. [Google Scholar] [CrossRef]
- Ku, C.Y.; Liu, Y.H.; Lin, H.Y.; Lu, S.C.; Lin, J.Y. Liver fatty acid-binding protein (L-FABP) promotes cellular angiogenesis and migration in hepatocellular carcinoma. Oncotarget 2016, 7, 18229–18246. [Google Scholar] [CrossRef]
- Jeong, C.Y.; Hah, Y.S.; Cho, B.I.; Lee, S.M.; Joo, Y.T.; Jung, E.J.; Jeong, S.H.; Lee, Y.J.; Choi, S.K.; Ha, W.S.; et al. Fatty acid-binding protein 5 promotes cell proliferation and invasion in human intrahepatic cholangiocarcinoma. Oncol. Rep. 2012, 28, 1283–1292. [Google Scholar] [CrossRef]
- Nakagawa, R.; Hiep, N.C.; Ouchi, H.; Sato, Y.; Harada, K. Expression of fatty-acid-binding protein 5 in intrahepatic and extrahepatic cholangiocarcinoma: The possibility of different energy metabolisms in anatomical location. Med. Mol. Morphol. 2020, 53, 42–49. [Google Scholar] [CrossRef]
- Ruiz de Gauna, M.; Biancaniello, F.; González-Romero, F.; Rodrigues, P.M.; Lapitz, A.; Gómez-Santos, B.; Olaizola, P.; Di Matteo, S.; Aurrekoetxea, I.; Labiano, I.; et al. Cholangiocarcinoma progression depends on the uptake and metabolization of extracellular lipids. Hepatology 2022, 76, 1617–1633. [Google Scholar] [CrossRef]
- Mead, J.R.; Irvine, S.A.; Ramji, D.P. Lipoprotein lipase: Structure, function, regulation, and role in disease. J. Mol. Med. 2002, 80, 753–769. [Google Scholar] [CrossRef]
- Wu, Z.; Ma, H.; Wang, L.; Song, X.; Zhang, J.; Liu, W.; Ge, Y.; Sun, Y.; Yu, X.; Wang, Z.; et al. Tumor suppressor ZHX2 inhibits NAFLD-HCC progression via blocking LPL-mediated lipid uptake. Cell Death Differ. 2020, 27, 1693–1708. [Google Scholar] [CrossRef]
- Che, L.; Paliogiannis, P.; Cigliano, A.; Pilo, M.G.; Chen, X.; Calvisi, D.F. Pathogenetic, Prognostic, and Therapeutic Role of Fatty Acid Synthase in Human Hepatocellular Carcinoma. Front. Oncol. 2019, 9, 1412. [Google Scholar] [CrossRef]
- Chen, J.; Ding, C.; Chen, Y.; Hu, W.; Lu, Y.; Wu, W.; Zhang, Y.; Yang, B.; Wu, H.; Peng, C.; et al. ACSL4 promotes hepatocellular carcinoma progression via c-Myc stability mediated by ERK/FBW7/c-Myc axis. Oncogenesis 2020, 9, 42. [Google Scholar] [CrossRef]
- Fullerton, M.D.; Galic, S.; Marcinko, K.; Sikkema, S.; Pulinilkunnil, T.; Chen, Z.P.; O’Neill, H.M.; Ford, R.J.; Palanivel, R.; O’Brien, M.; et al. Single phosphorylation sites in Acc1 and Acc2 regulate lipid homeostasis and the insulin-sensitizing effects of metformin. Nat. Med. 2013, 19, 1649–1654. [Google Scholar] [CrossRef]
- Icard, P.; Wu, Z.; Fournel, L.; Coquerel, A.; Lincet, H.; Alifano, M. ATP citrate lyase: A central metabolic enzyme in cancer. Cancer Lett. 2020, 471, 125–134. [Google Scholar] [CrossRef]
- Lai, K.K.Y.; Kweon, S.M.; Chi, F.; Hwang, E.; Kabe, Y.; Higashiyama, R.; Qin, L.; Yan, R.; Wu, R.P.; Lai, K.; et al. Stearoyl-CoA Desaturase Promotes Liver Fibrosis and Tumor Development in Mice via a Wnt Positive-Signaling Loop by Stabilization of Low-Density Lipoprotein-Receptor-Related Proteins 5 and 6. Gastroenterology 2017, 152, 1477–1491. [Google Scholar] [CrossRef]
- Li, L.; Pilo, G.M.; Li, X.; Cigliano, A.; Latte, G.; Che, L.; Joseph, C.; Mela, M.; Wang, C.; Jiang, L.; et al. Inactivation of fatty acid synthase impairs hepatocarcinogenesis driven by AKT in mice and humans. J. Hepatol. 2016, 64, 333–341. [Google Scholar] [CrossRef]
- Li, L.; Che, L.; Tharp, K.M.; Park, H.M.; Pilo, M.G.; Cao, D.; Cigliano, A.; Latte, G.; Xu, Z.; Ribback, S.; et al. Differential requirement for de novo lipogenesis in cholangiocarcinoma and hepatocellular carcinoma of mice and humans. Hepatology 2016, 63, 1900–1913. [Google Scholar] [CrossRef]
- Li, C.; Yang, W.; Zhang, J.; Zheng, X.; Yao, Y.; Tu, K.; Liu, Q. SREBP-1 has a prognostic role and contributes to invasion and metastasis in human hepatocellular carcinoma. Int. J. Mol. Sci. 2014, 15, 7124–7138. [Google Scholar] [CrossRef]
- Xiong, T.; Li, Z.; Huang, X.; Lu, K.; Xie, W.; Zhou, Z.; Tu, J. TO901317 inhibits the development of hepatocellular carcinoma by LXRα/Glut1 decreasing glycometabolism. Am. J. Physiol. Gastrointest. Liver Physiol. 2019, 316, G598–G607. [Google Scholar] [CrossRef] [PubMed]
- Visweswaran, M.; Arfuso, F.; Warrier, S.; Dharmarajan, A. Aberrant lipid metabolism as an emerging therapeutic strategy to target cancer stem cells. Stem Cells 2020, 38, 6–14. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.L.; Uthaya Kumar, D.B.; Punj, V.; Xu, J.; Sher, L.; Tahara, S.M.; Hess, S.; Machida, K. NANOG Metabolically Reprograms Tumor-Initiating Stem-like Cells through Tumorigenic Changes in Oxidative Phosphorylation and Fatty Acid Metabolism. Cell Metab. 2016, 23, 206–219. [Google Scholar] [CrossRef] [PubMed]
- Bansal, S.; Berk, M.; Alkhouri, N.; Partrick, D.A.; Fung, J.J.; Feldstein, A. Stearoyl-CoA desaturase plays an important role in proliferation and chemoresistance in human hepatocellular carcinoma. J. Surg. Res. 2014, 186, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.K.F.; Lau, E.Y.T.; Leung, D.H.W.; Lo, J.; Ho, N.P.Y.; Cheng, L.K.W.; Ma, S.; Lin, C.H.; Copland, J.A.; Ding, J.; et al. Stearoyl-CoA desaturase regulates sorafenib resistance via modulation of ER stress-induced differentiation. J. Hepatol. 2017, 67, 979–990. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Viscarra, J.; Kim, S.J.; Sul, H.S. Transcriptional regulation of hepatic lipogenesis. Nat. Rev. Mol. Cell Biol. 2015, 16, 678–689. [Google Scholar] [CrossRef] [PubMed]
- Gammella, E.; Recalcati, S.; Cairo, G. Dual Role of ROS as Signal and Stress Agents: Iron Tips the Balance in favor of Toxic Effects. Oxid. Med. Cell. Longev. 2016, 2016, 8629024. [Google Scholar] [CrossRef]
- Basak, T.; Kanwar, R.K. Iron imbalance in cancer: Intersection of deficiency and overload. Cancer Med. 2022, 11, 3837–3853. [Google Scholar] [CrossRef]
- Recalcati, S.; Gammella, E.; Buratti, P.; Cairo, G. Molecular regulation of cellular iron balance. IUBMB Life 2017, 69, 389–398. [Google Scholar] [CrossRef]
- Muto, Y.; Moroishi, T.; Ichihara, K.; Nishiyama, M.; Shimizu, H.; Eguchi, H.; Moriya, K.; Koike, K.; Mimori, K.; Mori, M.; et al. Disruption of FBXL5-mediated cellular iron homeostasis promotes liver carcinogenesis. J. Exp. Med. 2019, 216, 950–965. [Google Scholar] [CrossRef]
- Fonseca-Nunes, A.; Jakszyn, P.; Agudo, A. Iron and cancer risk--a systematic review and meta-analysis of the epidemiological evidence. Cancer Epidemiol. Biomark. Prev. 2014, 23, 12–31. [Google Scholar] [CrossRef] [PubMed]
- Torti, S.V.; Manz, D.H.; Paul, B.T.; Blanchette-Farra, N.; Torti, F.M. Iron and Cancer. Annu. Rev. Nutr. 2018, 38, 97–125. [Google Scholar] [CrossRef] [PubMed]
- Elmberg, M.; Hultcrantz, R.; Ekbom, A.; Brandt, L.; Olsson, S.; Olsson, R.; Lindgren, S.; Lööf, L.; Stål, P.; Wallerstedt, S.; et al. Cancer risk in patients with hereditary hemochromatosis and in their first-degree relatives. Gastroenterology 2003, 125, 1733–1741. [Google Scholar] [CrossRef] [PubMed]
- Fargion, S.; Valenti, L.; Fracanzani, A.L. Hemochromatosis gene (HFE) mutations and cancer risk: Expanding the clinical manifestations of hereditary iron overload. Hepatology 2010, 51, 1119–1121. [Google Scholar] [CrossRef]
- Sorrentino, P.; D’Angelo, S.; Ferbo, U.; Micheli, P.; Bracigliano, A.; Vecchione, R. Liver iron excess in patients with hepatocellular carcinoma developed on non-alcoholic steato-hepatitis. J. Hepatol. 2009, 50, 351–357. [Google Scholar] [CrossRef]
- Paganoni, R.; Lechel, A.; Vujic Spasic, M. Iron at the Interface of Hepatocellular Carcinoma. Int. J. Mol. Sci. 2021, 22, 4097. [Google Scholar] [CrossRef]
- Ahmad, S.S.; Basheer, F.T.; Idris, S.F.; Hariraj, R.; Mathialagan, R.; Douds, A. Cholangiocarcinoma presenting as hemobilia and recurrent iron-deficiency anemia: A case report. J. Med. Case Rep. 2010, 4, 133. [Google Scholar] [CrossRef]
- Dion, S.P.; Béliveau, F.; Morency, L.P.; Désilets, A.; Najmanovich, R.; Leduc, R. Functional diversity of TMPRSS6 isoforms and variants expressed in hepatocellular carcinoma cell lines. Sci. Rep. 2018, 8, 12562. [Google Scholar] [CrossRef]
- Park, S.; Jung, C.W.; Kim, K.; Kim, S.J.; Kim, W.S.; Jang, J.H. Iron deficient erythropoiesis might play key role in development of anemia in cancer patients. Oncotarget 2015, 6, 42803–42812. [Google Scholar] [CrossRef][Green Version]
- Roemhild, K.; von Maltzahn, F.; Weiskirchen, R.; Knüchel, R.; von Stillfried, S.; Lammers, T. Iron metabolism: Pathophysiology and pharmacology. Trends Pharmacol. Sci. 2021, 42, 640–656. [Google Scholar] [CrossRef]
- Shen, Y.; Li, X.; Zhao, B.; Xue, Y.; Wang, S.; Chen, X.; Yang, J.; Lv, H.; Shang, P. Iron metabolism gene expression and prognostic features of hepatocellular carcinoma. J. Cell Biochem. 2018, 119, 9178–9204. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.L.; Zhang, N.P.; Xu, R.C.; Zhang, G.C.; Liu, Z.Y.; Abuduwaili, W.; Wang, F.; Yu, X.N.; Shi, X.; Song, G.Q.; et al. Tumor cell-imposed iron restriction drives immunosuppressive polarization of tumor-associated macrophages. J. Transl. Med. 2021, 19, 347. [Google Scholar] [CrossRef] [PubMed]
- Jamnongkan, W.; Thanan, R.; Techasen, A.; Namwat, N.; Loilome, W.; Intarawichian, P.; Titapun, A.; Yongvanit, P. Upregulation of transferrin receptor-1 induces cholangiocarcinoma progression via induction of labile iron pool. Tumour. Biol. 2017, 39, 1010428317717655. [Google Scholar] [CrossRef] [PubMed]
- Sarcognato, S.; Sacchi, D.; Fabris, L.; Zanus, G.; Gringeri, E.; Niero, M.; Gallina, G.; Guido, M. Ferroptosis in Intrahepatic Cholangiocarcinoma. Front. Med. 2022, 9, 886229. [Google Scholar] [CrossRef]
- Raggi, C.; Gammella, E.; Correnti, M.; Buratti, P.; Forti, E.; Andersen, J.B.; Alpini, G.; Glaser, S.; Alvaro, D.; Invernizzi, P.; et al. Dysregulation of Iron Metabolism in Cholangiocarcinoma Stem-like Cells. Sci. Rep. 2017, 7, 17667. [Google Scholar] [CrossRef] [PubMed]
- Mancinelli, R.; Cutone, A.; Rosa, L.; Lepanto, M.S.; Onori, P.; Pannarale, L.; Franchitto, A.; Gaudio, E.; Valenti, P. Different iron-handling in inflamed small and large cholangiocytes and in small and large-duct type intrahepatic cholangiocarcinoma. Eur. J. Histochem. 2020, 64, 3156. [Google Scholar] [CrossRef] [PubMed]
- Hino, K.; Yanatori, I.; Hara, Y.; Nishina, S. Iron and liver cancer: An inseparable connection. FEBS J. 2022, 289, 7810–7829. [Google Scholar] [CrossRef] [PubMed]
- Liang, N.; Yang, T.; Huang, Q.; Yu, P.; Liu, C.; Chen, L.; Wang, Q.; Wang, G.; He, X. Mechanism of cancer stemness maintenance in human liver cancer. Cell Death Dis. 2022, 13, 394. [Google Scholar] [CrossRef] [PubMed]
- Recalcati, S.; Gammella, E.; Cairo, G. Dysregulation of iron metabolism in cancer stem cells. Free Radic. Biol. Med. 2019, 133, 216–220. [Google Scholar] [CrossRef]
- Basuli, D.; Tesfay, L.; Deng, Z.; Paul, B.; Yamamoto, Y.; Ning, G.; Xian, W.; McKeon, F.; Lynch, M.; Crum, C.P.; et al. Iron addiction: A novel therapeutic target in ovarian cancer. Oncogene 2017, 36, 4089–4099. [Google Scholar] [CrossRef]
- Kanojia, D.; Zhou, W.; Zhang, J.; Jie, C.; Lo, P.K.; Wang, Q.; Chen, H. Proteomic profiling of cancer stem cells derived from primary tumors of HER2/Neu transgenic mice. Proteomics 2012, 12, 3407–3415. [Google Scholar] [CrossRef] [PubMed]
- Lobello, N.; Biamonte, F.; Pisanu, M.E.; Faniello, M.C.; Jakopin, Ž.; Chiarella, E.; Giovannone, E.D.; Mancini, R.; Ciliberto, G.; Cuda, G.; et al. Ferritin heavy chain is a negative regulator of ovarian cancer stem cell expansion and epithelial to mesenchymal transition. Oncotarget 2016, 7, 62019–62033. [Google Scholar] [CrossRef] [PubMed]
- Schonberg, D.L.; Miller, T.E.; Wu, Q.; Flavahan, W.A.; Das, N.K.; Hale, J.S.; Hubert, C.G.; Mack, S.C.; Jarrar, A.M.; Karl, R.T.; et al. Preferential Iron Trafficking Characterizes Glioblastoma Stem-like Cells. Cancer Cell 2015, 28, 441–455. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Fu, X.; Wang, Y.; Liu, H.; Jiang, Y.; Zhao, Z.; You, F. Transferrin receptor regulates malignancies and the stemness of hepatocellular carcinoma-derived cancer stem-like cells by affecting iron accumulation. PLoS ONE 2020, 15, e0243812. [Google Scholar] [CrossRef]
- Oshima, N.; Yamada, Y.; Nagayama, S.; Kawada, K.; Hasegawa, S.; Okabe, H.; Sakai, Y.; Aoi, T. Induction of cancer stem cell properties in colon cancer cells by defined factors. PLoS ONE 2014, 9, e101735. [Google Scholar] [CrossRef]
- Thankamony, A.P.; Saxena, K.; Murali, R.; Jolly, M.K.; Nair, R. Cancer Stem Cell Plasticity—A Deadly Deal. Front. Mol. Biosci. 2020, 7, 79. [Google Scholar] [CrossRef]
- Nio, K.; Yamashita, T.; Kaneko, S. The evolving concept of liver cancer stem cells. Mol. Cancer 2017, 16, 4. [Google Scholar] [CrossRef]
- Gabay, M.; Li, Y.; Felsher, D.W. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb. Perspect. Med. 2014, 4, a014241. [Google Scholar] [CrossRef]
- Jagust, P.; de Luxán-Delgado, B.; Parejo-Alonso, B.; Sancho, P. Metabolism-Based Therapeutic Strategies Targeting Cancer Stem Cells. Front. Pharmacol. 2019, 10, 203. [Google Scholar] [CrossRef]
- French, R.; Pauklin, S. Epigenetic regulation of cancer stem cell formation and maintenance. Int. J. Cancer 2021, 148, 2884–2897. [Google Scholar] [CrossRef]
- Liu, L.; Liu, C.; Zhang, Q.; Shen, J.; Zhang, H.; Shan, J.; Duan, G.; Guo, D.; Chen, X.; Cheng, J.; et al. SIRT1-mediated transcriptional regulation of SOX2 is important for self-renewal of liver cancer stem cells. Hepatology 2016, 64, 814–827. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Huan, H.; Liu, C.; Luo, Y.; Shen, J.; Zhuo, Y.; Zhang, Z.; Qian, C. Deacetylation of β-catenin by SIRT1 regulates self-renewal and oncogenesis of liver cancer stem cells. Cancer Lett. 2019, 463, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Jinushi, M.; Chiba, S.; Yoshiyama, H.; Masutomi, K.; Kinoshita, I.; Dosaka-Akita, H.; Yagita, H.; Takaoka, A.; Tahara, H. Tumor-associated macrophages regulate tumorigenicity and anticancer drug responses of cancer stem/initiating cells. Proc. Natl. Acad. Sci. USA 2011, 108, 12425–12430. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Clauser, K.R.; Tam, W.L.; Fröse, J.; Ye, X.; Eaton, E.N.; Reinhardt, F.; Donnenberg, V.S.; Bhargava, R.; Carr, S.A.; et al. A breast cancer stem cell niche supported by juxtacrine signalling from monocytes and macrophages. Nat. Cell Biol. 2014, 16, 1105–1117. [Google Scholar] [CrossRef]
- Su, S.; Liu, Q.; Chen, J.; Chen, F.; He, C.; Huang, D.; Wu, W.; Lin, L.; Huang, W.; Zhang, J.; et al. A positive feedback loop between mesenchymal-like cancer cells and macrophages is essential to breast cancer metastasis. Cancer Cell 2014, 25, 605–620. [Google Scholar] [CrossRef]
- Wan, S.; Zhao, E.; Kryczek, I.; Vatan, L.; Sadovskaya, A.; Ludema, G.; Simeone, D.M.; Zou, W.; Welling, T.H. Tumor-associated macrophages produce interleukin 6 and signal via STAT3 to promote expansion of human hepatocellular carcinoma stem cells. Gastroenterology 2014, 147, 1393–1404. [Google Scholar] [CrossRef]
- Zhou, W.; Ke, S.Q.; Huang, Z.; Flavahan, W.; Fang, X.; Paul, J.; Wu, L.; Sloan, A.E.; McLendon, R.E.; Li, X.; et al. Periostin secreted by glioblastoma stem cells recruits M2 tumour-associated macrophages and promotes malignant growth. Nat. Cell Biol. 2015, 17, 170–182. [Google Scholar] [CrossRef]
- Lam, K.H.; Ma, S. Noncellular components in the liver cancer stem cell niche: Biology and potential clinical implications. Hepatology 2022. [Google Scholar] [CrossRef]
- Emami Nejad, A.; Najafgholian, S.; Rostami, A.; Sistani, A.; Shojaeifar, S.; Esparvarinha, M.; Nedaeinia, R.; Haghjooy Javanmard, S.; Taherian, M.; Ahmadlou, M.; et al. The role of hypoxia in the tumor microenvironment and development of cancer stem cell: A novel approach to developing treatment. Cancer Cell Int. 2021, 21, 62. [Google Scholar] [CrossRef]
- Cui, C.P.; Wong, C.C.; Kai, A.K.; Ho, D.W.; Lau, E.Y.; Tsui, Y.M.; Chan, L.K.; Cheung, T.T.; Chok, K.S.; Chan, A.C.Y.; et al. SENP1 promotes hypoxia-induced cancer stemness by HIF-1α deSUMOylation and SENP1/HIF-1α positive feedback loop. Gut 2017, 66, 2149–2159. [Google Scholar] [CrossRef]
- Bhuria, V.; Xing, J.; Scholta, T.; Bui, K.C.; Nguyen, M.L.T.; Malek, N.P.; Bozko, P.; Plentz, R.R. Hypoxia induced Sonic Hedgehog signaling regulates cancer stemness, epithelial-to-mesenchymal transition and invasion in cholangiocarcinoma. Exp. Cell Res. 2019, 385, 111671. [Google Scholar] [CrossRef]
- Yasuda, T.; Ishimoto, T.; Baba, H. Conflicting metabolic alterations in cancer stem cells and regulation by the stromal niche. Regen. Ther. 2021, 17, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Ling, S.; Shan, Q.; Zhan, Q.; Ye, Q.; Liu, P.; Xu, S.; He, X.; Ma, J.; Xiang, J.; Jiang, G.; et al. USP22 promotes hypoxia-induced hepatocellular carcinoma stemness by a HIF1α/USP22 positive feedback loop upon TP53 inactivation. Gut 2020, 69, 1322–1334. [Google Scholar] [CrossRef]
- Loong, J.H.; Wong, T.L.; Tong, M.; Sharma, R.; Zhou, L.; Ng, K.Y.; Yu, H.J.; Li, C.H.; Man, K.; Lo, C.M.; et al. Glucose deprivation-induced aberrant FUT1-mediated fucosylation drives cancer stemness in hepatocellular carcinoma. J. Clin. Investig. 2021, 131, e143377. [Google Scholar] [CrossRef]
- Hida, K.; Maishi, N.; Takeda, R.; Hida, Y. The Roles of Tumor Endothelial Cells in Cancer Metastasis. In Metastasis; Sergi, C.M., Ed.; Exon Publications: Brisbane, Australia, 2022. [Google Scholar]
- Lu, J.; Ye, X.; Fan, F.; Xia, L.; Bhattacharya, R.; Bellister, S.; Tozzi, F.; Sceusi, E.; Zhou, Y.; Tachibana, I.; et al. Endothelial cells promote the colorectal cancer stem cell phenotype through a soluble form of Jagged-1. Cancer Cell 2013, 23, 171–185. [Google Scholar] [CrossRef]
- Yan, G.N.; Yang, L.; Lv, Y.F.; Shi, Y.; Shen, L.L.; Yao, X.H.; Guo, Q.N.; Zhang, P.; Cui, Y.H.; Zhang, X.; et al. Endothelial cells promote stem-like phenotype of glioma cells through activating the Hedgehog pathway. J. Pathol. 2014, 234, 11–22. [Google Scholar] [CrossRef]
- Feng, C.; Yang, X.; Sun, J.; Luo, Q.; Song, G. [Effect of Conditioned Medium from Endothelial Cells on Cancer Stem Cell Phenotype of Hepatoma Cells]. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi 2015, 32, 1061–1066. [Google Scholar]
- Conigliaro, A.; Costa, V.; Lo Dico, A.; Saieva, L.; Buccheri, S.; Dieli, F.; Manno, M.; Raccosta, S.; Mancone, C.; Tripodi, M.; et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol. Cancer 2015, 14, 155. [Google Scholar] [CrossRef]
- Tang, K.H.; Ma, S.; Lee, T.K.; Chan, Y.P.; Kwan, P.S.; Tong, C.M.; Ng, I.O.; Man, K.; To, K.F.; Lai, P.B.; et al. CD133(+) liver tumor-initiating cells promote tumor angiogenesis, growth, and self-renewal through neurotensin/interleukin-8/CXCL1 signaling. Hepatology 2012, 55, 807–820. [Google Scholar] [CrossRef]
- Yang, M.; Liu, J.; Wang, F.; Tian, Z.; Ma, B.; Li, Z.; Wang, B.; Zhao, W. Lysyl oxidase assists tumor-initiating cells to enhance angiogenesis in hepatocellular carcinoma. Int J. Oncol. 2019, 54, 1398–1408. [Google Scholar] [CrossRef]
- Brivio, S.; Cadamuro, M.; Strazzabosco, M.; Fabris, L. Tumor reactive stroma in cholangiocarcinoma: The fuel behind cancer aggressiveness. World J. Hepatol. 2017, 9, 455–468. [Google Scholar] [CrossRef]
- Reina-Campos, M.; Moscat, J.; Diaz-Meco, M. Metabolism shapes the tumor microenvironment. Curr. Opin. Cell Biol. 2017, 48, 47–53. [Google Scholar] [CrossRef]
- Li, Y.; Wang, R.; Xiong, S.; Wang, X.; Zhao, Z.; Bai, S.; Wang, Y.; Zhao, Y.; Cheng, B. Cancer-associated fibroblasts promote the stemness of CD24. J. Mol. Med. 2019, 97, 243–255. [Google Scholar] [CrossRef]
- Liu, C.; Liu, L.; Chen, X.; Cheng, J.; Zhang, H.; Zhang, C.; Shan, J.; Shen, J.; Qian, C. LSD1 Stimulates Cancer-Associated Fibroblasts to Drive Notch3-Dependent Self-Renewal of Liver Cancer Stem-like Cells. Cancer Res. 2018, 78, 938–949. [Google Scholar] [CrossRef]
- Lin, Y.; Cai, Q.; Chen, Y.; Shi, T.; Liu, W.; Mao, L.; Deng, B.; Ying, Z.; Gao, Y.; Luo, H.; et al. CAFs shape myeloid-derived suppressor cells to promote stemness of intrahepatic cholangiocarcinoma through 5-lipoxygenase. Hepatology 2022, 75, 28–42. [Google Scholar] [CrossRef]
- Xia, H.; Huang, Z.; Xu, Y.; Yam, J.W.P.; Cui, Y. Reprogramming of central carbon metabolism in hepatocellular carcinoma. Biomed. Pharmacother. 2022, 153, 113485. [Google Scholar] [CrossRef]
- Li, J.; Eu, J.Q.; Kong, L.R.; Wang, L.; Lim, Y.C.; Goh, B.C.; Wong, A.L.A. Targeting Metabolism in Cancer Cells and the Tumour Microenvironment for Cancer Therapy. Molecules 2020, 25, 4831. [Google Scholar] [CrossRef]
- Greten, F.R.; Grivennikov, S.I. Inflammation and Cancer: Triggers, Mechanisms, and Consequences. Immunity 2019, 51, 27–41. [Google Scholar] [CrossRef]
- Simón, L.; Sanhueza, S.; Gaete-Ramírez, B.; Varas-Godoy, M.; Quest, A.F.G. Role of the Pro-Inflammatory Tumor Microenvironment in Extracellular Vesicle-Mediated Transfer of Therapy Resistance. Front. Oncol. 2022, 12, 897205. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, X.; Wang, L.; Zhang, C. Interplay between inflammatory tumor microenvironment and cancer stem cells. Oncol. Lett. 2018, 16, 679–686. [Google Scholar] [CrossRef]
- Wang, X.; Sun, W.; Shen, W.; Xia, M.; Chen, C.; Xiang, D.; Ning, B.; Cui, X.; Li, H.; Li, X.; et al. Long non-coding RNA DILC regulates liver cancer stem cells via IL-6/STAT3 axis. J. Hepatol. 2016, 64, 1283–1294. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, J.N.; Zeng, T.T.; He, F.; Chen, S.P.; Ma, S.; Bi, J.; Zhu, X.F.; Guan, X.Y. CD133+ liver cancer stem cells resist interferon-gamma-induced autophagy. BMC Cancer 2016, 16, 15. [Google Scholar] [CrossRef] [PubMed]
- Müller, L.; Tunger, A.; Plesca, I.; Wehner, R.; Temme, A.; Westphal, D.; Meier, F.; Bachmann, M.; Schmitz, M. Bidirectional Crosstalk Between Cancer Stem Cells and Immune Cell Subsets. Front. Immunol. 2020, 11, 140. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wen, H.; Zhou, C.; Su, Q.; Lin, Y.; Xie, Y.; Huang, Y.; Qiu, Q.; Lin, J.; Huang, X.; et al. TNF-α derived from M2 tumor-associated macrophages promotes epithelial-mesenchymal transition and cancer stemness through the Wnt/β-catenin pathway in SMMC-7721 hepatocellular carcinoma cells. Exp. Cell Res. 2019, 378, 41–50. [Google Scholar] [CrossRef]
- Wei, R.; Zhu, W.W.; Yu, G.Y.; Wang, X.; Gao, C.; Zhou, X.; Lin, Z.F.; Shao, W.Q.; Wang, S.H.; Lu, M.; et al. S100 calcium-binding protein A9 from tumor-associated macrophage enhances cancer stem cell-like properties of hepatocellular carcinoma. Int. J. Cancer 2021, 148, 1233–1244. [Google Scholar] [CrossRef]
- Pickup, M.; Novitskiy, S.; Moses, H.L. The roles of TGFβ in the tumour microenvironment. Nat. Rev. Cancer 2013, 13, 788–799. [Google Scholar] [CrossRef]
- Papadaki, S.; Magklara, A. Regulation of Metabolic Plasticity in Cancer Stem Cells and Implications in Cancer Therapy. Cancers 2022, 14, 5912. [Google Scholar] [CrossRef]
- Shen, Y.A.; Chen, C.C.; Chen, B.J.; Wu, Y.T.; Juan, J.R.; Chen, L.Y.; Teng, Y.C.; Wei, Y.H. Potential Therapies Targeting Metabolic Pathways in Cancer Stem Cells. Cells 2021, 10, 1772. [Google Scholar] [CrossRef]
- Saito, T.; Chiba, T.; Yuki, K.; Zen, Y.; Oshima, M.; Koide, S.; Motoyama, T.; Ogasawara, S.; Suzuki, E.; Ooka, Y.; et al. Metformin, a diabetes drug, eliminates tumor-initiating hepatocellular carcinoma cells. PLoS ONE 2013, 8, e70010. [Google Scholar] [CrossRef]
- Sun, L.; Wang, Y.; Wang, L.; Yao, B.; Chen, T.; Li, Q.; Liu, Z.; Liu, R.; Niu, Y.; Song, T.; et al. Resolvin D1 prevents epithelial-mesenchymal transition and reduces the stemness features of hepatocellular carcinoma by inhibiting paracrine of cancer-associated fibroblast-derived COMP. J. Exp. Clin. Cancer Res. 2019, 38, 170. [Google Scholar] [CrossRef]
- Xiong, S.; Wang, R.; Chen, Q.; Luo, J.; Wang, J.; Zhao, Z.; Li, Y.; Wang, Y.; Wang, X.; Cheng, B. Cancer-associated fibroblasts promote stem cell-like properties of hepatocellular carcinoma cells through IL-6/STAT3/Notch signaling. Am. J. Cancer Res. 2018, 8, 302–316. [Google Scholar] [PubMed]
- De Gregorio, E.; Colell, A.; Morales, A.; Marí, M. Relevance of SIRT1-NF-κB Axis as Therapeutic Target to Ameliorate Inflammation in Liver Disease. Int. J. Mol. Sci. 2020, 21, 3858. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Ma, S.; Tong, M. Metabolic Plasticity of Cancer Stem Cells in Response to Microenvironmental Cues. Cancers 2022, 14, 5345. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Zhao, Y.; Yan, H.; Yang, Y.; Shen, S.; Dai, X.; Ji, X.; Ji, F.; Gong, X.G.; Li, L.; et al. Single tumor-initiating cells evade immune clearance by recruiting type II macrophages. Genes Dev. 2017, 31, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Zhao, Z.; Song, J.; Lan, X.; Lu, S.; Chen, M.; Wang, Z.; Chen, W.; Fan, X.; Wu, F.; et al. Interactions between interleukin-6 and myeloid-derived suppressor cells drive the chemoresistant phenotype of hepatocellular cancer. Exp. Cell Res. 2017, 351, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Mima, K.; Okabe, H.; Ishimoto, T.; Hayashi, H.; Nakagawa, S.; Kuroki, H.; Watanabe, M.; Beppu, T.; Tamada, M.; Nagano, O.; et al. CD44s regulates the TGF-β-mediated mesenchymal phenotype and is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Res. 2012, 72, 3414–3423. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Li, Q.J.; Feng, Y.; Zhang, Y.; Markowitz, G.J.; Ning, S.; Deng, Y.; Zhao, J.; Jiang, S.; Yuan, Y.; et al. TGF-β-miR-34a-CCL22 signaling-induced Treg cell recruitment promotes venous metastases of HBV-positive hepatocellular carcinoma. Cancer Cell 2012, 22, 291–303. [Google Scholar] [CrossRef]
- Li, X.; Zhang, Y.; Ma, W.; Fu, Q.; Liu, J.; Yin, G.; Chen, P.; Dai, D.; Chen, W.; Qi, L.; et al. Enhanced glucose metabolism mediated by CD147 contributes to immunosuppression in hepatocellular carcinoma. Cancer Immunol. Immunother. 2020, 69, 535–548. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Correnti, M.; Binatti, E.; Gammella, E.; Invernizzi, P.; Recalcati, S. The Emerging Role of Tumor Microenvironmental Stimuli in Regulating Metabolic Rewiring of Liver Cancer Stem Cells. Cancers 2023, 15, 5. https://doi.org/10.3390/cancers15010005
Correnti M, Binatti E, Gammella E, Invernizzi P, Recalcati S. The Emerging Role of Tumor Microenvironmental Stimuli in Regulating Metabolic Rewiring of Liver Cancer Stem Cells. Cancers. 2023; 15(1):5. https://doi.org/10.3390/cancers15010005
Chicago/Turabian StyleCorrenti, Margherita, Eleonora Binatti, Elena Gammella, Pietro Invernizzi, and Stefania Recalcati. 2023. "The Emerging Role of Tumor Microenvironmental Stimuli in Regulating Metabolic Rewiring of Liver Cancer Stem Cells" Cancers 15, no. 1: 5. https://doi.org/10.3390/cancers15010005
APA StyleCorrenti, M., Binatti, E., Gammella, E., Invernizzi, P., & Recalcati, S. (2023). The Emerging Role of Tumor Microenvironmental Stimuli in Regulating Metabolic Rewiring of Liver Cancer Stem Cells. Cancers, 15(1), 5. https://doi.org/10.3390/cancers15010005






