Robust Performance of the Novel Research-Use-Only Idylla GeneFusion Assay Using a Diverse Set of Pathological Samples with a Proposed 1-Day Workflow for Advanced NSCLC Evaluation
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results
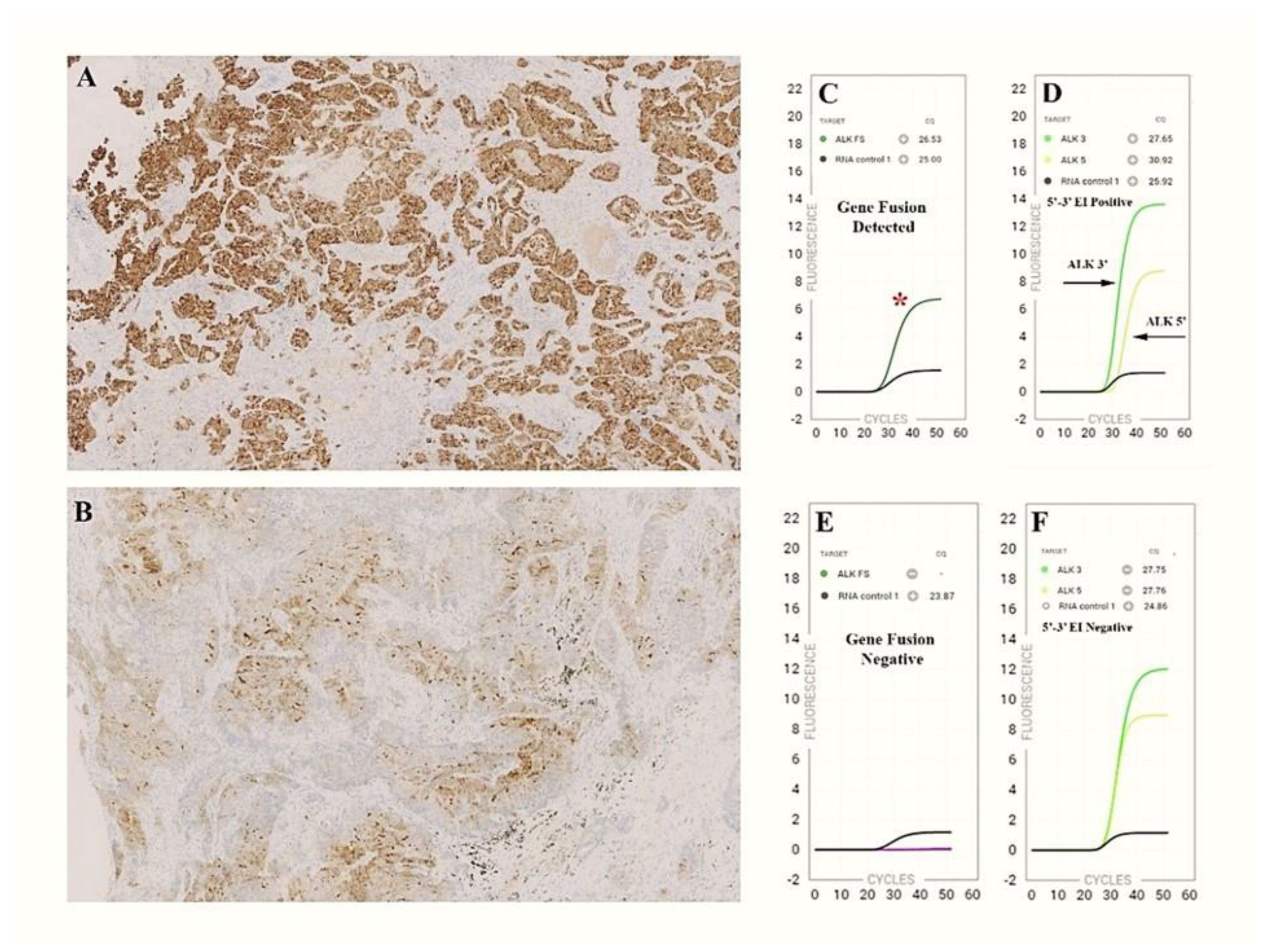
3.1. ALK Evaluation
3.1.1. ALK Cohort
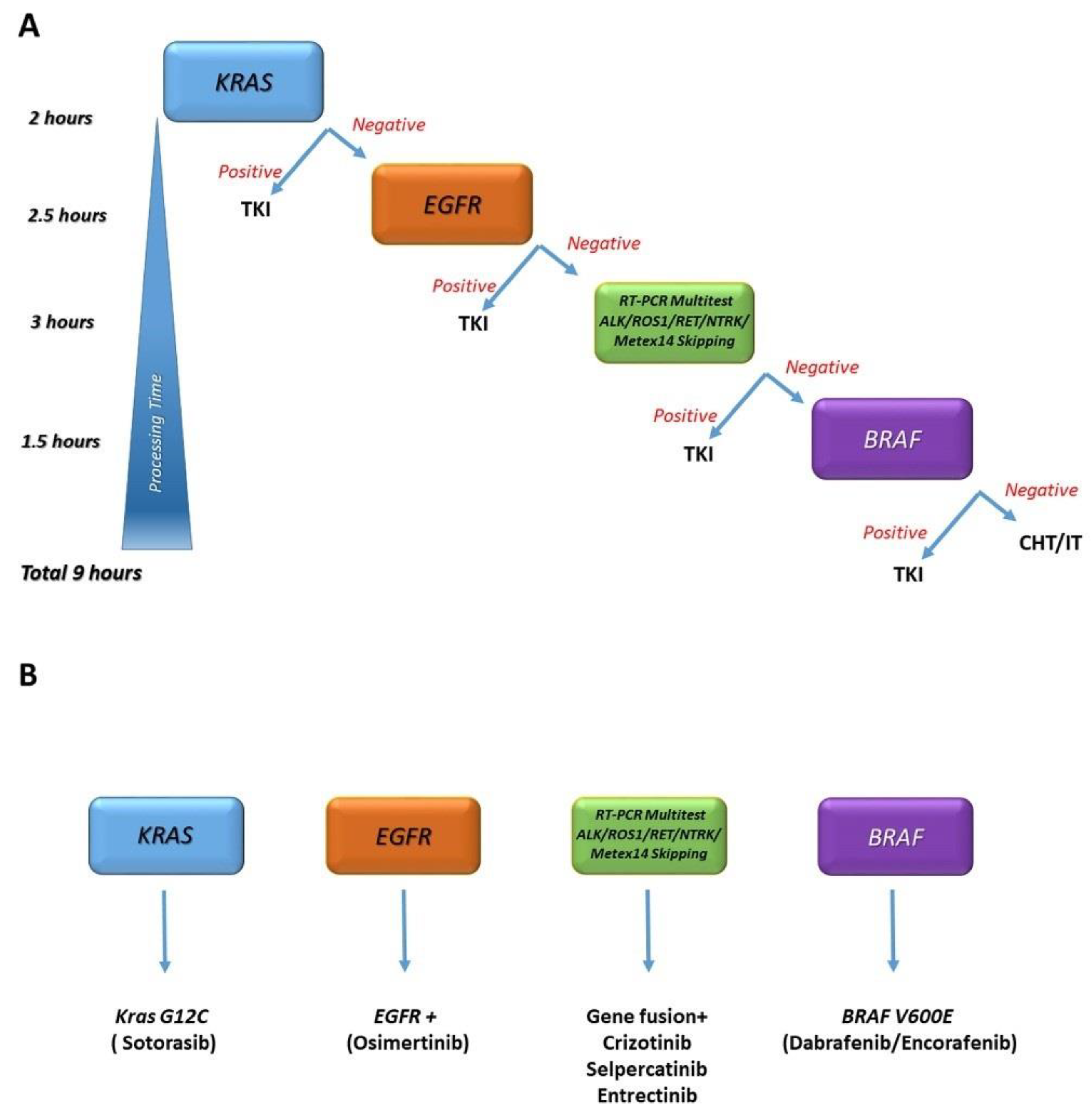
3.1.2. ALK Detection Workflow
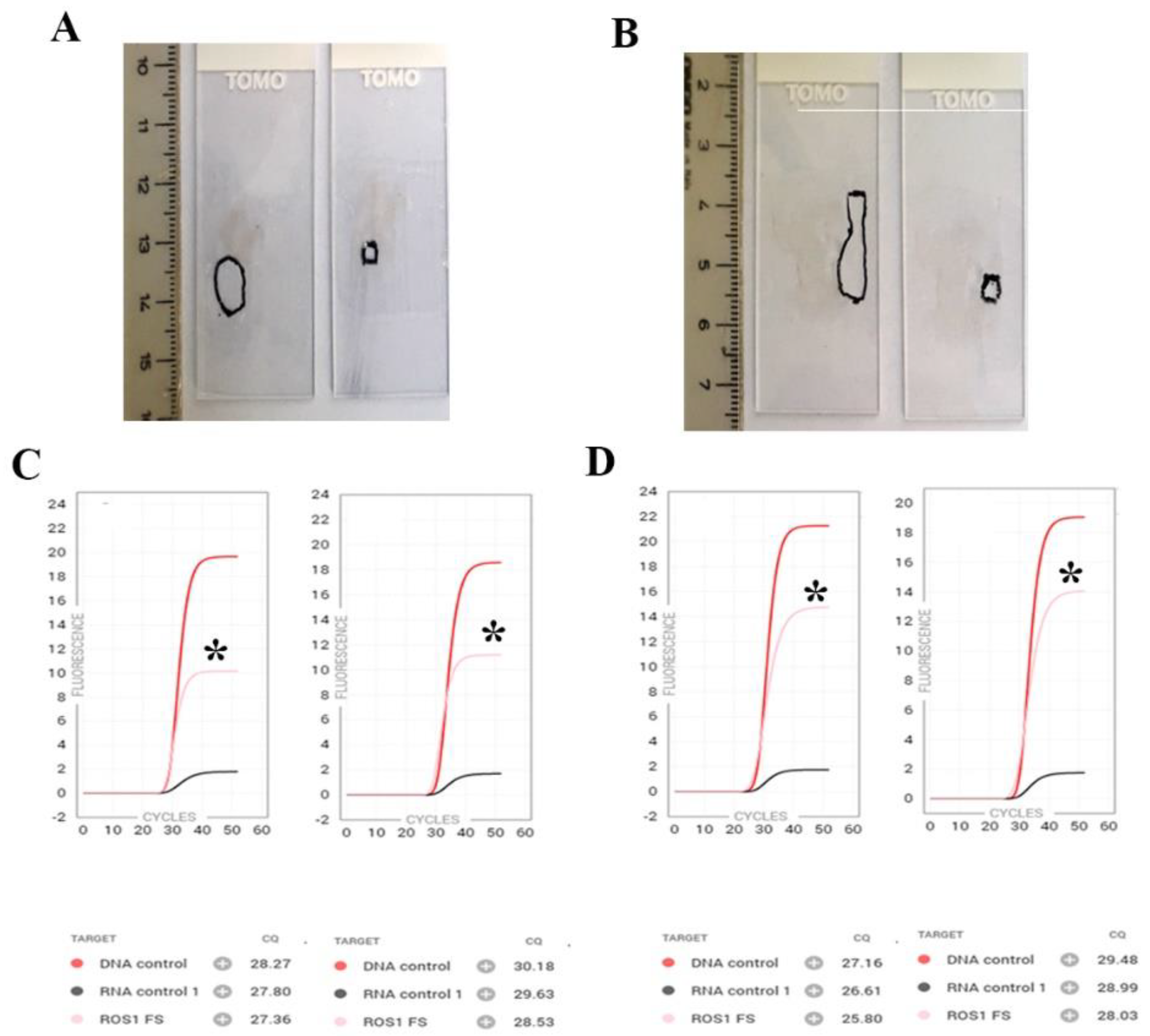
3.2. ROS1/RET/NTRK/MET ex14 Skipping Evaluation
3.2.1. ROS1/RET/NTRK/MET ex14 Skipping Cohort
3.2.2. ROS1/RET/NTRK/MET ex14 Skipping Workflow
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kris, M.G.; Johnson, B.E.; Berry, L.D.; Kwiatkowski, D.J.; Iafrate, A.J.; Wistuba, I.I.; Varella-Garcia, M.; Franklin, W.A.; Aronson, S.L.; Su, P.F.; et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 2014, 311, 1998–2006. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.E.; Lee, B.; Hong, M.; Song, J.Y.; Jung, K.; Lira, M.E.; Mao, M.; Han, J.; Kim, J.; Choi, Y.L. Comprehensive analysis of RET and ROS1 rearrangement in lung adenocarcinoma. Mod. Pathol. 2015, 28, 468–479. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Zhang, Y.; Li, Y.; Hu, H.; Wang, L.; Li, H.; Wang, R.; Ye, T.; Luo, X.; Zhang, Y.; et al. ALK, ROS1 and RET fusions in 1139 lung adenocarcinomas: A comprehensive study of common and fusion pattern-specific clinicopathologic, histologic and cytologic features. Lung Cancer 2014, 84, 121–126. [Google Scholar] [CrossRef]
- Howlader, N.; Forjaz, G.; Mooradian, M.J.; Meza, R.; Kong, C.Y.; Cronin, K.A.; Mariotto, A.B.; Lowy, D.R.; Feuer, E.J. The Effect of Advances in Lung-Cancer Treatment on Population Mortality. N. Engl. J. Med. 2020, 383, 640–649. [Google Scholar] [CrossRef] [PubMed]
- Jordan, E.J.; Kim, H.R.; Arcila, M.E.; Barron, D.; Chakravarty, D.; Gao, J.; Chang, M.T.; Ni, A.; Kundra, R.; Jonsson, P.; et al. Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging Therapies. Cancer Discov. 2017, 7, 596–609. [Google Scholar] [CrossRef]
- Nozaki, Y.; Yamamoto, H.; Iwasaki, T.; Sato, M.; Jiromaru, R.; Hongo, T.; Yasumatsu, R.; Oda, Y. Clinicopathological features and immunohistochemical utility of NTRK-, ALK-, and ROS1-rearranged papillary thyroid carcinomas and anaplastic thyroid carcinomas. Hum. Pathol. 2020, 106, 82–92. [Google Scholar] [CrossRef]
- Solomon, J.P.; Hechtman, J.F. Detection of NTRK Fusions: Merits and Limitations of Current Diagnostic Platforms. Cancer Res. 2019, 79, 3163–3168. [Google Scholar] [CrossRef]
- Yang, S.R.; Aypar, U.; Rosen, E.Y.; Mata, D.A.; Benayed, R.; Mullaney, K.; Jayakumaran, G.; Zhang, Y.; Frosina, D.; Drilon, A.; et al. A Performance Comparison of Commonly Used Assays to Detect RET Fusions. Clin. Cancer Res. 2021, 27, 1316–1328. [Google Scholar] [CrossRef]
- Park, G.; Kim, T.H.; Lee, H.O.; Lim, J.A.; Won, J.K.; Min, H.S.; Lee, K.E.; Park, D.J.; Park, Y.J.; Park, W.Y. Standard immunohistochemistry efficiently screens for anaplastic lymphoma kinase rearrangements in differentiated thyroid cancer. Endocr. Relat. Cancer 2015, 22, 55–63. [Google Scholar] [CrossRef]
- Memorial Sloan Kettering Cancer Center, West Harrison, N.Y. Mod. Healthc. 2015, 45, 14–15.
- Mosele, F.; Remon, J.; Mateo, J.; Westphalen, C.B.; Barlesi, F.; Lolkema, M.P.; Normanno, N.; Scarpa, A.; Robson, M.; Meric-Bernstam, F.; et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2020, 31, 1491–1505. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R. Applications of artificial intelligence multiomics in precision oncology. J. Cancer Res. Clin. Oncol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Williams, H.L.; Walsh, K.; Diamond, A.; Oniscu, A.; Deans, Z.C. Validation of the Oncomine() focus panel for next-generation sequencing of clinical tumour samples. Virchows Arch. 2018, 473, 489–503. [Google Scholar] [CrossRef]
- Aguado, C.; Gimenez-Capitan, A.; Roman, R.; Rodriguez, S.; Jordana-Ariza, N.; Aguilar, A.; Cabrera-Galvez, C.; Rivas-Corredor, C.; Lianes, P.; Viteri, S.; et al. RNA-Based Multiplexing Assay for Routine Testing of Fusion and Splicing Variants in Cytological Samples of NSCLC Patients. Diagnostics 2020, 11, 15. [Google Scholar] [CrossRef] [PubMed]
- Evangelista, A.F.; Zanon, M.F.; Carloni, A.C.; de Paula, F.E.; Morini, M.A.; Ferreira-Neto, M.; Soares, I.C.; Miziara, J.E.; de Marchi, P.; Scapulatempo-Neto, C.; et al. Detection of ALK fusion transcripts in FFPE lung cancer samples by NanoString technology. BMC Pulm. Med. 2017, 17, 86. [Google Scholar] [CrossRef]
- Lira, M.E.; Choi, Y.L.; Lim, S.M.; Deng, S.; Huang, D.; Ozeck, M.; Han, J.; Jeong, J.Y.; Shim, H.S.; Cho, B.C.; et al. A single-tube multiplexed assay for detecting ALK, ROS1, and RET fusions in lung cancer. J. Mol. Diagn. 2014, 16, 229–243. [Google Scholar] [CrossRef]
- WHO Classification of Tumours Editorial Board. Thoracic Tumours, 5th ed.; In-Ternational Agency for Research on Cancer: Lyon, France, 2021. [Google Scholar]
- Lira, M.E.; Kim, T.M.; Huang, D.; Deng, S.; Koh, Y.; Jang, B.; Go, H.; Lee, S.H.; Chung, D.H.; Kim, W.H.; et al. Multiplexed gene expression and fusion transcript analysis to detect ALK fusions in lung cancer. J. Mol. Diagn. 2013, 15, 51–61. [Google Scholar] [CrossRef]
- Liu, F.; Wei, Y.; Zhang, H.; Jiang, J.; Zhang, P.; Chu, Q. NTRK Fusion in Non-Small Cell Lung Cancer: Diagnosis, Therapy, and TRK Inhibitor Resistance. Front. Oncol. 2022, 12, 864666. [Google Scholar] [CrossRef]
- Teishikata, T.; Shiraishi, K.; Shinno, Y.; Kobayashi, Y.; Kashima, J.; Ishiyama, T.; Yoshida, T.; Mori, T.; Yatabe, Y. An Alert to Possible False Positives With a Commercial Assay for MET Exon 14 Skipping. J. Thorac. Oncol. 2021, 16, 2133–2138. [Google Scholar] [CrossRef]
- Subramanian, J.; Tawfik, O. Detection of MET exon 14 skipping mutations in non-small cell lung cancer: Overview and community perspective. Expert. Rev. Anticancer Ther. 2021, 21, 877–886. [Google Scholar] [CrossRef]
- Depoilly, T.; Garinet, S.; van Kempen, L.C.; Schuuring, E.; Clave, S.; Bellosillo, B.; Ercolani, C.; Buglioni, S.; Siemanowski, J.; Merkelbach-Bruse, S.; et al. Multicenter Evaluation of the Idylla GeneFusion in Non-Small-Cell Lung Cancer. J. Mol. Diagn. 2021, 24, 1021–1030. [Google Scholar] [CrossRef] [PubMed]
- Sorber, L.; Van Dorst, B.; Bellon, E.; Zwaenepoel, K.; Lambin, S.; De Winne, K.; Lardon, F.; Pauwels, P.; Siozopoulou, V. NTRK Gene Fusion Detection in a Pan-Cancer Setting Using the Idylla GeneFusion Assay. J. Mol. Diagn. 2022, 24, 750–759. [Google Scholar] [CrossRef] [PubMed]
- Ukkola, I.; Nummela, P.; Kero, M.; Tammio, H.; Tuominen, J.; Kairisto, V.; Kallajoki, M.; Haglund, C.; Peltomaki, P.; Kytola, S.; et al. Gene fusions and oncogenic mutations in MLH1 deficient and BRAFV600E wild-type colorectal cancers. Virchows Arch. 2022, 480, 807–817. [Google Scholar] [CrossRef]
- Chu, Y.H.; Barbee, J.; Yang, S.R.; Chang, J.C.; Liang, P.; Mullaney, K.; Chan, R.; Salazar, P.; Benayed, R.; Offin, M.; et al. Clinical Utility and Performance of an Ultrarapid Multiplex RNA-Based Assay for Detection of ALK, ROS1, RET, and NTRK1/2/3 Rearrangements and MET Exon 14 Skipping Alterations. J. Mol. Diagn. 2022, 24, 642–654. [Google Scholar] [CrossRef] [PubMed]
- Fujii, T.; Uchiyama, T.; Matsuoka, M.; Myojin, T.; Sugimoto, S.; Nitta, Y.; Okabe, F.; Sugimoto, A.; Sekita-Hatakeyama, Y.; Morita, K.; et al. Evaluation of DNA and RNA quality from archival formalin-fixed paraffin-embedded tissue for next-generation sequencing - Retrospective study in Japanese single institution. Pathol. Int. 2019, 70, 602–611. [Google Scholar] [CrossRef]
- Ambrosini-Spaltro, A.; Farnedi, A.; Calistri, D.; Rengucci, C.; Prisinzano, G.; Chiadini, E.; Capelli, L.; Angeli, D.; Bennati, C.; Valli, M.; et al. The role of next-generation sequencing in detecting gene fusions with known and unknown partners: A single-center experience with methodologies’ integration. Hum. Pathol. 2022, 123, 20–30. [Google Scholar] [CrossRef]
- Boppudi, S.; Scheil-Bertram, S.; Faust, E.; Annamneedi, A.; Fisseler-Eckoff, A. Assessing and Evaluating the Scope and Constraints of Idylla Molecular Assays by Using Different Source Materials in Routine Diagnostic Settings. Int. J. Mol. Sci. 2022, 23, 12515. [Google Scholar] [CrossRef]
- De Marchi, F.; Haley, L.; Fryer, H.; Ibrahim, J.; Beierl, K.; Zheng, G.; Gocke, C.D.; Eshleman, J.R.; Belchis, D.; Illei, P.; et al. Clinical Validation of Coexisting Activating Mutations Within EGFR, Mitogen-Activated Protein Kinase, and Phosphatidylinositol 3-Kinase Pathways in Lung Cancers. Arch. Pathol. Lab. Med. 2019, 143, 174–182. [Google Scholar] [CrossRef]
| Sample ID | Gender | Age | Histology | Block Age at Testing (Months) | Specimen | Tumor Cell Content | Reference Method | ALK Specific Fusion | ALK 3′5′ Imbalance | Idylla Overall Result | Comparison to Reference | Cq RNA Controls | Cq DNA Controls |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| #1 #1-B | M | 47 | ADC ADC | 3 2 | biopsy re-biopsy | >50% >50% | IHC pos IHC pos | Y Y | Y Y | pos pos | C C | 26.6 27.0 | 27.6 29.4 |
| #2 | F | 65 | ADC | 1 | biopsy | >50% | IHC pos | Y | Y | pos | C | 29 | 29.9 |
| #3 | F | 87 | ADC | 1 | biopsy | >50% | IHC pos | N | Y | pos | C | 27.7 | 28.5 |
| #4 | M | 75 | LCNEC | 13 | surgical | >50% | IHC dubious | N | N | neg | D | 26 | 26.2 |
| #5 | F | 66 | ADC | 26 | biopsy | >50% | IHC pos | Y | Y | pos | C | 27.8 | 29.8 |
| #5-B | after 3 weeks | >50% | Not performed | Y | Y | pos | 28 | 29.8 | |||||
| #6 | M | 54 | ADC | 25 | pleural biopsy | >50% | IHC pos | Y | Y | pos | C | 26.3 | 28.9 |
| #7 | F | 77 | ADC | 31 | biopsy | >50% | IHC pos | Y | Y | pos | C | 30.5 | 31.6 |
| #8 | F | 48 | ADC | 24 | biopsy | >50% | IHC pos | Y | Y | pos | C | 30.1 | 31.3 |
| #9 | F | 52 | ADC | 29 | biopsy | >50% | IHC pos | Y | Y | pos | C | 29.1 | 31.3 |
| #10 | M | 69 | ADC | 12 | biopsy | >50% | IHC pos | Y | Y | pos | C | 27.5 | 30.4 |
| #11 | M | 64 | ADC | 17 | pleural biopsy | >50% | IHC pos | Y | Y | pos | C | 27 | 29.9 |
| #12 | M | 41 | ADC | 27 | pleural biopsy | >50% | IHC pos | Y | Y | pos | C | 29.3 | 31.8 |
| #13 | M | 60 | ADC | 19 | surgical | >50% | IHC pos | N | Y | pos | C | 27.3 | 28.5 |
| #14 | M | 72 | ADC+SCLC | 1 | biopsy | >50% | IHC dubious | N | N | neg | D | 26.3 | 27.1 |
| #15 | M | 52 | ADC | 3 | surgical | >50% | IHC pos | Y | Y | pos | C | 25.3 | 28.5 |
| #15-B | 4 | surgical (metastatic lymph node) | >50% | Not performed | Y | Y | pos | 28.4 | 30 | ||||
| #16 | M | 44 | ADC | 15 | surgical | >50% | IHC pos | Y | Y | pos | C | 27.2 | 28.7 |
| #16-B | 15 | surgical (metastatic lymph node) | >50% | Not performed | Y | Y | pos | 27.3 | 29 | ||||
| #17 | F | 69 | ADC | 123 | surgical | >50% | FISH pos | Y | Y | pos | C | 29.6 | 27.2 |
| #17-B | 123 | surgical (metastatic lymph node) | >50% | Not performed | Y | Y | pos | 28.9 | 28.9 | ||||
| #17-C | ADC | 1 | re-biopsy | >50% | IHC pos | Y | Y | pos | C | 28.6 | 31.2 | ||
| #18 | M | 71 | ADC | 1 | biopsy | >30% | IHC pos | Y | Y | pos | C | 28 | 28.2 |
| #18-B | 1 | pleural biopsy | >50% | Not performed | Y | Y | pos | 27.5 | 28.1 | ||||
| #19 | F | 85 | ADC | 3 | biopsy | 50% | Amoy-DX pos | Y | Y | pos | C | 27 | 27.7 |
| #20 | M | 57 | ADC | 13 | biopsy | >50% | IHC pos | Y | Y | pos | C | 29 | 31.3 |
| #21 | F | 38 | ADC | 84 | biopsy | >50% | IHC pos | Y | Y | pos | C | 27.3 | 28.5 |
| #22 | M | 51 | ADC | 85 | biopsy | >50% | IHC pos | Y | Y | pos | C | 28.4 | 29.7 |
| Sample ID | Gender | Age | Histology | Block Age at Testing (months) | Specimen | Tumor Cell Content | Reference Method/Gene Target | Gene-Specific Fusion | Gene 3′-5′ Imbalance | Idylla Overall Result | Other Method | Comparison to Reference | Cq RNA Controls | Cq DNA Controls |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| #23 | F | 80 | ADC | 28 | pleural biopsy | >50% | FISH/ROS1 Pos | Y | N | Y | None | C | 30.4 | 31.5 |
| #24 | F | 63 | ADC | 2 | cell block | 30% | FISH/ROS1 Pos | Y | N | Y | None | C | 29.8 | 30.9 |
| #25 | F | 47 | ADC | 20 | biopsy | >50% | FISH/ROS1 Pos | N | N | N | None | D | 29.2 | 29.8 |
| #26 | M | 77 | ADC | 19 | biopsy | >50% | FISH/ROS1 Pos | N | N | N | None | D | 30.2 | 31.1 |
| #27 | F | 72 | ADC | 44 | surgical | >50% | FISH/ROS1 Pos | Y | N | Y | None | C | 27.6 | 27.2 |
| #27-B | small md | Y | N | Y | None | C | 30 | 29.5 | ||||||
| #28 | F | 75 | ADC | 74 | surgical | >50% | FISH/ROS1 Pos | Y | N | Y | None | C | 28.8 | 28.3 |
| #28-B | small md | Y | N | Y | None | C | 30.3 | 30.2 | ||||||
| #29 | F | 49 | ADC | 48 | pleural biopsy | >50% | FISH/ROS1 Pos | Y | N | Y | NGS/SDC4(2)-ROS1(32) | C | 28.2 | 30.9 |
| #29-B | 48 | cell block | 30% | Y | N | Y | None | 33 | 31.4 | |||||
| #29-C | 1 | pleural biopsy | >50% | Y | N | Y | NGS/SDC4(2)-ROS1(32) | 27.1 | 30.1 | |||||
| #29-D | after 60 days | Y | N | Y | None | 27.5 | 29.7 | |||||||
| #30 | M | 74 | ADC | 1 | biopsy | >50% | IHC/ROS1 Pos | Y | N | Y | None | C | 31.2 | 33.5 |
| #31 | F | 71 | ADC | 4 | biopsy | 20% | AmoyDx/RET Pos | Y | Y | Y | None | C | 29 | 30 |
| #32 | M | 49 | ADC | 3 | biopsy | >50% | AmoyDx/RET Pos | Y | Y | Y | None | C | 28 | 27.8 |
| #33 | F | 75 | ADC | 3 | biopsy | 40% | AmoyDx/RET Pos | Y | Y | Y | NGS/KIF5B(16)-RET(12) | C | 27.5 | 27.1 |
| #33-B | 1 | cell block | 20% | Y | Y | Y | None | 28.1 | 27.7 | |||||
| #33-C | after 30days | Y | Y | Y | None | 29.4 | 29.8 | |||||||
| #34 | M | 69 | ADC | 23 | surgical | >50% | NGS/MET Pos | N | N | N | EasyPGX/Negative | D | 26.6 | 27.8 |
| #35 | F | 81 | ADC | 11 | surgical | >50% | EasyPgx/MET Pos | Y | N/A | Y | None | C | 26.1 | 26.6 |
| #36 | F | 71 | ADC | 7 | surgical | >50% | EasyPgx/MET Pos | Y | N/A | Y | None | C | 27.3 | 27.8 |
| #36-B | after 49days | Y | N/A | Y | None | C | 27.9 | 27.1 | ||||||
| #37 | M | 31 | PGS-carcinoma | 14 | surgical | >50 | NGS/ETV6(5) - NTRK3(15) | N/A | NTRK/3 | Y | None | C | 28.5 | 28.7 |
| #38 | M | 57 | PGS- carcinoma | 46 | surgical | >50% | IHC PanNTRK/NTRK Pos | N/A | NTRK/3 | Y | None | C | 28.1 | 28.9 |
| #38-B | after 36 days | N/A | NTRK/3 | Y | None | C | 30.5 | 29.6 | ||||||
| #39 | M | 55 | ADC | 35 | pleural biopsy | >50% | IHC/ROS1 Pos | Y | N | Y | FISH/ROS1 | C | 30.2 | 31.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leone, A.; Muscarella, L.A.; Graziano, P.; Tornese, A.; Grillo, L.R.; Di Lorenzo, A.; Bronzini, M.; Scarpino, S.; Sparaneo, A.; Rossi, G. Robust Performance of the Novel Research-Use-Only Idylla GeneFusion Assay Using a Diverse Set of Pathological Samples with a Proposed 1-Day Workflow for Advanced NSCLC Evaluation. Cancers 2023, 15, 292. https://doi.org/10.3390/cancers15010292
Leone A, Muscarella LA, Graziano P, Tornese A, Grillo LR, Di Lorenzo A, Bronzini M, Scarpino S, Sparaneo A, Rossi G. Robust Performance of the Novel Research-Use-Only Idylla GeneFusion Assay Using a Diverse Set of Pathological Samples with a Proposed 1-Day Workflow for Advanced NSCLC Evaluation. Cancers. 2023; 15(1):292. https://doi.org/10.3390/cancers15010292
Chicago/Turabian StyleLeone, Alvaro, Lucia Anna Muscarella, Paolo Graziano, Andrea Tornese, Lucia Rosalba Grillo, Angela Di Lorenzo, Monica Bronzini, Stefania Scarpino, Angelo Sparaneo, and Giulio Rossi. 2023. "Robust Performance of the Novel Research-Use-Only Idylla GeneFusion Assay Using a Diverse Set of Pathological Samples with a Proposed 1-Day Workflow for Advanced NSCLC Evaluation" Cancers 15, no. 1: 292. https://doi.org/10.3390/cancers15010292
APA StyleLeone, A., Muscarella, L. A., Graziano, P., Tornese, A., Grillo, L. R., Di Lorenzo, A., Bronzini, M., Scarpino, S., Sparaneo, A., & Rossi, G. (2023). Robust Performance of the Novel Research-Use-Only Idylla GeneFusion Assay Using a Diverse Set of Pathological Samples with a Proposed 1-Day Workflow for Advanced NSCLC Evaluation. Cancers, 15(1), 292. https://doi.org/10.3390/cancers15010292







