Simple Summary
Short, dysfunctional telomeres represent the genetic biomarkers of cancer. Studies in early-stage non-small-cell lung cancer (NSCLC) have shown that telomere length and telomerase levels are correlated with survival. In patients with advanced NSCLC, telomere status has not yet been investigated, and its clinical significance remains unknown. We studied telomere length and the expression of telomerase and shelterin genes in a cohort of 79 patients with advanced NSCLC, and evaluated these parameters as potential prognostic and predictive factors. Telomere shortening, high levels of telomerase and aberrant expression of shelterin genes TRF2, RAP1 and TIN2 were significantly correlated with shorter survival. Furthermore, a worse response to immunotherapy was observed in patients with shorter telomeres. The determination of telomere parameters in advanced NSCLC could be useful for individualized treatment decisions.
Abstract
Telomere length appears to correlate with survival in early non-small-cell lung cancer (NSCLC), but the prognostic impact of telomere status in advanced NSCLC remains undetermined. Our purpose was to evaluate telomere parameters as prognostic and predictive biomarkers in advanced NSCLC. In 79 biopsies obtained before treatment, we analyzed the telomere length and expression of TERT and shelterin complex genes (TRF1, TRF2, POT1, TPP1, RAP1, and TIN2), using quantitative PCR. Non-responders to first-line chemotherapy were characterized by shorter telomeres and low RAP1 expression (p = 0.0035 and p = 0.0069), and tended to show higher TERT levels (p = 0.058). In multivariate analysis, short telomeres were associated with reduced event-free (EFS, p = 0.0023) and overall survival (OS, p = 0.00041). TERT and TRF2 overexpression correlated with poor EFS (p = 0.0069 and p = 0.00041) and OS (p = 0.0051 and p = 0.007). Low RAP1 and TIN2 expression-levels were linked to reduced EFS (p = 0.00032 and p = 0.0069) and OS (p = 0.000051 and p = 0.02). Short telomeres were also associated with decreased survival after nivolumab therapy (p = 0.097). Evaluation of telomere status in advanced NSCLC emerges as a useful biomarker that allows for the selection of patient groups with different clinical evolutions, to establish personalized treatment.
1. Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide [1]. Non-small-cell lung cancer (NSCLC) is the most frequent lung cancer, accounting for 85% of all cases. NSCLC is frequently diagnosed at an advanced stage, and the overall prognosis is poor. Nevertheless, treatment responses and survival times are different in individual patients, which could be explained by biological heterogeneity of NSCLC [2]. This underlines the importance of biomarkers to identify subjects at higher risk of drug resistance and disease progression, who would benefit from more specific therapies. Furthermore, therapies targeting immune checkpoints have changed the treatment of NSCLC, enabling longer survival for patients with advanced disease. However, progression rates reported with nivolumab are above 30% [3,4], and approximately 14% of cases have hyperprogressive disease [5]. An early switch to salvage treatment in these patients should be considered, but no predictive factor has yet been identified.
Dysfunctional telomeres represent the genetic biomarkers of cancer [6,7]. Telomeres are nucleoprotein complexes essential for the protection of chromosomal ends. Due to incomplete DNA replication, telomeres shorten with every cell division [8]. Critical telomere shortening contributes to genomic instability and promotes cancerogenesis [9]. To maintain shortened telomeres, cancer cells activate telomerase, a specialized ribonucleoprotein that regenerates telomeric DNA, which enables survival and the limitless proliferation of tumors [10]. Upregulation of TERT (telomerase reverse transcriptase) gene leads to telomerase activation in cancers [11]. Telomeric DNA is protected by the shelterin protein complex, which regulates telomere length (TL) and prevents the inappropriate DNA damage response at chromosomal ends [12]. Shelterin contains six proteins: POT1 (protection of telomeres 1), TRF1 and TRF2 (telomeric repeat-binding factor 1 and 2), TIN2 (TRF1-interacting nuclear protein 2), RAP1 (repressor/activator protein 1) and TPP1 (TIN2 and POT1-interacting protein). Severe telomere shortening and abnormal expression of TERT and shelterin-complex genes were previously correlated with cancer prognosis [6,13,14].
Although relatively rare, telomere studies in NSCLC have shown that TL and telomerase levels were correlated with survival [15,16]. However, these clinical correlations have been evaluated only for early-stage surgically resected NSCLC. Moreover, only one study explored the prognostic value of shelterin gene expression in early-stage tumors [17]. In patients with advanced NSCLC, telomere status has not yet been investigated and its clinical significance remains unknown. In the present study, we measured TL and the expression of TERT and shelterin genes in a cohort of 79 patients with advanced NSCLC, and evaluated these parameters as potential prognostic and predictive factors.
2. Materials and Methods
2.1. Patients and Samples
This retrospective study was performed on 79 patients with histologically proven NSCLC from the Pulmonology Department of the University Hospital of Clermont-Ferrand. For this cohort, we collected the data on demographic characteristics, performance and smoking status, histological diagnosis, mutation status, TNM stage, treatment modalities, tumor-response evaluation, and follow-up. Tumor tissue samples were obtained using fibroscopy or transparietal biopsy and snap frozen until molecular analysis. Each sample was analyzed using histology, for diagnostic purposes, which also enabled us to verify the adequate tumor-cell content (>30%).
2.2. DNA and RNA Extraction
DNA and RNA were simultaneously extracted from snap-frozen tumor biopsies with the AllPrep DNA/RNA/miRNA Universal Kit (Qiagen, Courtaboeuf, France), in accordance with the manufacturer’s instructions.
2.3. Assessment of Telomere Length Using Quantitative PCR
Average telomere length in tumor DNA was evaluated with quantitative real-time PCR in a LightCycler 480 System (Roche Diagnostics, Meylan, France) using SYBR Green I technology (SYBR Green Kit, Roche Diagnostics), as described elsewhere [18]. This method measures the template amounts of telomere repeat (T) and a reference single-copy gene (S), which are then used to determine the relative telomere length as the T/S ratio [19]. Glyceraldehyde-3-phosphate dehydrogenase gene was used as the reference gene. Each PCR series also included normal DNA control. The T/S ratio of tumor samples was normalized relative to the ratio obtained in the control DNA. The normalized T/S ratio of 1 corresponds to the average length of telomeres which is identical to that of the normal control DNA, whereas ratios T/S < 1 indicate different degrees of telomeric shortening.
2.4. Quantitative RT-PCR for TERT and Shelterin Complex GENE expression
Total RNA was converted to complementary DNA by reverse transcription, using Superscript II reverse transcriptase (Invitrogen, Cergy-Pontoise, France), in accordance with the manufacturer’s instructions. The expression of TERT and the shelterin complex genes TRF1, TRF2, POT1, TPP1, RAP1, and TIN2 were quantified using real-time RT-PCR in a LightCycler 480 System (Roche Diagnostics, Meylan, France), as described previously [18,20]. The normalized copy numbers (NCN) were expressed as the ratio of the numbers of transcript copies of the target and control (beta2-microglobulin) genes, multiplied by 100.
2.5. Statistical Analysis
Statistical analysis was performed using SEM software V1 [21]. The Student’s, Kruskal–Wallis, and Mann–Whitney tests were used for comparisons between the telomere data and clinical parameters. Event-free survival (EFS) was defined as the time between the date of diagnosis and the date of occurrence of the first event. The events were death from any cause, or clinical and/or radiological progression evaluated according to RECIST criteria [22]. Overall survival (OS) was defined as the time between the date of diagnosis and the date of death from any cause. Survival analysis was performed using the univariate Cox regression models, and survival curves were established in accordance with the Kaplan–Meier method and compared using the log-rank test. Multivariate Cox regression models were used to test the independent prognostic value of the telomere parameters. A sensitivity analysis was used to determine the best cutoff values associated with EFS and OS for the telomere parameters. To be precise, for each parameter value, EFS and OS were compared between cases that were below and above this value, using the Cox proportional hazards regression. The proportional hazard hypothesis was studied using Schoenfeld’s test. A sensitivity analysis was realized to find the best cutoff value for each parameter. An example of the best cutoff determination for TL is presented in Figure S1. An unsupervised hierarchical clustering was performed with SEM software [21]. Distances between clusters were calculated using 1-Pearson’s-correlation-coefficient values, and the dendrogram was constructed in accordance with to Ward’s algorithm.
3. Results
3.1. Patient Characteristics
The characteristics of the study cohort are presented in Table 1. Male smokers (active smokers or ex-smokers) represented the majority of the population. Adenocarcinoma was the most frequent type (58.2%) followed by squamous-cell carcinoma (31.7%). Eight patients had other histological types (six large-cell neuroendocrine carcinomas and two large-cell lung undifferentiated carcinomas). Two-thirds of NSCLC (70.8%) were metastatic at diagnosis. Thirteen patients had a KRAS-activating mutation, and four patients an EGFR mutation. Twenty-one patients with early NSCLC received primary local treatment with surgery (eleven patients), concomitant or sequential radiochemotherapy (eight patients) or radiotherapy alone (two patients). In advanced NSCLC, the first-line regimen was platinum-based chemotherapy (cisplatin or carboplatin) for fifty-two patients, and anti-EGFR tyrosine-kinase-inhibitor therapy for four patients with mutated EGFR.

Table 1.
Demographic and disease characteristics of the NSCLC cohort (n = 79).
3.2. Telomere Parameters in Early and Advanced NSCLC
We compared telomere characteristics at diagnosis between early (n = 21) and advanced NSCLC (n = 56) patients. TL was significantly shorter in advanced than in early NSCLC tumors (median T/S ratios: 0.4 vs. 0.59; p = 0.025). Expression levels of RAP1 and TPP1 were significantly lower in advanced NSCLC cases (median NCN: 11.65 vs. 30.74; p = 0.0020 and 33.7 vs. 106.3; p = 0.012, respectively). TIN2 and TRF1 tended to be more weakly expressed in advanced NSCLC cases (68.2 vs. 218.7; p = 0.054 and 49.5 vs. 177.6; p = 0.070, respectively). There was no significant difference in expression levels of TERT, TRF2, and POT1 between early and advanced NSCLC cases.
3.3. Association of Telomere Parameters with the Response to First-Line Therapy in Advanced NSCLC
Fifty-six patients with advanced NSCLC were treated with first-line chemotherapy. A scanner assessment of response was performed after three months of treatment. TL was significantly shorter (p = 0.0035, Figure 1a) and TERT expression tended to be higher (p = 0.058, Figure 1b) in cases with progressive disease, in comparison to patients who responded to therapy or had stable disease. The expression of all shelterin complex genes except TRF2 tended to be lower in the group of patients who progressed after first-line chemotherapy (Figure 1c–h). In particular, a very significant decrease was observed for RAP1 expression (p = 0.0069, Figure 1g).
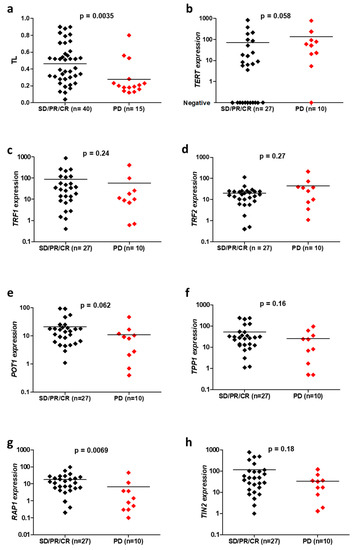
Figure 1.
Comparison of telomeric parameters, including telomere length (TL) (a), TERT (b), TRF1 (c), TRF2 (d), POT1 (e), TPP1 (f), RAP1 (g) and TIN2 (h) expressions, depending on the response to the first-line chemotherapy for advanced NSCLC, in accordance with RECIST 1.1 criteria: progressive disease (PD) versus stable disease (SD), partial response (PR) or complete response (CR).
3.4. Association of Telomere Parameters with Survival in Advanced NSCLC
For 56 patients who received first-line chemotherapy for advanced NSCLC, the associations of telomere parameters with EFS and OS were tested in univariate analyses, and the results are summarized in Table 2.

Table 2.
Univariate analyses for event-free and overall survival in the advanced NSCLC cohort.
Advanced NSCLC with short telomeres (<0.23) had significantly reduced EFS and OS, with a median EFS and OS of 4 months vs. 9 and 25 months, respectively, in the subgroup with longer telomeres (Figure 2a; p < 10−3). High TERT expression (>18.4) negatively affected EFS, with a median EFS and OS of 5 months compared with 10 and 32 months, respectively, in patients with low TERT expression (Figure 2b; p = 0.0018 and p = 0.00035). High tumor TRF2 expression (>29) was significantly associated with reduced survival, with a median EFS and OS of 2 and 4 months vs. 8 and 29 months, respectively, in cases showing low TRF2 expression (Figure 2c; p < 10−6 and p = 0.000071). Patients with low RAP1 expression in their tumors (<1.4) showed a median EFS and OS of 4 months, whereas the median EFS and OS were 8 and 32 months, respectively, when RAP1 expression was high (Figure 2d; p = 0.00023 and p = 0.0000018). Patients with low tumor TIN2 expression (<36) also had a significantly worse clinical evolution (an EFS of 5 and OS of 6 months) compared with patients with high TIN2 (Figure 2e; EFS of 11 and OS of 32 months; p = 0.015 and p = 0.0018).
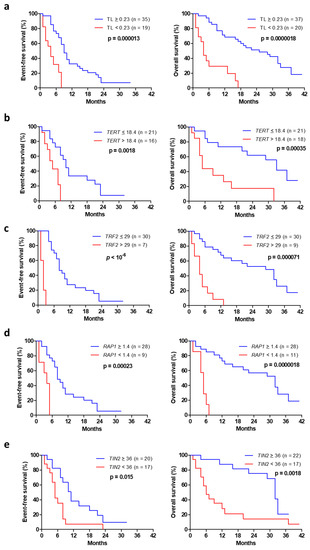
Figure 2.
Event-free (left) and overall (right) survival from the date of diagnosis in advanced NSCLC as a function of telomere length (a), TERT expression (b), TRF2 expression (c), RAP1 expression (d) and TIN2 expression (e).
A multivariate analysis, including age and performance status, indicated that short telomeres (<0.23), high TERT (>18.4), high TRF2 (>29) and low levels of RAP1 (<1.4) and TIN2 (<36) expression remained significant prognostic factors (Table 3).

Table 3.
Multivariate analyses for EFS and OS in the advanced NSCLC cohort.
In addition, an unsupervised hierarchical-clustering analysis was performed, to identify patient subgroups in accordance with the distribution of the telomeric markers (Figure S2 and Figure 3a). Survival analysis showed that the profiles of the telomeric biomarkers were highly correlated with EFS and OS (Figure 3b,c; p < 10−6). In particular, cluster #1 patients with negative telomeric markers had a longer EFS and OS (a median of 14 and 34 months). In cluster #2, the main features were low TIN2 and high TERT, and patients had an intermediate prognosis (an EFS of 7.4 months and OS of 16.7 months). In clusters #3 and #4, tumors had multiple telomeric markers, and patient survival was considerably shortened (an EFS of 4.3 and 2.2 months and an OS of 5.5 and 2.5 months, respectively).

Figure 3.
Telomeric markers combined together (a), statistically correlated with event-free (b) and overall survival (c).
3.5. Correlation of Telomere Length with Survival in Advanced NSCLC Treated with Immunotherapy
In advanced NSCLC, nivolumab was given after the failure of at least one prior platinum-based chemotherapy regimen. Most patients received nivolumab in the second-line setting. To assess the impact of TL on the survival of patients treated with nivolumab, OS was defined as death from any cause, and determined from the date of the first infusion of nivolumab. In advanced NSCLC treated with nivolumab, short telomeres (<0.23) negatively affected overall survival, with a median OS of 4 months vs. 12 months when TL was ≥0.23 (Figure 4: HR = 2.33 [0.89–5.88], p = 0.097).
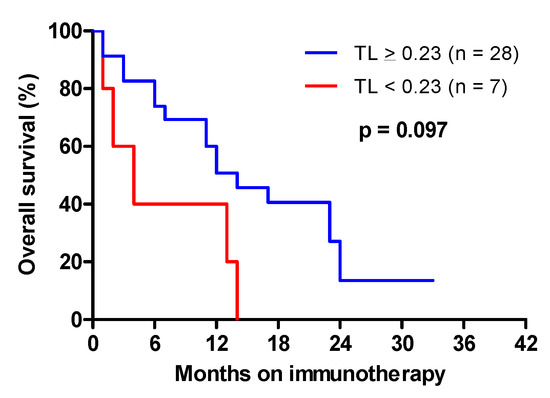
Figure 4.
Overall survival from the date of the initiation of immunotherapy in advanced NSCLC as a function of telomere length.
4. Discussion
Telomere attrition is one of the initiating events in lung cancerogenesis [23]. Subsequent activation of telomerase activity favors cancer-cell immortality and tumor progression. Short telomeres and the presence of telomerase activity in tumor tissue were significantly associated with reduced disease-free survival after curative surgery in early-stage NSCLC [15,16,24]. However, it is unknown whether telomere parameters may have prognostic value in advanced NSCLC. To address this issue, we assessed the impact of tumor telomere characteristics on treatment response and survival in advanced NSCLC patients, who represented the large majority of our cohort.
We found that telomeres were significantly shorter in advanced than in localized stages, which is in line with a previous report showing that telomeres were significantly shorter in the IIIB-IV stage than in the I-IIIA tumors [24]. We correlated telomere status with treatment response and survival for patients at advanced stages. At diagnosis, we identified approximately 35% of patients whose tumors displayed very short telomeres. In this sub-group, the response to first-line chemotherapy was considerably worse, and EFS, as well as OS, was significantly shorter than in patients with longer telomeres in their tumors. Thus, enhanced telomere shortening was strongly associated with tumor resistance and a worse prognosis in advanced NSCLC.
Among lung-cancer cases, 85% of tumors express telomerase independently of the disease stage [16]. We did not observe any significant difference in TERT expression levels between early and advanced cases. In early, resectable NSCLC, the presence of telomerase activity was shown to negatively affect survival after surgery [24,25]. In advanced NSCLC, we found that the EFS and OS of patients with higher TERT levels were significantly worse. Also, higher TERT expression was detected in patients who were resistant to first-line chemotherapy. Thus, telomerase overexpression is strongly associated with tumor aggressiveness in advanced NSCLC.
Expression levels of shelterin complex genes were previously studied in lung cancer, but only TRF1, TRF2, POT1, and RAP1 mRNA were measured in early NSCLC, and no consistent pattern of expression was identified [17,24,26]. We examined the expression of all six core members of shelterin in early and advanced disease, and found significantly lower expression of RAP1 and TPP1 and reduced expression of TIN2 and TRF1 in advanced NSCLC. No change was found for TRF2 and POT1 expression. Interestingly, RAP1 expression was significantly lower in patients who did not respond to chemotherapy. Moreover, we found that expression levels of TRF2, RAP1, and TIN2 were significantly correlated with the survival of patients with advanced NSCLC.
High TRF2 expression was significantly associated with shorter PFS and OS. TRF2 has been increasingly recognized as involved in telomere maintenance and DNA damage response (DDR) [27]. Short telomeres are perceived as double-strand DNA breaks and induce DDR mediated by p53/ATM (ataxia-telangiectasia mutated) proteins. TRF2 has been shown to inhibit the action of the ATM protein [28,29]. Increased expression of TRF2 in response to short telomeres is a factor of poor prognosis, because ATM-dependent apoptosis may be more strongly inhibited, and this allows the cell to survive and cancer to progress [30]. High expression of TRF2 is associated with poor prognosis in other cancers such as hepatocellular carcinomas or advanced-stage cervical cancers [31,32].
Low expression of RAP1 at diagnosis was a poor-prognostic factor, with reduced EFS and OS. RAP1 protects the telomere ends from the attacks by the DNA repair mechanisms similarly to other proteins of the shelterin complex, but by different mechanisms [33]. In particular, RAP1 plays an important role in the repression of aberrant homology-directed DNA repair at the telomeres, which prevents tumor cells from further telomere loss and genomic rearrangements [34]. Altered RAP1 expression has been found in multiple types of human cancer, including NSCLC [35,36,37,38]. In early NSCLC, low RAP1 expression has been previously correlated with short survival [17]. Our observation shows that the decreased RAP1 expression is correlated with the resistance to first-line chemotherapy and poor survival, in advanced NSCLC.
We found that low TIN2 expression was associated with reduced EFS and OS. To our knowledge, this is the first study showing the prognostic impact of low TIN2 in NSCLC. TIN2 binds to TRF1, TRF2, and TPP1, thus forming a bridge between the double-stranded telomeric DNA-related proteins and those related to single-stranded sequences [12]. The TRF1-TIN2 interaction is mediated by the TRFH domain and a pattern specific for the C-terminal region of TIN2, while the N-terminal region of TIN2 associates with the hinge domain of TRF2 [39]. When these interactions are simultaneous, TIN2 connects TRF1 to TRF2, and this link contributes to the stabilization of TRF2 on telomeres. TIN2 also recruits TPP1 (and thus POT1) via a third interaction site located in its N-terminal region [40]. TIN2 is a key component of the shelterin complex, and depletion of TIN2 has a powerful destabilizing effect [41]. TIN2 deficiency in aging mice was shown to lead to telomere fragility, accumulation of DNA damage at chromosomal ends and an enhanced lymphoma formation [42]. This mechanism may be relevant to the development of human malignancies. As compared to normal tissue, TIN2 transcription levels are lower in tumor tissue in breast cancer [43], gastric cancer [44] and chronic lymphocytic leukemia [20]. Low TIN2 expression was also shown to be a factor of poor prognosis in chronic lymphocytic leukemia [45,46,47].
Immune checkpoint inhibitors have changed the management of treatment of advanced NSCLC, with improved survival and better tolerance compared with standard chemotherapy. Nivolumab is a humanized monoclonal antibody against programmed death 1 (PD-1), approved as second-line treatment in patients with advanced NSCLC [48]. Current tests to predict the response to immunotherapy such as tumor PD-L1 overexpression or mutation burden have so far yielded inconsistent results [49]. We found that in patients treated with nivolumab, short telomeres negatively affected survival. To our knowledge, this is the first evidence that telomere length could be a prognostic factor and, possibly, a response predictor in advanced NSCLC treated with immunotherapy. Recently, genomic instability involving arm and whole-chromosome copy-number aberration has been proposed as a predictive biomarker for cancer immunotherapy [50]. Telomere dysfunctions can lead to the formation of cancer with a heavily rearranged genome [9]. Indeed, the unprotected chromosome ends generate end-to-end fusions and dicentric chromosomes, leading to many forms of genome instability, including global chromosomal copy-number alterations [51]. Therefore, the presence of short telomeres is consistent with enhanced genome instability of this particular type, which could lead to a poor response to nivolumab in advanced NSCLC.
Patients with newly diagnosed advanced NSCLC are currently tested for the presence of actionable mutations in tumor DNA [52]. In patients with no identified driver, RNA-based testing is considered, to search for druggable fusion genes. Including the telomeric biomarkers into the clinical-routine setting would be relatively easy, since the tests can be performed on very small amounts of already available DNA and RNA samples. The analytical performance of the tests has to be certified in accordance with medical laboratory quality-requirements. The tests can be recommended for all patients, to provide prognostic and predictive information. In addition, telomere targeting has been proposed for prolonging disease control of therapy-resistant NSCLC patients, and clinical benefits from telomerase inhibition seem to be greater in patients with tumors showing a more pronounced telomere dysfunction [53,54].
5. Conclusions
We have shown that short telomere-length, high levels of TERT and TRF2, and low expression of RAP1 and TIN2 are significantly associated with poor EFS and OS in advanced NSCLC. Moreover, enhanced telomere shortening in tumors could be a biomarker for worse survival in patients treated with nivolumab. Thus, evaluation of telomere status emerges as a useful molecular tool that allows for the selection of groups of NSCLC patients with different clinical evolutions, to establish personalized therapy protocols.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers15010290/s1, Figure S1: The best cutoff determination for telomere length values. Forest-plot for hazard ratios (HR) and 95% confidence intervals (95% CI) associated with the overall survival (OS) for each value of telomere length (between 0.17 and 0.39). This sensitivity analysis shows that the best cutoff of TL associated with the OS is 0.23. Figure S2: An unsupervised hierarchical clustering was performed by combining telomere length (TL) and TERT, TRF2, RAP1 and TIN2 gene expression as dichotomic parameters relative to best cutoff values used for survival analysis. Negative prognostic factors (short TL, high TERT and TRF2, low RAP1 and TIN2) are shown in red and their absence is indicated in green. The different color intensities of dichotomic parameters reflect normalized values and not original binary (0/1) values.
Author Contributions
Conceptualization, E.F., P.M. and A.T.; methodology, E.F., L.V. and A.T.; software, B.P.; validation, E.F., L.V., P.M. and A.T.; formal analysis, E.F., L.V., P.M. and A.T.; investigation, E.F., L.V., G.J., H.J., S.B., J.-O.B., B.P., A.C., F.P.-L., F.C., P.M. and A.T.; resources, E.F., B.P., P.M. and A.T.; data curation, E.F., B.P., P.M. and A.T.; writing—original draft preparation, E.F., B.P., P.M. and A.T.; writing—review and editing, E.F., L.V., G.J., H.J., S.B., J.-O.B., B.P., A.C., F.P.-L., F.C., P.M. and A.T. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was approved by the institutional review board (IRB00008526, Comité de Protection des Personnes SUD-EST VI, CHU Clermont-Ferrand).
Informed Consent Statement
This retrospective monocentric study was performed on samples remaining after routine diagnostic tests. Written informed consent was obtained from patients to use the residual samples for research purposes. All samples and data were anonymized.
Data Availability Statement
The data supporting reported results are available from the corresponding author on request.
Acknowledgments
The authors are grateful to Farida Godeau and Delphine Voisin for technical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the Global Cancer Incidence and Mortality in 2018: GLOBOCAN Sources and Methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef] [PubMed]
- Zito Marino, F.; Bianco, R.; Accardo, M.; Ronchi, A.; Cozzolino, I.; Morgillo, F.; Rossi, G.; Franco, R. Molecular Heterogeneity in Lung Cancer: From Mechanisms of Origin to Clinical Implications. Int. J. Med. Sci. 2019, 16, 981–989. [Google Scholar] [CrossRef] [PubMed]
- Borghaei, H.; Paz-Ares, L.; Horn, L.; Spigel, D.R.; Steins, M.; Ready, N.E.; Chow, L.Q.; Vokes, E.E.; Felip, E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 373, 1627–1639. [Google Scholar] [CrossRef]
- Brahmer, J.; Reckamp, K.L.; Baas, P.; Crinò, L.; Eberhardt, W.E.E.; Poddubskaya, E.; Antonia, S.; Pluzanski, A.; Vokes, E.E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 373, 123–135. [Google Scholar] [CrossRef]
- Ferrara, R.; Mezquita, L.; Texier, M.; Lahmar, J.; Audigier-Valette, C.; Tessonnier, L.; Mazieres, J.; Zalcman, G.; Brosseau, S.; Le Moulec, S.; et al. Hyperprogressive Disease in Patients with Advanced Non-Small Cell Lung Cancer Treated With PD-1/PD-L1 Inhibitors or With Single-Agent Chemotherapy. JAMA Oncol. 2018, 4, 1543–1552. [Google Scholar] [CrossRef] [PubMed]
- Bisoffi, M.; Heaphy, C.M.; Griffith, J.K. Telomeres: Prognostic Markers for Solid Tumors. Int. J. Cancer 2006, 119, 2255–2260. [Google Scholar] [CrossRef]
- Gaspar, T.B.; Sá, A.; Lopes, J.M.; Sobrinho-Simões, M.; Soares, P.; Vinagre, J. Telomere Maintenance Mechanisms in Cancer. Genes 2018, 9, 241. [Google Scholar] [CrossRef]
- Bernadotte, A.; Mikhelson, V.M.; Spivak, I.M. Markers of Cellular Senescence. Telomere Shortening as a Marker of Cellular Senescence. Aging 2016, 8, 3–11. [Google Scholar] [CrossRef]
- Maciejowski, J.; de Lange, T. Telomeres in Cancer: Tumour Suppression and Genome Instability. Nat. Rev. Mol. Cell Biol. 2017, 18, 175–186. [Google Scholar] [CrossRef]
- Kim, N.W.; Piatyszek, M.A.; Prowse, K.R.; Harley, C.B.; West, M.D.; Ho, P.L.; Coviello, G.M.; Wright, W.E.; Weinrich, S.L.; Shay, J.W. Specific Association of Human Telomerase Activity with Immortal Cells and Cancer. Science 1994, 266, 2011–2015. [Google Scholar] [CrossRef]
- Leão, R.; Apolónio, J.D.; Lee, D.; Figueiredo, A.; Tabori, U.; Castelo-Branco, P. Mechanisms of Human Telomerase Reverse Transcriptase (HTERT) Regulation: Clinical Impacts in Cancer. J. Biomed. Sci. 2018, 25, 22. [Google Scholar] [CrossRef] [PubMed]
- Palm, W.; de Lange, T. How Shelterin Protects Mammalian Telomeres. Annu. Rev. Genet. 2008, 42, 301–334. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Wang, R.-L.; Liu, J.-J.; Zhou, J.; Li, X.; Hu, W.-W.; Jiang, W.-J.; Hao, N.-B. The Prognostic Significance of HTERT Overexpression in Cancers: A Systematic Review and Meta-Analysis. Medicine 2018, 97, e11794. [Google Scholar] [CrossRef] [PubMed]
- Cookson, J.C.; Laughton, C.A. The Levels of Telomere-Binding Proteins in Human Tumours and Therapeutic Implications. Eur. J. Cancer 2009, 45, 536–550. [Google Scholar] [CrossRef]
- Jeon, H.-S.; Choi, Y.Y.; Choi, J.E.; Lee, W.K.; Lee, E.; Yoo, S.S.; Lee, S.Y.; Lee, J.; Cha, S.I.; Kim, C.H.; et al. Telomere Length of Tumor Tissues and Survival in Patients with Early Stage Non-Small Cell Lung Cancer. Mol. Carcinog. 2014, 53, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Marcelo, T.; Gómez, A.; Pascua, I.; de Juan, C.; Head, J.; Hernando, F.; Jarabo, J.-R.; Calatayud, J.; Torres-García, A.-J.; Iniesta, P. Telomere Length and Telomerase Activity in Non-Small Cell Lung Cancer Prognosis: Clinical Usefulness of a Specific Telomere Status. J. Exp. Clin. Cancer Res. 2015, 34, 78. [Google Scholar] [CrossRef]
- Lin, X.; Gu, J.; Lu, C.; Spitz, M.R.; Wu, X. Expression of Telomere-Associated Genes as Prognostic Markers for Overall Survival in Patients with Non-Small Cell Lung Cancer. Clin. Cancer Res. 2006, 12, 5720–5725. [Google Scholar] [CrossRef]
- Véronèse, L.; Tournilhac, O.; Callanan, M.; Prie, N.; Kwiatkowski, F.; Combes, P.; Chauvet, M.; Davi, F.; Gouas, L.; Verrelle, P.; et al. Telomeres and Chromosomal Instability in Chronic Lymphocytic Leukemia. Leukemia 2013, 27, 490–493. [Google Scholar] [CrossRef]
- Cawthon, R.M. Telomere Measurement by Quantitative PCR. Nucleic Acids Res. 2002, 30, e47. [Google Scholar] [CrossRef]
- Poncet, D.; Belleville, A.; t’kint de Roodenbeke, C.; Roborel de Climens, A.; Ben Simon, E.; Merle-Beral, H.; Callet-Bauchu, E.; Salles, G.; Sabatier, L.; Delic, J.; et al. Changes in the Expression of Telomere Maintenance Genes Suggest Global Telomere Dysfunction in B-Chronic Lymphocytic Leukemia. Blood 2008, 111, 2388–2391. [Google Scholar] [CrossRef]
- Kwiatkowski, F.; Girard, M.; Hacene, K.; Berlie, J. Sem: A suitable statistical software adaptated for research in oncology. Bull. Cancer 2000, 87, 715–721. [Google Scholar]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New Response Evaluation Criteria in Solid Tumours: Revised RECIST Guideline (Version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef] [PubMed]
- Lantuejoul, S.; Raynaud, C.; Salameire, D.; Gazzeri, S.; Moro-Sibilot, D.; Soria, J.-C.; Brambilla, C.; Brambilla, E. Telomere Maintenance and DNA Damage Responses during Lung Carcinogenesis. Clin. Cancer Res. 2010, 16, 2979–2988. [Google Scholar] [CrossRef] [PubMed]
- Frías, C.; García-Aranda, C.; De Juan, C.; Morán, A.; Ortega, P.; Gómez, A.; Hernando, F.; López-Asenjo, J.-A.; Torres, A.-J.; Benito, M.; et al. Telomere Shortening Is Associated with Poor Prognosis and Telomerase Activity Correlates with DNA Repair Impairment in Non-Small Cell Lung Cancer. Lung Cancer Amst. Neth. 2008, 60, 416–425. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.-Q.; Cutz, J.-C.; Liu, N.; Lau, D.; Shepherd, F.A.; Squire, J.A.; Tsao, M.-S. Amplification of Telomerase (HTERT) Gene Is a Poor Prognostic Marker in Non-Small-Cell Lung Cancer. Br. J. Cancer 2006, 94, 1452–1459. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.-P.; Ko, J.-L.; Shai, S.-E.; Lee, L.-W. Modulation of Telomere Shelterin by TRF1 and TRF2 Interacts with Telomerase to Maintain the Telomere Length in Non-Small Cell Lung Cancer. Lung Cancer Amst. Neth. 2007, 58, 310–316. [Google Scholar] [CrossRef]
- Rizzo, A.; Iachettini, S.; Salvati, E.; Zizza, P.; Maresca, C.; D’Angelo, C.; Benarroch-Popivker, D.; Capolupo, A.; Del Gaudio, F.; Cosconati, S.; et al. SIRT6 Interacts with TRF2 and Promotes Its Degradation in Response to DNA Damage. Nucleic Acids Res. 2017, 45, 1820–1834. [Google Scholar] [CrossRef]
- van Gent, D.C.; Hoeijmakers, J.H.; Kanaar, R. Chromosomal Stability and the DNA Double-Stranded Break Connection. Nat. Rev. Genet. 2001, 2, 196–206. [Google Scholar] [CrossRef]
- Khanna, K.K.; Jackson, S.P. DNA Double-Strand Breaks: Signaling, Repair and the Cancer Connection. Nat. Genet. 2001, 27, 247–254. [Google Scholar] [CrossRef]
- Lavin, M.F.; Kozlov, S. ATM Activation and DNA Damage Response. Cell Cycle 2007, 6, 931–942. [Google Scholar] [CrossRef]
- Kim, H.; Yoo, J.E.; Cho, J.Y.; Oh, B.-K.; Yoon, Y.-S.; Han, H.-S.; Lee, H.S.; Jang, J.J.; Jeong, S.H.; Kim, J.W.; et al. Telomere Length, TERT and Shelterin Complex Proteins in Hepatocellular Carcinomas Expressing “Stemness”-Related Markers. J. Hepatol. 2013, 59, 746–752. [Google Scholar] [CrossRef]
- Ozden, S.; Tiber, P.M.; Ozgen, Z.; Ozyurt, H.; Serakinci, N.; Orun, O. Expression of TRF2 and Its Prognostic Relevance in Advanced Stage Cervical Cancer Patients. Biol. Res. 2014, 47, 61. [Google Scholar] [CrossRef] [PubMed]
- Patel, T.N.; Vasan, R.; Gupta, D.; Patel, J.; Trivedi, M. Shelterin Proteins and Cancer. Asian Pac. J. Cancer Prev. 2015, 16, 3085–3090. [Google Scholar] [CrossRef]
- Rai, R.; Chen, Y.; Lei, M.; Chang, S. TRF2-RAP1 Is Required to Protect Telomeres from Engaging in Homologous Recombination-Mediated Deletions and Fusions. Nat. Commun. 2016, 7, 10881. [Google Scholar] [CrossRef] [PubMed]
- Braig, M.; Pällmann, N.; Preukschas, M.; Steinemann, D.; Hofmann, W.; Gompf, A.; Streichert, T.; Braunschweig, T.; Copland, M.; Rudolph, K.L.; et al. A ‘telomere-Associated Secretory Phenotype’ Cooperates with BCR-ABL to Drive Malignant Proliferation of Leukemic Cells. Leukemia 2014, 28, 2028–2039. [Google Scholar] [CrossRef]
- Aoude, L.G.; Pritchard, A.L.; Robles-Espinoza, C.D.; Wadt, K.; Harland, M.; Choi, J.; Gartside, M.; Quesada, V.; Johansson, P.; Palmer, J.M.; et al. Nonsense Mutations in the Shelterin Complex Genes ACD and TERF2IP in Familial Melanoma. J. Natl. Cancer Inst. 2015, 107, dju408. [Google Scholar] [CrossRef]
- Massion, P.P.; Zou, Y.; Chen, H.; Jiang, A.; Coulson, P.; Amos, C.I.; Wu, X.; Wistuba, I.; Wei, Q.; Shyr, Y.; et al. Smoking-Related Genomic Signatures in Non-Small Cell Lung Cancer. Am. J. Respir. Crit. Care Med. 2008, 178, 1164–1172. [Google Scholar] [CrossRef]
- Sweet-Cordero, A.; Tseng, G.C.; You, H.; Douglass, M.; Huey, B.; Albertson, D.; Jacks, T. Comparison of Gene Expression and DNA Copy Number Changes in a Murine Model of Lung Cancer. Genes. Chromosomes Cancer 2006, 45, 338–348. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, Y.; van Overbeek, M.; Donigian, J.R.; Baciu, P.; de Lange, T.; Lei, M. A Shared Docking Motif in TRF1 and TRF2 Used for Differential Recruitment of Telomeric Proteins. Science 2008, 319, 1092–1096. [Google Scholar] [CrossRef]
- Frescas, D.; de Lange, T. TRF2-Tethered TIN2 Can Mediate Telomere Protection by TPP1/POT1. Mol. Cell. Biol. 2014, 34, 1349–1362. [Google Scholar] [CrossRef]
- Ye, J.Z.-S.; Donigian, J.R.; van Overbeek, M.; Loayza, D.; Luo, Y.; Krutchinsky, A.N.; Chait, B.T.; de Lange, T. TIN2 Binds TRF1 and TRF2 Simultaneously and Stabilizes the TRF2 Complex on Telomeres. J. Biol. Chem. 2004, 279, 47264–47271. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, K.; Illing, A.; Leithäuser, F.; Baisantry, A.; Quintanilla-Martinez, L.; Rudolph, K.L. Gene Dosage Reductions of Trf1 and/or Tin2 Induce Telomere DNA Damage and Lymphoma Formation in Aging Mice. Leukemia 2016, 30, 749–753. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Salhab, M.; Jiang, W.G.; Newbold, R.F.; Mokbel, K. The Expression of Gene Transcripts of Telomere-Associated Genes in Human Breast Cancer: Correlation with Clinico-Pathological Parameters and Clinical Outcome. Breast Cancer Res. Treat. 2008, 109, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M.; Tsuji, N.; Nakamura, M.; Moriai, R.; Kobayashi, D.; Yagihashi, A.; Watanabe, N. Down-Regulation of TRF1, TRF2 and TIN2 Genes Is Important to Maintain Telomeric DNA for Gastric Cancers. Anticancer Res. 2002, 22, 3303–3307. [Google Scholar]
- Guièze, R.; Pages, M.; Véronèse, L.; Combes, P.; Lemal, R.; Gay-Bellile, M.; Chauvet, M.; Callanan, M.; Kwiatkowski, F.; Pereira, B.; et al. Telomere Status in Chronic Lymphocytic Leukemia with TP53 Disruption. Oncotarget 2016, 7, 56976–56985. [Google Scholar] [CrossRef][Green Version]
- Augereau, A.; T’kint de Roodenbeke, C.; Simonet, T.; Bauwens, S.; Horard, B.; Callanan, M.; Leroux, D.; Jallades, L.; Salles, G.; Gilson, E.; et al. Telomeric Damage in Early Stage of Chronic Lymphocytic Leukemia Correlates with Shelterin Dysregulation. Blood 2011, 118, 1316–1322. [Google Scholar] [CrossRef]
- Ishdorj, G.; Kost, S.E.F.; Beiggi, S.; Zang, Y.; Gibson, S.B.; Johnston, J.B. A Novel Spliced Variant of the TIN2 Shelterin Is Present in Chronic Lymphocytic Leukemia. Leuk. Res. 2017, 59, 66–74. [Google Scholar] [CrossRef]
- Somasundaram, A.; Socinski, M.A.; Villaruz, L.C. Immune Checkpoint Blockade in Lung Cancer. Discov. Med. 2016, 22, 55–65. [Google Scholar]
- Bodor, J.N.; Boumber, Y.; Borghaei, H. Biomarkers for Immune Checkpoint Inhibition in Non–Small Cell Lung Cancer (NSCLC). Cancer 2020, 126, 260–270. [Google Scholar] [CrossRef]
- Davoli, T.; Uno, H.; Wooten, E.C.; Elledge, S.J. Tumor Aneuploidy Correlates with Markers of Immune Evasion and with Reduced Response to Immunotherapy. Science 2017, 355, eaaf8399. [Google Scholar] [CrossRef]
- Roger, L.; Jones, R.E.; Heppel, N.H.; Williams, G.T.; Sampson, J.R.; Baird, D.M. Extensive Telomere Erosion in the Initiation of Colorectal Adenomas and Its Association with Chromosomal Instability. J. Natl. Cancer Inst. 2013, 105, 1202–1211. [Google Scholar] [CrossRef] [PubMed]
- Aisner, D.L.; Riely, G.J. Non–Small Cell Lung Cancer: Recommendations for Biomarker Testing and Treatment. J. Natl. Compr. Cancer Netw. 2021, 19, 610–613. Available online: https://jnccn.org/configurable/content/journals$002fjnccn$002f19$002f5.5$002farticlep610.xml?t:ac=journals%24002fjnccn%24002f19%24002f5.5%24002farticle-p610.xml (accessed on 14 December 2022). [CrossRef]
- Chiappori, A.A.; Kolevska, T.; Spigel, D.R.; Hager, S.; Rarick, M.; Gadgeel, S.; Blais, N.; Von Pawel, J.; Hart, L.; Reck, M.; et al. A Randomized Phase II Study of the Telomerase Inhibitor Imetelstat as Maintenance Therapy for Advanced Non-Small-Cell Lung Cancer. Ann. Oncol. 2015, 26, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Mender, I.; LaRanger, R.; Luitel, K.; Peyton, M.; Girard, L.; Lai, T.-P.; Batten, K.; Cornelius, C.; Dalvi, M.P.; Ramirez, M.; et al. Telomerase-Mediated Strategy for Overcoming Non-Small Cell Lung Cancer Targeted Therapy and Chemotherapy Resistance. Neoplasia 2018, 20, 826–837. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).