A Survey of Naturally Occurring Molecules as New Endoplasmic Reticulum Stress Activators with Selective Anticancer Activity
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemical Library and Reagents
2.2. Cell Culture Conditions
2.3. Monolayer and 3D Cell Viability Assays
2.4. Evaluation of the Mitochondrial Membrane Potential (ΔΨm)
2.5. RNA Sample Preparation, Conversion to cDNA and RT-qPCR Analysis
2.6. Cytosolic Ca2+ Level Determination
2.7. Caspase-3 and -4 Activities
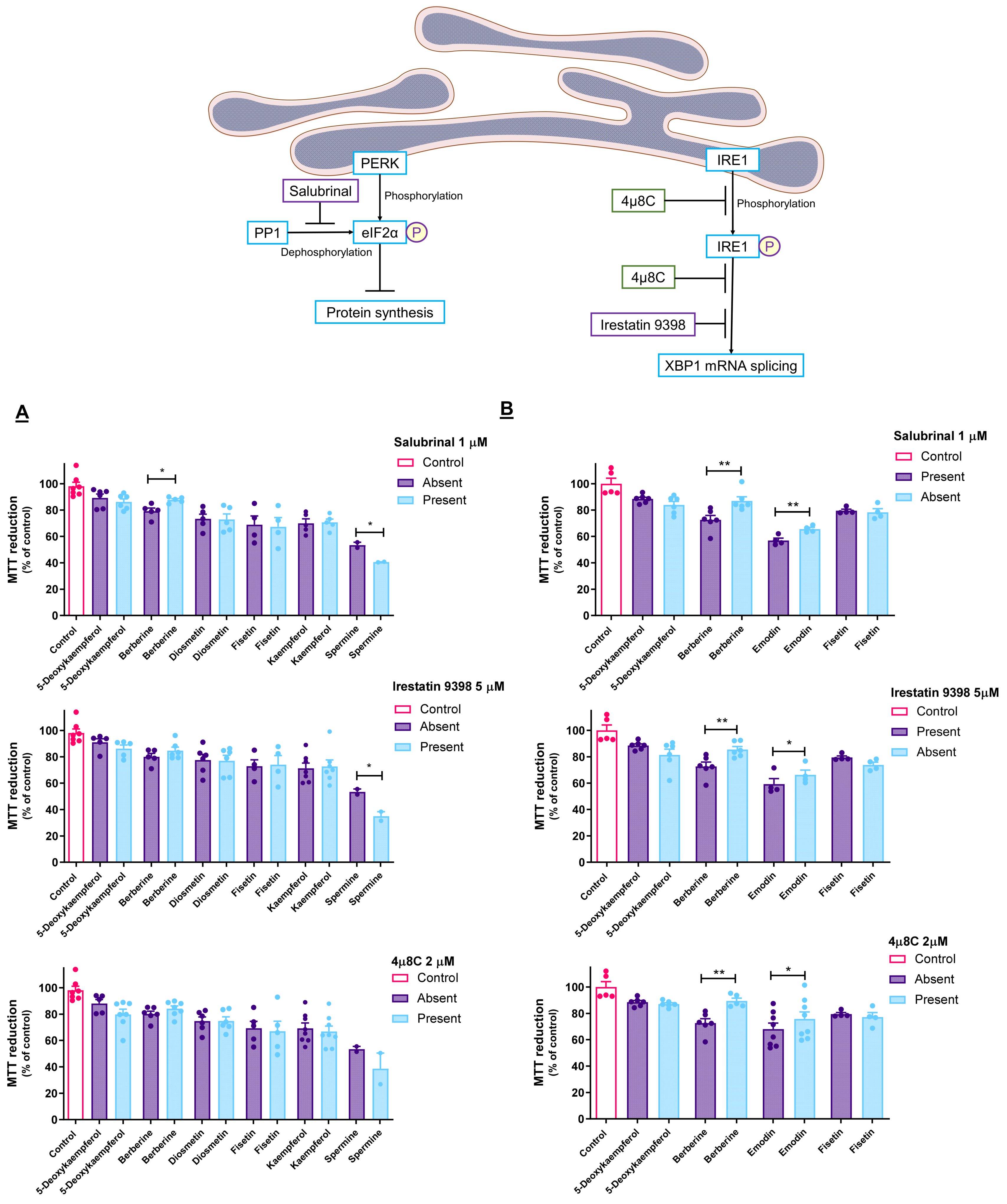
2.8. Study of the Involvement of the PERK and IRE-1 Branches of the UPR on Cell Viability
2.9. Statistical Analysis
3. Results
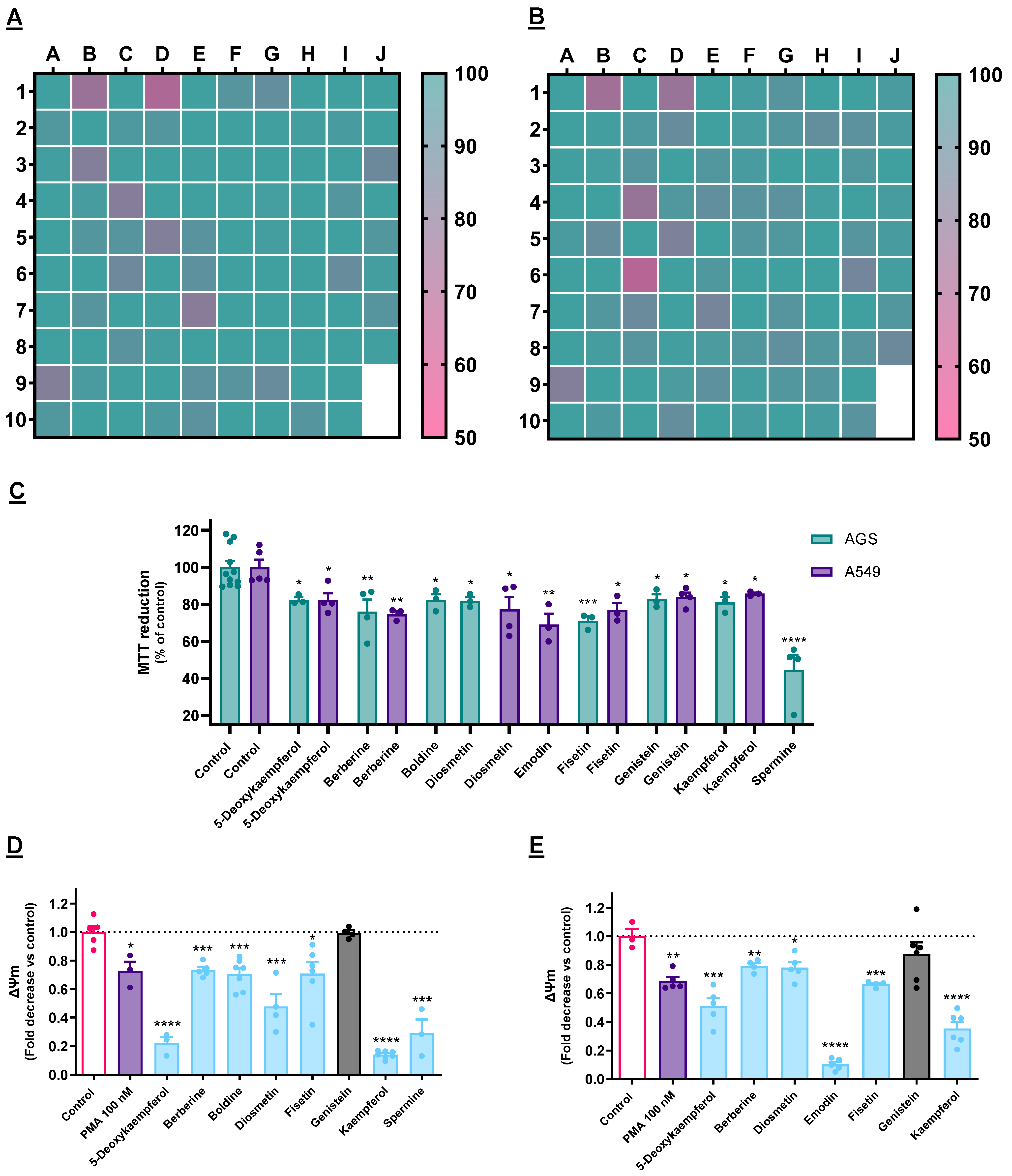
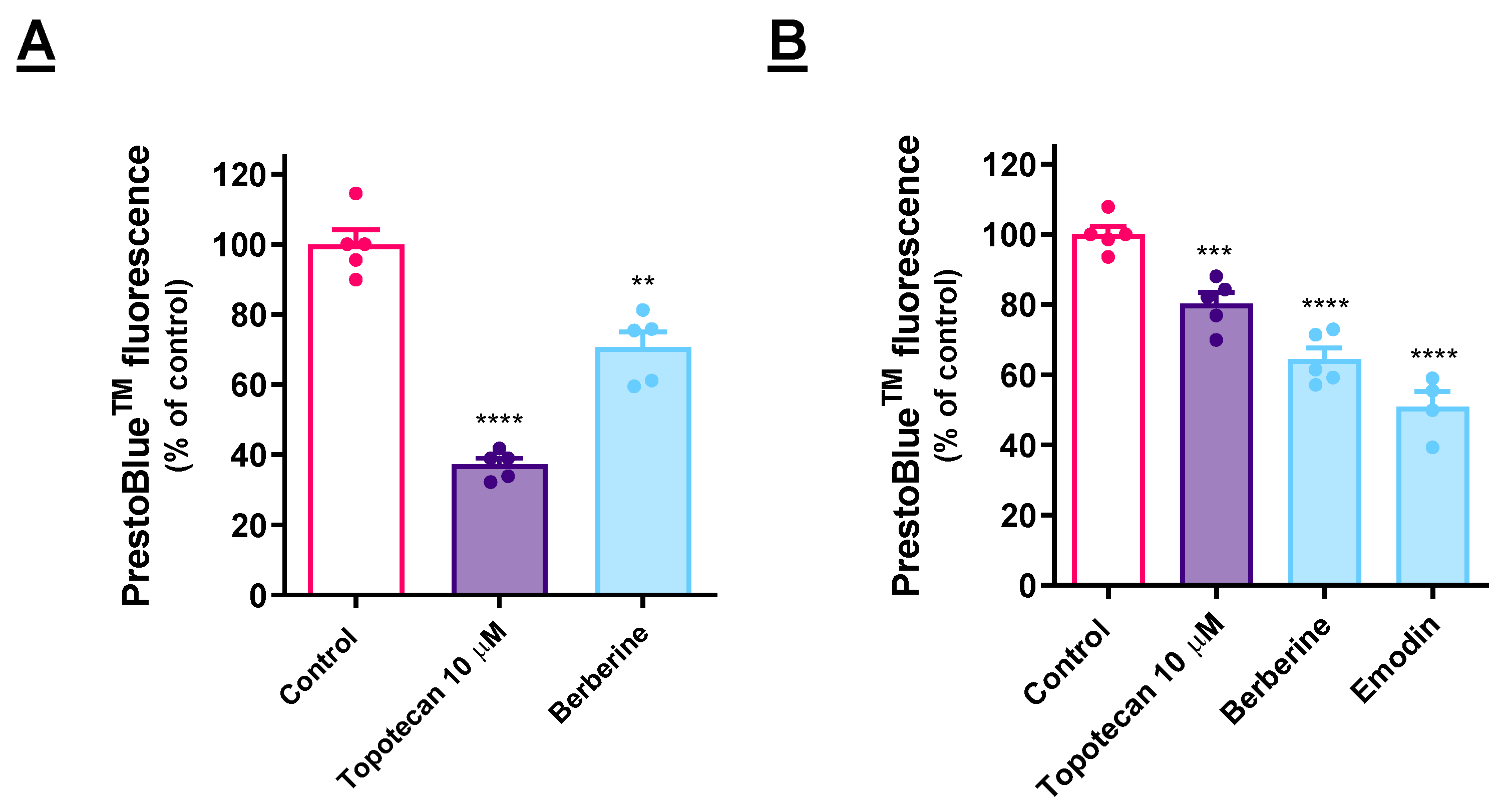
3.1. Cytotoxicity Evaluation
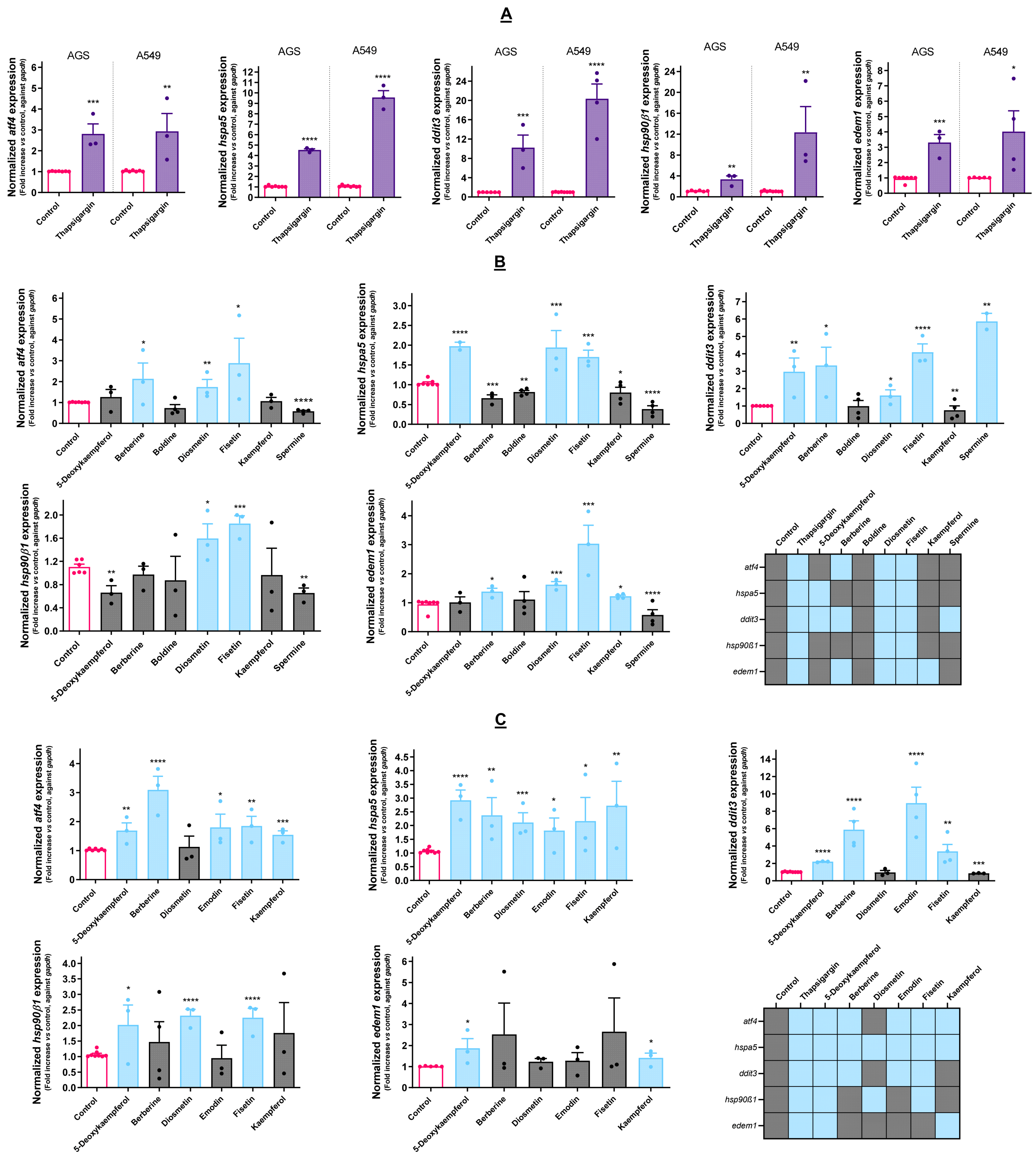
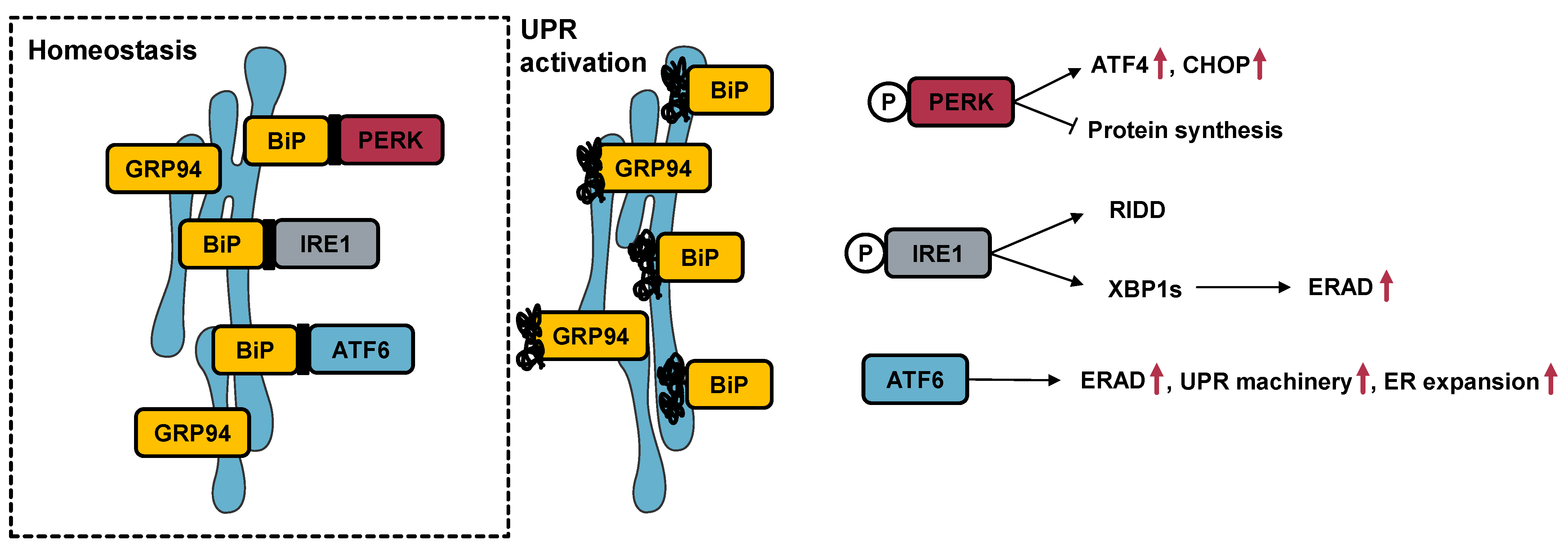
3.2. Effect of Cytotoxic Molecules on UPR Activation
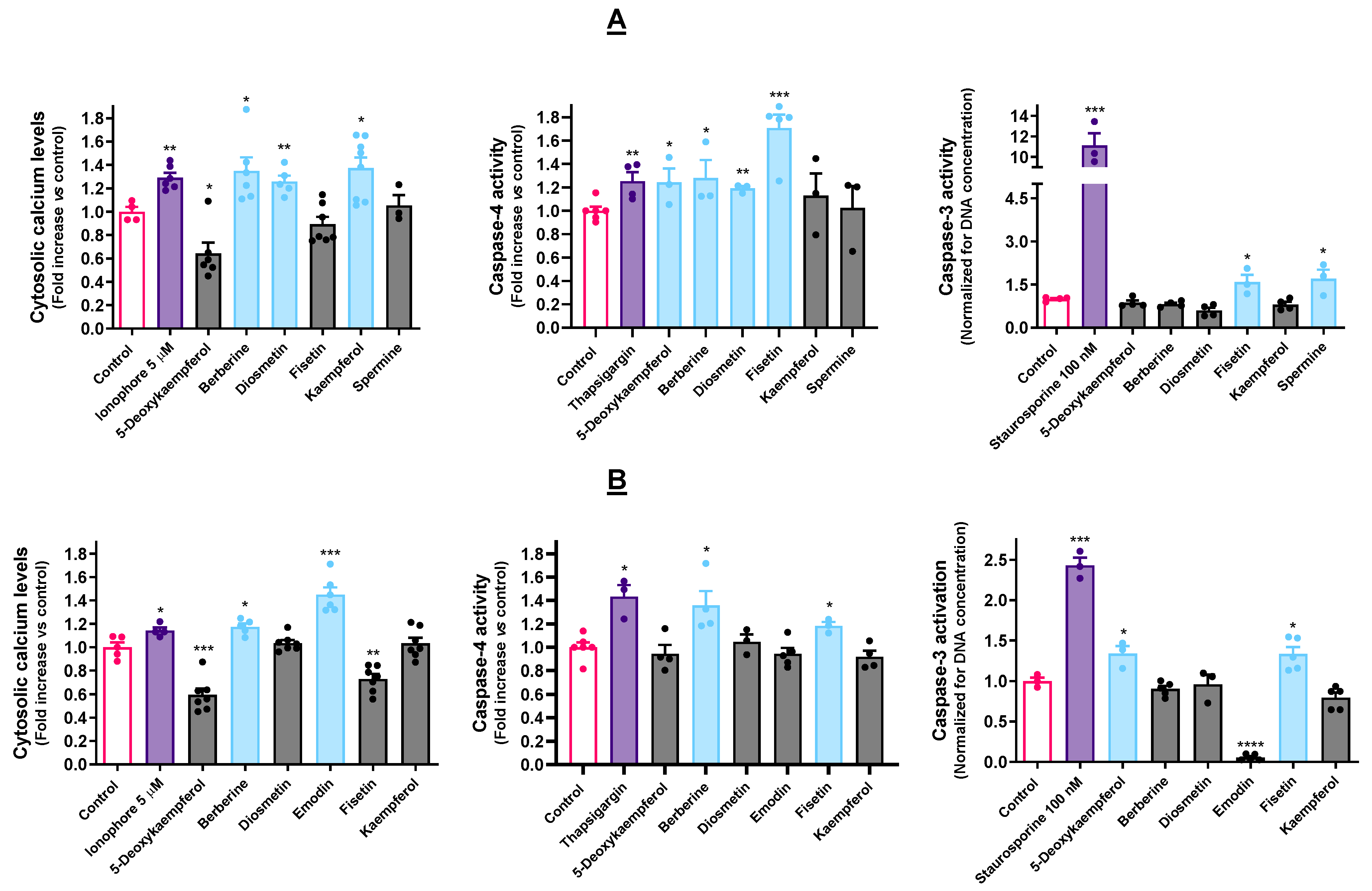
3.3. Selected Molecules Impact Ca2+ Homeostasis and Caspase Activity
3.4. Pharmacological Modulation of UPR Impacts Cytotoxicity of ER Stressors
3.5. Cytotoxic Effect of ER Stressors Is Retained in 3D Models
4. Discussion
4.1. Selection of Cytotoxic Molecules towards Cancer Cells
4.2. The Selected Molecules Are Capable of Triggering UPR Activation
4.3. ER Stressors Disturb Ca2+ Homeostasis and Elicit Caspase Activation
4.4. Cytotoxic Molecules Disturb ER Homeostasis at Different Levels
4.5. Cytotoxic Effect of ER Stressors Is Retained in 3D Models
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Miller, K.D.; Nogueira, L.; Mariotto, A.B.; Rowland, J.H.; Yabroff, K.R.; Alfano, C.M.; Jemal, A.; Kramer, J.L.; Siegel, R.L. Cancer treatment and survivorship statistics, 2019. CA Cancer J. Clin. 2019, 69, 363–385. [Google Scholar] [CrossRef] [PubMed]
- Global Burden of Disease Cancer, C.; Fitzmaurice, C.; Dicker, D.; Pain, A.; Hamavid, H.; Moradi-Lakeh, M.; MacIntyre, M.F.; Allen, C.; Hansen, G.; Woodbrook, R.; et al. The Global Burden of Cancer 2013. JAMA Oncol. 2015, 1, 505–527. [Google Scholar]
- Bravo, R.; Parra, V.; Gatica, D.; Rodriguez, A.E.; Torrealba, N.; Paredes, F.; Wang, Z.V.; Zorzano, A.; Hill, J.A.; Jaimovich, E.; et al. Endoplasmic reticulum and the unfolded protein response: Dynamics and metabolic integration. Int. Rev. Cell Mol. Biol. 2013, 301, 215–290. [Google Scholar] [PubMed]
- Bravo-Sagua, R.; Rodriguez, A.E.; Kuzmicic, J.; Gutierrez, T.; Lopez-Crisosto, C.; Quiroga, C.; Díaz-Elizondo, J.; Chiong, M.; Gillette, T.G.; Rothermel, B.A.; et al. Cell death and survival through the endoplasmic reticulum-mitochondrial axis. Curr. Mol. Med. 2013, 13, 317–329. [Google Scholar] [CrossRef]
- Hetz, C. The unfolded protein response: Controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 2012, 13, 89–102. [Google Scholar] [CrossRef]
- da Silva, D.C.; Valentão, P.; Andrade, P.B.; Pereira, D.M. Endoplasmic reticulum stress signaling in cancer and neurodegenerative disorders: Tools and strategies to understand its complexity. Pharmacol. Res. 2020, 155, 104702. [Google Scholar] [CrossRef]
- Wang, M.; Kaufman, R.J. The impact of the endoplasmic reticulum protein-folding environment on cancer development. Nat. Rev. Cancer 2014, 14, 581–597. [Google Scholar] [CrossRef]
- Kim, C.; Kim, B. Anti-cancer natural products and their bioactive compounds inducing ER stress-mediated apoptosis: A review. Nutrients 2018, 10, 1021. [Google Scholar] [CrossRef]
- King, A.P.; Wilson, J.J. Endoplasmic reticulum stress: An arising target for metal-based anticancer agents. Chem. Soc. Rev. 2020, 49, 8113–8136. [Google Scholar] [CrossRef]
- Pereira, D.M.; Valentao, P.; Correia-da-Silva, G.; Teixeira, N.; Andrade, P.B. Translating endoplasmic reticulum biology into the clinic: A role for ER-targeted natural products? Nat. Prod. Rep. 2015, 32, 705–722. [Google Scholar] [CrossRef]
- Cheimonidi, C.; Samara, P.; Polychronopoulos, P.; Tsakiri, E.N.; Nikou, T.; Myrianthopoulos, V.; Sakellaropoulos, T.; Zoumpourlis, V.; Mikros, E.; Papassideri, I.; et al. Selective cytotoxicity of the herbal substance acteoside against tumor cells and its mechanistic insights. Redox Biol. 2018, 16, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, V.; Andrade, P.B.; Valentão, P.; Pereira, D.M. Benzoquinones from Cyperus spp. trigger IRE1α-independent and PERK-dependent ER stress in human stomach cancer cells and are novel proteasome inhibitors. Phytomedicine 2019, 63, 153017. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Evans, P.A. A concise, efficient and scalable total synthesis of thapsigargin and nortrilobolide from (R)-(−)-carvone. J. Am. Chem. Soc. 2017, 139, 6046–6049. [Google Scholar] [CrossRef] [PubMed]
- Lytton, J.; Westlin, M.; Hanley, M.R. Thapsigargin inhibits the sarcoplasmic or endoplasmic reticulum Ca-ATPase family of calcium pumps. J. Biol. Chem. 1991, 266, 17067–17071. [Google Scholar] [CrossRef]
- Thastrup, O.; Cullen, P.J.; Drøbak, B.K.; Hanley, M.R.; Dawson, A.P. Thapsigargin, a tumor promoter, discharges intracellular Ca2+ stores by specific inhibition of the endoplasmic reticulum Ca2(+)-ATPase. Proc. Natl. Acad. Sci. USA 1990, 87, 2466–2470. [Google Scholar] [CrossRef]
- Atta, H.M. Biochemical studies on antibiotic production from Streptomyces sp.: Taxonomy, fermentation, isolation and biological properties. J. Saudi Chem. Soc. 2015, 19, 12–22. [Google Scholar] [CrossRef]
- Wang, H.; Wang, X.; Ke, Z.-J.; Comer, A.L.; Xu, M.; Frank, J.A.; Zhang, Z.; Shi, X.; Luo, J. Tunicamycin-induced unfolded protein response in the developing mouse brain. Toxicol. Appl. Pharmacol. 2015, 283, 157–167. [Google Scholar] [CrossRef]
- Hamada, J.; Onuma, H.; Ochi, F.; Hirai, H.; Takemoto, K.; Miyoshi, A.; Matsushita, M.; Kadota, Y.; Ohashi, J.; Kawamura, R.; et al. Endoplasmic reticulum stress induced by tunicamycin increases resistin messenger ribonucleic acid through the pancreatic endoplasmic reticulum eukaryotic initiation factor 2α kinase–activating transcription factor 4–CAAT/enhancer binding protein-α homologous protein pathway in THP-1 human monocytes. J. Diabetes Investig. 2016, 7, 312–323. [Google Scholar]
- Berning, L.; Scharf, L.; Aplak, E.; Stucki, D.; von Montfort, C.; Reichert, A.S.; Stahl, W.; Brenneisen, P. In vitro selective cytotoxicity of the dietary chalcone cardamonin (CD) on melanoma compared to healthy cells is mediated by apoptosis. PLoS ONE 2019, 14, e0222267. [Google Scholar] [CrossRef]
- Kim, J.H.; Jung, C.H.; Jang, B.-H.; Go, H.Y.; Park, J.-H.; Choi, Y.-K.; Hong, S.I.; Shin, Y.C.; Ko, S.-G. Selective cytotoxic effects on human cancer cell lines of phenolic-rich ethyl-acetate fraction from Rhus verniciflua stokes. Am. J. Chin. Med. 2009, 37, 609–620. [Google Scholar] [CrossRef]
- Oakes, S.A. Endoplasmic reticulum stress signaling in cancer cells. Am. J. Pathol. 2020, 190, 934–946. [Google Scholar] [CrossRef] [PubMed]
- Oakes, S.A. Endoplasmic reticulum proteostasis: A key checkpoint in cancer. Am. J. Physiol. Cell Physiol. 2017, 312, C93–C102. [Google Scholar] [CrossRef] [PubMed]
- Correia da Silva, D.; Valentão, P.; Andrade, P.B.; Pereira, D.M. A pipeline for natural small molecule inhibitors of endoplasmic reticulum stress. Front. Pharmacol. 2022, 13, 956154. [Google Scholar] [CrossRef]
- Correia da Silva, D.; Andrade, P.B.; Valentão, P.; Pereira, D.M. Neurotoxicity of the steroidal alkaloids tomatine and tomatidine is RIP1 kinase- and caspase-independent and involves the eIF2α branch of the endoplasmic reticulum. J. Steroid Biochem. Mol. Biol. 2017, 171, 178–186. [Google Scholar] [CrossRef]
- Wang, Y.; Biswas, G.; Prabu, S.K.; Avadhani, N.G. Modulation of mitochondrial metabolic function by phorbol 12-myristate 13-acetate through increased mitochondrial translocation of protein kinase Cα in C2C12 myocytes. Biochem. Pharmacol. 2006, 72, 881–892. [Google Scholar] [CrossRef]
- Badisa, R.B.; Darling-Reed, S.F.; Joseph, P.; Cooperwood, J.S.; Latinwo, L.M.; Goodman, C.B. Selective cytotoxic activities of two novel synthetic drugs on human breast carcinoma MCF-7 cells. Anticancer Res. 2009, 29, 2993–2996. [Google Scholar] [PubMed]
- Gupta, S.; Cuffe, L.; Szegezdi, E.; Logue, S.E.; Neary, C.; Healy, S.; Samali, A. Mechanisms of ER stress-mediated mitochondrial membrane permeabilization. Int. J. Cell Biol. 2010, 2010, 170215. [Google Scholar] [CrossRef]
- Rzymski, T.; Milani, M.; Pike, L.; Buffa, F.; Mellor, H.R.; Winchester, L.; Pires, I.; Hammond, E.; Ragoussis, I.; Harris, A.L. Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene 2010, 29, 4424–4435. [Google Scholar] [CrossRef]
- Hu, H.; Tian, M.; Ding, C.; Yu, S. The C/EBP homologous protein (CHOP) transcription factor functions in endoplasmic reticulum stress-induced apoptosis and microbial infection. Front. Immunol. 2019, 9, 3083. [Google Scholar] [CrossRef]
- Hetz, C.; Zhang, K.; Kaufman, R.J. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell Biol. 2020, 21, 421–438. [Google Scholar] [CrossRef]
- Marzec, M.; Eletto, D.; Argon, Y. GRP94: An HSP90-like protein specialized for protein folding and quality control in the endoplasmic reticulum. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2012, 1823, 774–787. [Google Scholar] [CrossRef] [PubMed]
- Kim, R.S.; Hasegawa, D.; Goossens, N.; Tsuchida, T.; Athwal, V.; Sun, X.; Robinson, C.L.; Bhattacharya, D.; Chou, H.-I.; Zhang, D.Y.; et al. The XBP1 arm of the unfolded protein response induces fibrogenic activity in hepatic stellate cells through autophagy. Sci. Rep. 2016, 6, 39342. [Google Scholar] [CrossRef] [PubMed]
- Määttänen, P.; Gehring, K.; Bergeron, J.J.M.; Thomas, D.Y. Protein quality control in the ER: The recognition of misfolded proteins. Semin. Cell Dev. Biol. 2010, 21, 500–511. [Google Scholar] [CrossRef] [PubMed]
- van Ziel, A.M.; Scheper, W. The UPR in neurodegenerative disease: Not just an inside job. Biomolecules 2020, 10, 1090. [Google Scholar] [CrossRef]
- Krebs, J.; Agellon, L.B.; Michalak, M. Ca2+ homeostasis and endoplasmic reticulum (ER) stress: An integrated view of calcium signaling. Biochem. Biophys. Res. Commun. 2015, 460, 114–121. [Google Scholar] [CrossRef]
- Kraus, A.; Michalak, M. Endoplasmic Reticulum Dynamics and Calcium Signaling. In New Comprehensive Biochemistry; Krebs, J., Michalak, M., Eds.; Elsevier: Amsterdam, The Netherlands, 2007; Volume 41. [Google Scholar]
- Duchen, M.R. Mitochondria and calcium: From cell signalling to cell death. J. Physiol. 2000, 529 Pt 1, 57–68. [Google Scholar] [CrossRef]
- Srivastava, S.P.; Davies, M.V.; Kaufman, R.J. Calcium depletion from the endoplasmic reticulum activates the double-stranded RNA-dependent protein kinase (PKR) to inhibit protein synthesis. J. Biol. Chem. 1995, 270, 16619–16624. [Google Scholar] [CrossRef]
- Li, Y.; Yan, Y.; Liu, F.; Wang, M.; Feng, F.; Xiao, Y. Effects of calcium ionophore A23187 on the apoptosis of hepatic stellate cells stimulated by transforming growth factor-β(1). Cell. Mol. Biol. Lett. 2018, 23, 1–9. [Google Scholar] [CrossRef]
- Boya, P.; Cohen, I.; Zamzami, N.; Vieira, H.L.A.; Kroemer, G. Endoplasmic reticulum stress-induced cell death requires mitochondrial membrane permeabilization. Cell Death Differ. 2002, 9, 465–467. [Google Scholar] [CrossRef]
- Hitomi, J.; Katayama, T.; Eguchi, Y.; Kudo, T.; Taniguchi, M.; Koyama, Y.; Manabe, T.; Yamagishi, S.; Bando, Y.; Imaizumi, K.; et al. Involvement of caspase-4 in endoplasmic reticulum stress-induced apoptosis and AB-induced cell death. J. Cell Biol. 2004, 165, 347–356. [Google Scholar] [CrossRef]
- Momoi, T. Caspases involved in ER stress-mediated cell death. J. Chem. Neuroanat. 2004, 28, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Obeng, E.A.; Boise, L.H. Caspase-12 and caspase-4 are not required for caspase-dependent endoplasmic reticulum stress-induced apoptosis. J. Biol. Chem. 2005, 280, 29578–29587. [Google Scholar] [CrossRef]
- Stepczynska, A.; Lauber, K.; Engels, I.H.; Janssen, O.; Kabelitz, D.; Wesselborg, S.; Schulze-Osthoff, K. Staurosporine and conventional anticancer drugs induce overlapping, yet distinct pathways of apoptosis and caspase activation. Oncogene 2001, 20, 1193–1202. [Google Scholar] [CrossRef] [PubMed]
- Musaogullari, A.; Mandato, A.; Chai, Y.-C. Role of glutathione depletion and reactive oxygen species generation on caspase-3 activation: A study with the kinase inhibitor staurosporine. Front. Physiol. 2020, 11, 998. [Google Scholar] [CrossRef]
- Yang, R.-Q.; Guo, P.-F.; Ma, Z.; Chang, C.; Meng, Q.-N.; Gao, Y.; Khan, I.; Wang, X.-B.; Cui, Z.-J. Effects of simvastatin on iNOS and caspase-3 levels and oxidative stress following smoke inhalation injury. Mol. Med. Rep. 2020, 22, 3405–3417. [Google Scholar] [CrossRef] [PubMed]
- Egger, L.; Madden, D.T.; Rhême, C.; Rao, R.V.; Bredesen, D.E. Endoplasmic reticulum stress-induced cell death mediated by the proteasome. Cell Death Differ. 2007, 14, 1172–1180. [Google Scholar] [CrossRef]
- Mekahli, D.; Bultynck, G.; Parys, J.B.; De Smedt, H.; Missiaen, L. Endoplasmic reticulum calcium depletion and disease. Cold Spring Harb. Perspect. Biol. 2011, 3, a004317. [Google Scholar] [CrossRef] [PubMed]
- Rozpędek, W.; Pytel, D.; Mucha, B.; Leszczyńska, H.; Diehl, J.A.; Majsterek, I. The role of the PERK/eIF2α/ATF4/CHOP signaling pathway in tumor progression during endoplasmic reticulum stress. Curr. Mol. Med. 2016, 16, 533–544. [Google Scholar] [CrossRef]
- Matsuoka, M.; Komoike, Y. Experimental evidence shows salubrinal, an eIF2α dephosphorylation inhibitor, reduces xenotoxicant-induced cellular damage. Int. J. Mol. Sci. 2015, 16, 16275–16287. [Google Scholar] [CrossRef]
- Maurel, M.; Chevet, E.; Tavernier, J.; Gerlo, S. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem. Sci. 2014, 39, 245–254. [Google Scholar] [CrossRef]
- Cross, B.C.S.; Bond, P.J.; Sadowski, P.G.; Jha, B.K.; Zak, J.; Goodman, J.M.; Silverman, R.H.; Neubert, T.A.; Baxendale, I.R.; Ron, D.; et al. The molecular basis for selective inhibition of unconventional mRNA splicing by an IRE1-binding small molecule. Proc. Natl. Acad. Sci. USA 2012, 109, E869–E878. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Niwa, M.; Koong, A.C. Targeting the IRE1α-XBP1 branch of the unfolded protein response in human diseases. Semin. Cancer Biol. 2015, 33, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Feldman, D.; Koong, A.C. Irestatin, a potent inhibitor of IRE1α and the unfolded protein response, is a hypoxia-selective cytotoxin and impairs tumor growth. J. Clin. Oncol. 2007, 25 (18_suppl), 3514. [Google Scholar] [CrossRef]
- Ron, D.; Walter, P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 2007, 8, 519–529. [Google Scholar] [CrossRef]
- Vandewynckel, Y.-P.; Laukens, D.; Geerts, A.; Bogaerts, E.; Paridaens, A.; Verhelst, X.; Janssens, S.; Heindryckx, F.; Van Vlierberghe, H. The paradox of the unfolded protein response in cancer. Anticancer Res. 2013, 33, 4683–4694. [Google Scholar] [PubMed]
- Feldman, D.E.; Chauhan, V.; Koong, A.C. The unfolded protein response: A novel component of the hypoxic stress response in tumors. Mol. Cancer Res. 2005, 3, 597–605. [Google Scholar] [CrossRef]
- Fang, Y.; Eglen, R.M. Three-Dimensional Cell Cultures in Drug Discovery and Development. SLAS Discov. Adv. Sci. Drug Discov. 2017, 22, 456–472. [Google Scholar] [CrossRef]
- Sun, T.; Jackson, S.; Haycock, J.W.; MacNeil, S. Culture of skin cells in 3D rather than 2D improves their ability to survive exposure to cytotoxic agents. J. Biotechnol. 2006, 122, 372–381. [Google Scholar] [CrossRef]
- Edmondson, R.; Broglie, J.J.; Adcock, A.F.; Yang, L. Three-dimensional cell culture systems and their applications in drug discovery and cell-based biosensors. Assay Drug Dev. Technol. 2014, 12, 207–218. [Google Scholar] [CrossRef]
- Fennema, E.; Rivron, N.; Rouwkema, J.; van Blitterswijk, C.; de Boer, J. Spheroid culture as a tool for creating 3D complex tissues. Trends Biotechnol. 2013, 31, 108–115. [Google Scholar] [CrossRef]
- Lv, D.; Hu, Z.; Lu, L.; Lu, H.; Xu, X. Three-dimensional cell culture: A powerful tool in tumor research and drug discovery. Oncol. Lett. 2017, 14, 6999–7010. [Google Scholar] [CrossRef] [PubMed]
| Gene | Accession Number | Primers | Annealing Temperature (°C) | Amplicon Length (bp) |
|---|---|---|---|---|
| GAPDH (GAPDH) | NM_002046.6 | F: AGGTCGGAGTCAACGGATTT | 60 | 157 |
| R: TGGAATTTGCCATGGGTGGA | ||||
| HSPA5 (GRP78) | NM_005347.4 | F: ACTCCTGAAGGGGAACGTCT | 59.5 | 161 |
| R: TTTTCAACCACCTTGAACGGC | ||||
| DDIT33 (CHOP) | NM_001195053.1 | F: AAGTCTAAGGCACTGAGCGT | 59 | 93 |
| R: TTGAACACTCTCTCCTCAGGT | ||||
| HSP90β1 (GRP94) | NM_003299.2 | F: GCTCTATGTGCGCCGTGTAT | 60.5 | 91 |
| R: ATCTGAGTCCACCACACCCTT | ||||
| ATF4 (ATF4) | NM_001675.4 | F: ACAACAGCAAGGAGGATGCC | 60 | 135 |
| R: CCAACGTGGTCAGAAGGTCA | ||||
| EDEM1 (EDEM1) | NM_014674.2 | F: GCGGGGACCCTTCAAATCT | 60 | 117 |
| R: CGGCTTTCTGGAACTCGGAT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Correia da Silva, D.; Valentão, P.; Pereira, D.M. A Survey of Naturally Occurring Molecules as New Endoplasmic Reticulum Stress Activators with Selective Anticancer Activity. Cancers 2023, 15, 293. https://doi.org/10.3390/cancers15010293
Correia da Silva D, Valentão P, Pereira DM. A Survey of Naturally Occurring Molecules as New Endoplasmic Reticulum Stress Activators with Selective Anticancer Activity. Cancers. 2023; 15(1):293. https://doi.org/10.3390/cancers15010293
Chicago/Turabian StyleCorreia da Silva, Daniela, Patrícia Valentão, and David M. Pereira. 2023. "A Survey of Naturally Occurring Molecules as New Endoplasmic Reticulum Stress Activators with Selective Anticancer Activity" Cancers 15, no. 1: 293. https://doi.org/10.3390/cancers15010293
APA StyleCorreia da Silva, D., Valentão, P., & Pereira, D. M. (2023). A Survey of Naturally Occurring Molecules as New Endoplasmic Reticulum Stress Activators with Selective Anticancer Activity. Cancers, 15(1), 293. https://doi.org/10.3390/cancers15010293









