Honeycomb-like Structured Film, a Novel Therapeutic Device, Suppresses Tumor Growth in an In Vivo Ovarian Cancer Model
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Cultures
2.2. Preparation of HCFs
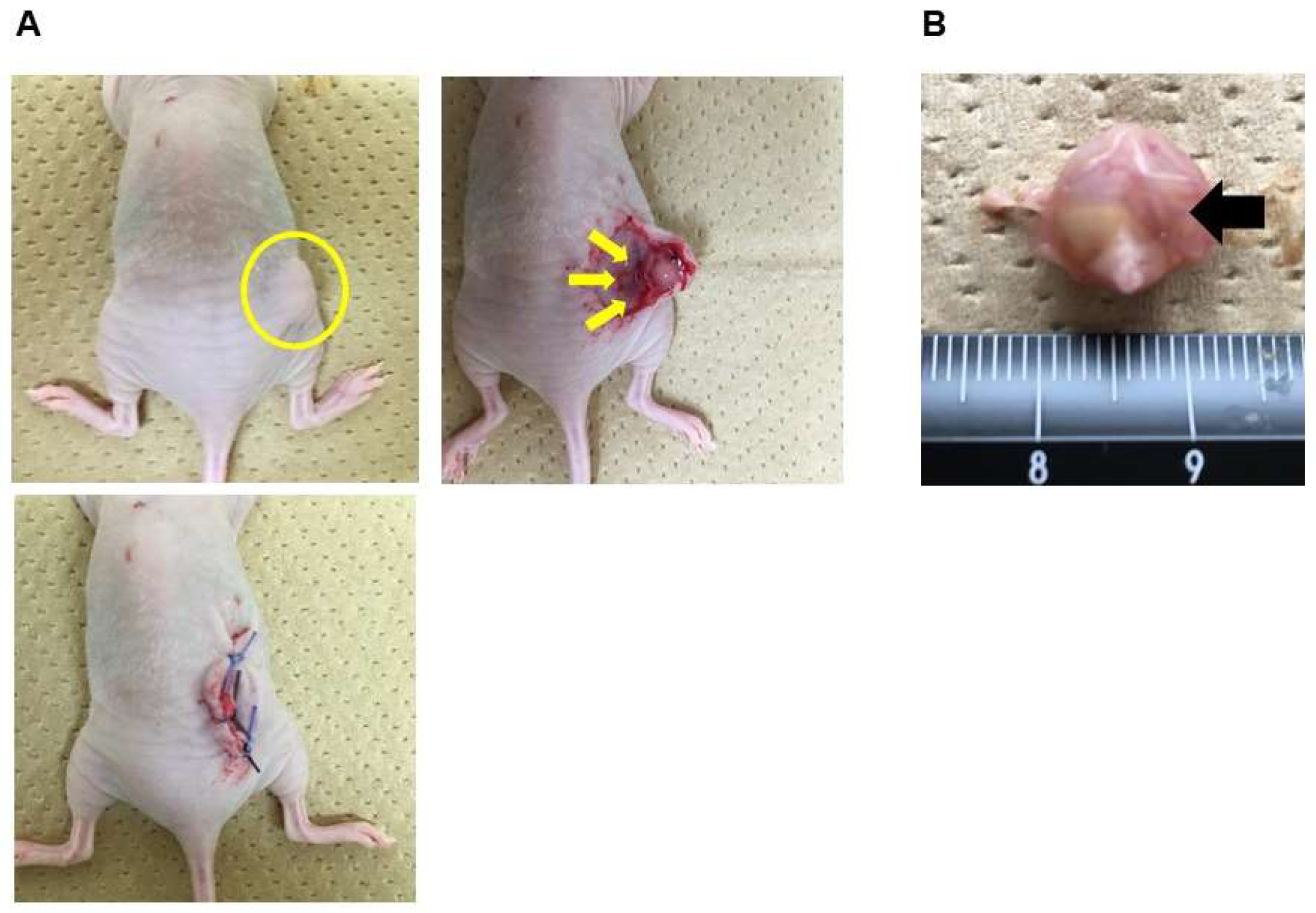
2.3. Subcutaneous Xenograft Model and Treatment of HCFs in vivo
2.4. RNA Sequencing and RNA-Sequencing Data Analysis
2.5. Real-Time PCR Analysis
2.6. Scanning Electron Microscopy (SEM) Observation
2.7. Confocal Laser-Scanning Microscopy (CLSM) Observation
2.8. Statistical Analysis
3. Results
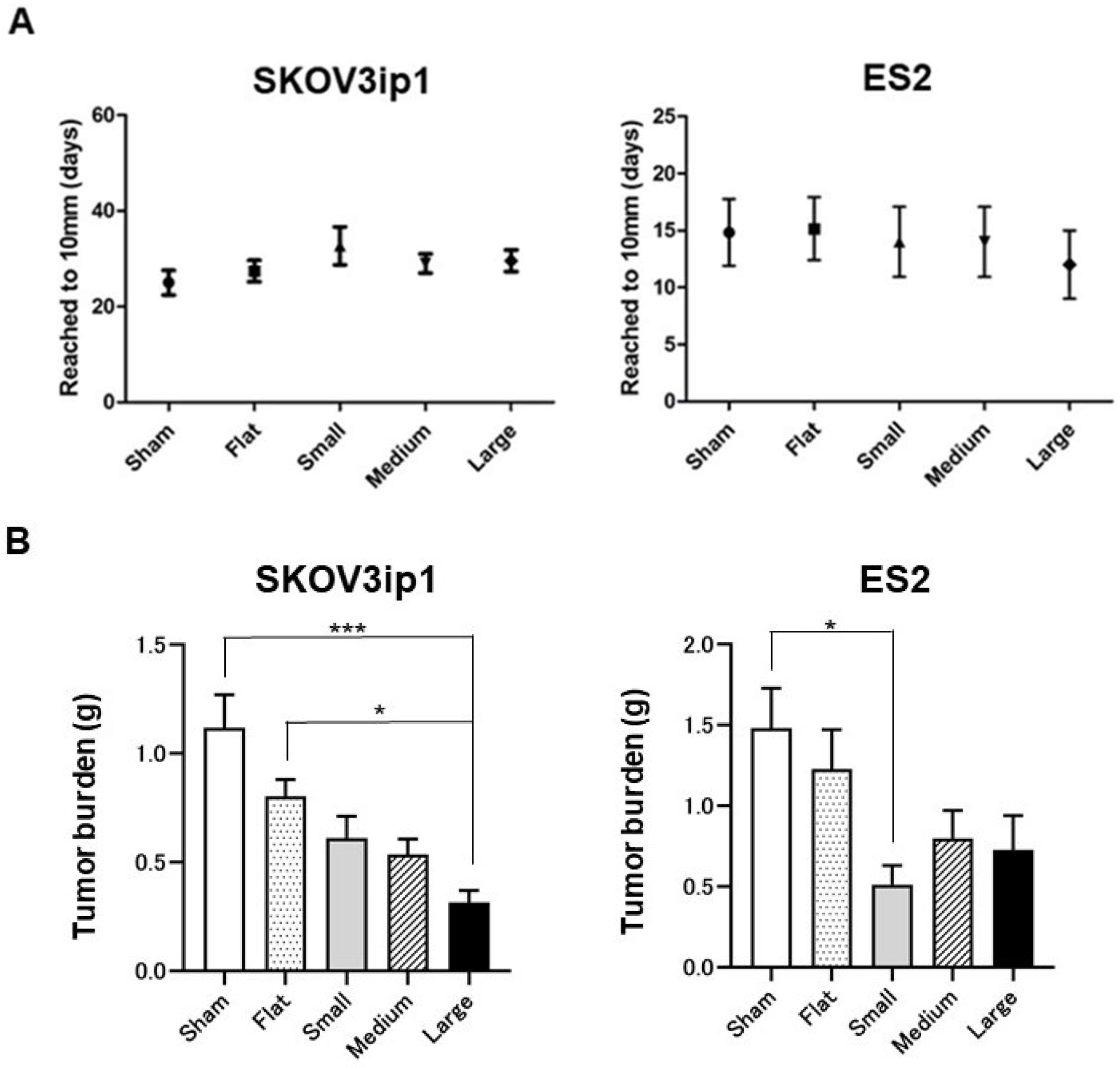
3.1. Effects of HCFs on Tumor Growth in Ovarian Cancer
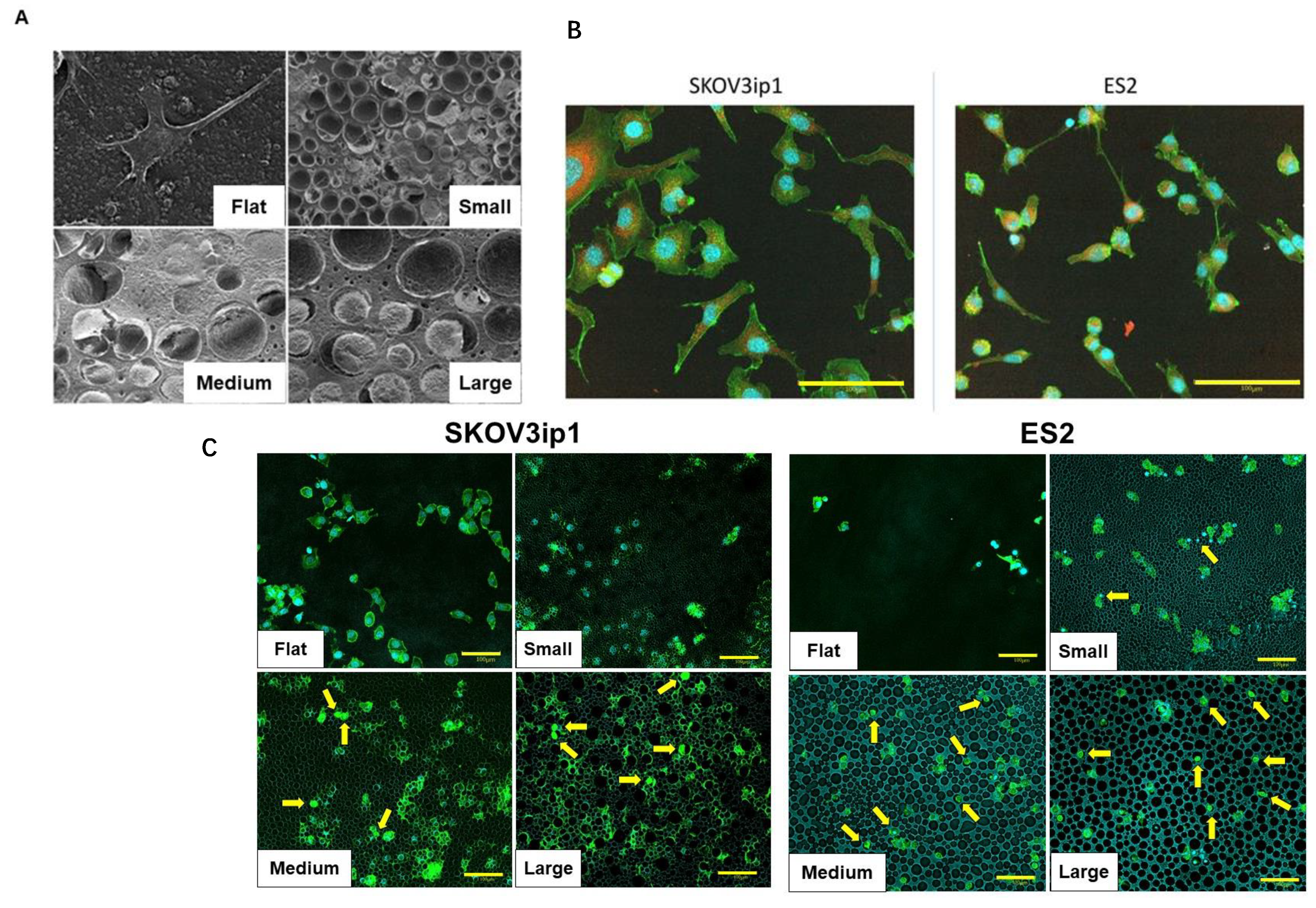
3.2. Morphological Changes in Ovarian Cancer Cells Cultured on HCFs
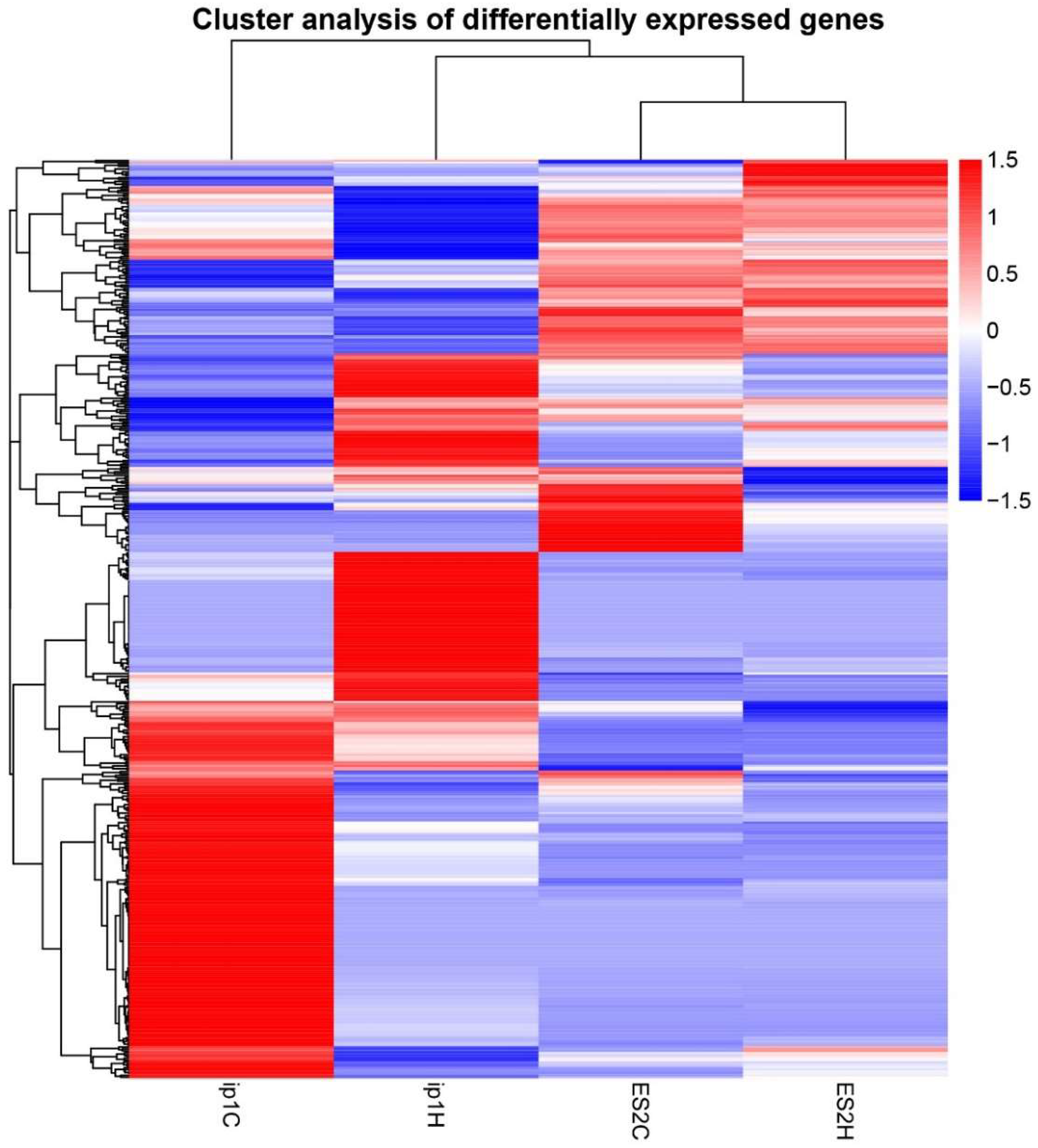
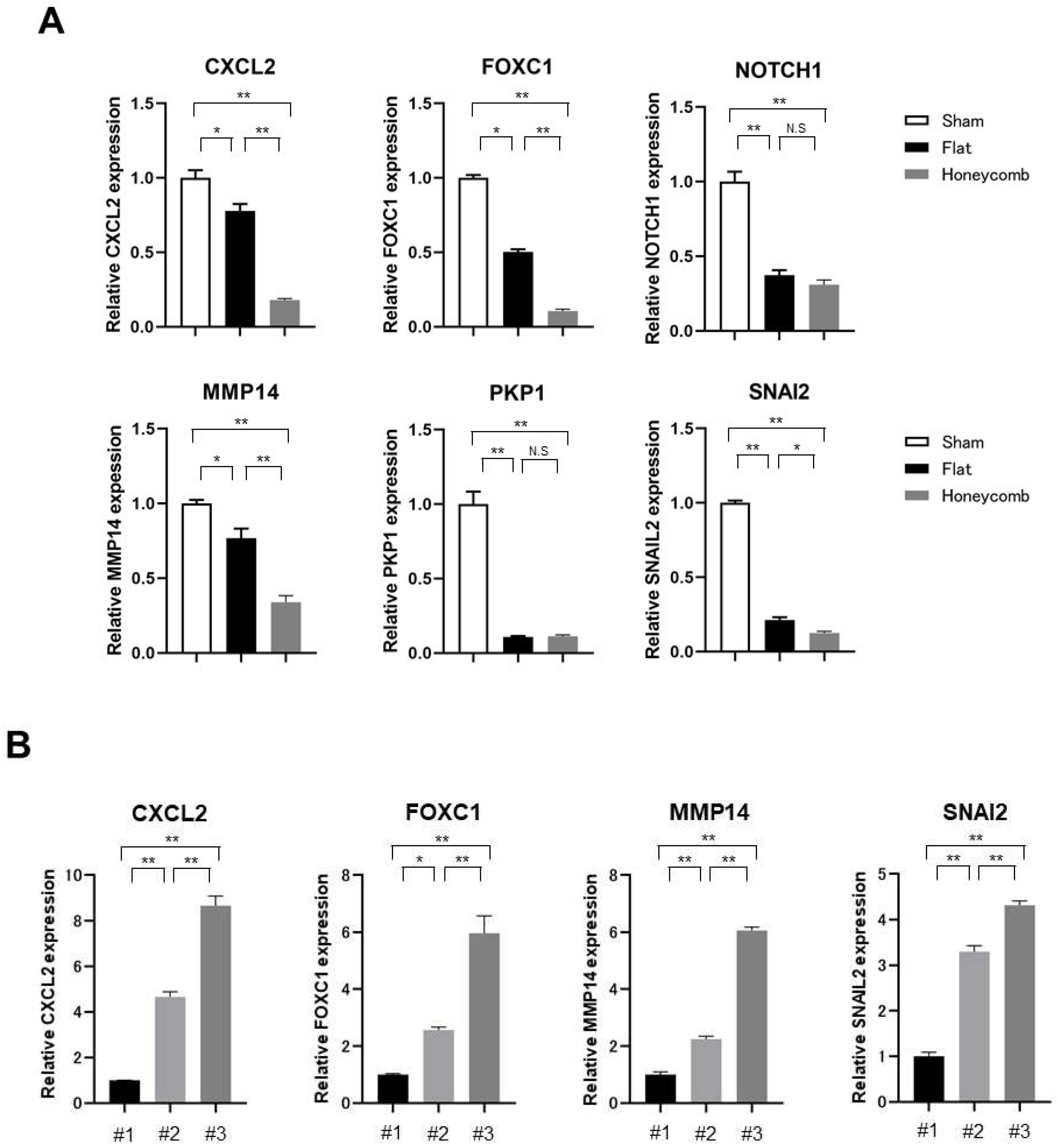
3.3. The Mechanism of Tumorigenic Inhibitory Effect by HCFs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- The Surveillance, Epidemiology, and End Results (SEER) Program. Available online: https://seer.cancer.gov/statfacts/html/ovary.html (accessed on 29 July 2022).
- Lheureux, S.; Braunstein, M.; Oza, A.M. Epithelial ovarian cancer: Evolution of management in the era of precision medicine. CA Cancer J. Clin. 2019, 69, 280–304. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.J.; Hodeib, M.; Chang, J.; Bristow, R.E. Survival impact of complete cytoreduction to no gross residual disease for advanced-stage ovarian cancer: A meta-analysis. Gynecol. Oncol. 2013, 130, 493–498. [Google Scholar] [CrossRef] [PubMed]
- Wallace, S.; Kumar, A.; Mc Gree, M.; Weaver, A.; Mariani, A.; Langstraat, C.; Dowdy, S.; Bakkum-Gamez, J.; Cliby, W. Efforts at maximal cytoreduction improve survival in ovarian cancer patients, even when complete gross resection is not feasible. Gynecol. Oncol. 2017, 145, 21–26. [Google Scholar] [CrossRef]
- Derlatka, P.; Sienko, J.; Grabowska-Derlatka, L.; Palczewski, P.; Danska-Bidzinska, A.; Bidzinski, M.; Czajkowski, K. Results of optimal debulking surgery with bowel resection in patients with advanced ovarian cancer. World J. Surg. Oncol. 2016, 14, 58. [Google Scholar] [CrossRef] [PubMed]
- Giorda, G.; Gadducci, A.; Lucia, E.; Sorio, R.; Bounous, V.E.; Sopracordevole, F.; Tinelli, A.; Baldassarre, G.; Campagnutta, E. Prognostic role of bowel involvement in optimally cytoreduced advanced ovarian cancer: A retrospective study. J. Ovarian Res. 2014, 7, 72. [Google Scholar] [CrossRef] [PubMed]
- Dowdy, S.C.; Loewen, R.T.; Aletti, G.; Feitoza, S.S.; Cliby, W. Assessment of outcomes and morbidity following diaphragmatic peritonectomy for women with ovarian carcinoma. Gynecol. Oncol. 2008, 109, 303–307. [Google Scholar] [CrossRef] [PubMed]
- Eisenkop, S.M.; Spirtos, N.M.; Lin, W.C. Splenectomy in the context of primary cytoreductive operations for advanced epithelial ovarian cancer. Gynecol. Oncol. 2006, 100, 344–348. [Google Scholar] [CrossRef]
- Greggi, S.; Falcone, F.; Carputo, R.; Raspagliesi, F.; Scaffa, C.; Laurelli, G.; Giorda, G.; Di Vagno, G.; Petruzzelli, F.; Cormio, G.; et al. Primary surgical cytoreduction in advanced ovarian cancer: An outcome analysis within the MITO (Multicentre Italian Trials in Ovarian Cancer and Gynecologic Malignancies) Group. Gynecol. Oncol. 2016, 140, 425–429. [Google Scholar] [CrossRef]
- Xu, Z.; Becerra, A.Z.; Justiniano, C.F.; Aquina, C.T.; Fleming, F.J.; Boscoe, F.P.; Schymura, M.J.; Sinno, A.K.; Chaoul, J.; Morrow, G.R.; et al. Complications and Survivorship Trends after Primary Debulking Surgery for Ovarian Cancer. J. Surg. Res. 2020, 246, 34–41. [Google Scholar] [CrossRef]
- Soo Hoo, S.; Marriott, N.; Houlton, A.; Nevin, J.; Balega, J.; Singh, K.; Yap, J.; Sethuram, R.; Elattar, A.; Luesley, D.; et al. Patient-Reported Outcomes After Extensive (Ultraradical) Surgery for Ovarian Cancer: Results from a Prospective Longitudinal Feasibility Study. Int. J. Gynecol. Cancer 2015, 25, 1599–1607. [Google Scholar] [CrossRef]
- Hollister, S.J. Porous scaffold design for tissue engineering. Nat. Mater. 2005, 4, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Takebayashi, M.; Miyama, M.; Nishida, J.; Shimomura, M. Design of novel biointerfaces (II). Fabrication of self-organized porous polymer film with highly uniform pores. Biomed. Mater. Eng. 2004, 14, 439–446. [Google Scholar] [PubMed]
- Dalby, M.J.; Gadegaard, N.; Tare, R.; Andar, A.; Riehle, M.O.; Herzyk, P.; Wilkinson, C.D.; Oreffo, R.O. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat. Mater. 2007, 6, 997–1003. [Google Scholar] [CrossRef]
- Tanaka, M.; Takayama, A.; Ito, E.; Sunami, H.; Yamamoto, S.; Shimomura, M. Effect of pore size of self-organized honeycomb-patterned polymer films on spreading, focal adhesion, proliferation, and function of endothelial cells. J. Nanosci. Nanotechnol. 2007, 7, 763–772. [Google Scholar] [CrossRef] [PubMed]
- McMillan, J.R.; Akiyama, M.; Tanaka, M.; Yamamoto, S.; Goto, M.; Abe, R.; Sawamura, D.; Shimomura, M.; Shimizu, H. Small-diameter porous poly (epsilon-caprolactone) films enhance adhesion and growth of human cultured epidermal keratinocyte and dermal fibroblast cells. Tissue Eng. 2007, 13, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Nishikawa, K.; Okubo, H.; Kamachi, H.; Kawai, T.; Matsushita, M.; Todo, S.; Shimomura, M. Control of hepatocyte adhesion and function on self-organized honeycomb-patterned polymer film. Colloids Surf. A 2006, 284–285, 464–469. [Google Scholar] [CrossRef]
- Tsuruma, A.; Tanaka, M.; Yamamoto, S.; Shimomura, M. Control of neural stem cell differentiation on honeycomb films. Colloids Surf. A 2008, 313–314, 536–540. [Google Scholar] [CrossRef]
- Tanaka, M. Design of novel 2D and 3D biointerfaces using self-organization to control cell behavior. Biochim. Biophys. Acta 2011, 1810, 251–258. [Google Scholar] [CrossRef]
- Yu, D.; Wolf, J.K.; Scanlon, M.; Price, J.E.; Hung, M.C. Enhanced c-erbB-2/neu expression in human ovarian cancer cells correlates with more severe malignancy that can be suppressed by E1A. Cancer Res. 1993, 53, 891–898. [Google Scholar]
- Li, J.; Xu, C.; Lee, H.J.; Ren, S.; Zi, X.; Zhang, Z.; Wang, H.; Yu, Y.; Yang, C.; Gao, X.; et al. A genomic and epigenomic atlas of prostate cancer in Asian populations. Nature 2020, 580, 93–99. [Google Scholar] [CrossRef]
- Geiger, B.; Spatz, J.P.; Bershadsky, A.D. Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 2009, 10, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Tytell, J.D.; Ingber, D.E. Mechanotransduction at a distance: Mechanically coupling the extracellular matrix with the nucleus. Nat. Rev. Mol. Cell Biol. 2009, 10, 75–82. [Google Scholar] [CrossRef] [PubMed]
- Del Rio, A.; Perez-Jimenez, R.; Liu, R.; Roca-Cusachs, P.; Fernandez, J.M.; Sheetz, M.P. Stretching single talin rod molecules activates vinculin binding. Science 2009, 323, 638–641. [Google Scholar] [CrossRef] [PubMed]
- Jablonska, J.; Wu, C.F.; Andzinski, L.; Leschner, S.; Weiss, S. CXCR2-mediated tumor-associated neutrophil recruitment is regulated by IFN-β. Int. J. Cancer 2014, 134, 1346–1358. [Google Scholar] [CrossRef] [PubMed]
- Ray, P.S.; Wang, J.; Qu, Y.; Sim, M.S.; Shamonki, J.; Bagaria, S.P.; Ye, X.; Liu, B.; Elashoff, D.; Hoon, D.S.; et al. FOXC1 is a potential prognostic biomarker with functional significance in basal-like breast cancer. Cancer Res. 2010, 70, 3870–3876. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Cain-Hom, C.; Choy, L.; Hagenbeek, T.J.; de Leon, G.P.; Chen, Y.; Finkle, D.; Venook, R.; Wu, X.; Ridgway, J.; et al. Therapeutic antibody targeting of individual Notch receptors. Nature 2010, 464, 1052–1057. [Google Scholar] [CrossRef]
- Pahwa, S.; Stawikowski, M.J.; Fields, G.B. Monitoring and Inhibiting MT1-MMP during Cancer Initiation and Progression. Cancers 2014, 6, 416–435. [Google Scholar] [CrossRef]
- McGrath, J.A.; McMillan, J.R.; Shemanko, C.S.; Runswick, S.K.; Leigh, I.M.; Lane, E.B.; Garrod, D.R.; Eady, R.A. Mutations in the plakophilin 1 gene result in ectodermal dysplasia/skin fragility syndrome. Nat. Genet. 1997, 17, 240–244. [Google Scholar] [CrossRef]
- Hajra, K.M.; Chen, D.Y.; Fearon, E.R. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002, 62, 161–168. [Google Scholar]
- Nicolas, A.; Geiger, B.; Safran, S.A. Cell mechanosensitivity controls the anisotropy of focal adhesions. Proc. Natl. Acad. Sci. USA 2004, 101, 12520–12525. [Google Scholar] [CrossRef]
- Shemesh, T.; Geiger, B.; Bershadsky, A.D.; Kozlov, M.M. Focal adhesions as mechanosensors: A physical mechanism. Proc. Natl. Acad. Sci. USA 2005, 102, 12383–12388. [Google Scholar] [CrossRef] [PubMed]
- Vergote, I.; Tropé, C.G.; Amant, F.; Kristensen, G.B.; Ehlen, T.; Johnson, N.; Verheijen, R.H.; van der Burg, M.E.; Lacave, A.J.; Panici, P.B.; et al. European Organization for Research and Treatment of Cancer-Gynaecological Cancer Group; NCIC Clinical Trials Group. Neoadjuvant chemotherapy or primary surgery in stage IIIC or IV ovarian cancer. N. Engl. J. Med. 2010, 363, 943–953. [Google Scholar] [CrossRef] [PubMed]
- Kehoe, S.; Hook, J.; Nankivell, M.; Jayson, G.C.; Kitchener, H.; Lopes, T.; Luesley, D.; Perren, T.; Bannoo, S.; Mascarenhas, M.; et al. Primary chemotherapy versus primary surgery for newly diagnosed advanced ovarian cancer (CHORUS): An open-label, randomised, controlled, non-inferiority trial. Lancet 2015, 386, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Onda, T.; Satoh, T.; Ogawa, G.; Saito, T.; Kasamatsu, T.; Nakanishi, T.; Mizutani, T.; Takehara, K.; Okamoto, A.; Ushijima, K.; et al. Japan Clinical Oncology Group. Comparison of survival between primary debulking surgery and neoadjuvant chemotherapy for stage III/IV ovarian, tubal and peritoneal cancers in phase III randomised trial. Eur. J. Cancer 2020, 130, 114–125. [Google Scholar] [CrossRef] [PubMed]
- Fagotti, A.; Ferrandina, M.G.; Vizzielli, G.; Pasciuto, T.; Fanfani, F.; Gallotta, V.; Margariti, P.A.; Chiantera, V.; Costantini, B.; Gueli Alletti, S.; et al. Randomized trial of primary debulking surgery versus neoadjuvant chemotherapy for advanced epithelial ovarian cancer (SCORPION-NCT01461850). Int. J. Gynecol. Cancer 2020, 30, 1657–1664. [Google Scholar] [CrossRef]
- Moore, K.; Colombo, N.; Scambia, G.; Kim, B.G.; Oaknin, A.; Friedlander, M.; Lisyanskaya, A.; Floquet, A.; Leary, A.; Sonke, G.S.; et al. Maintenance Olaparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2018, 379, 2495–2505. [Google Scholar] [CrossRef]
- González-Martín, A.; Pothuri, B.; Vergote, I.; DePont Christensen, R.; Graybill, W.; Mirza, M.R.; McCormick, C.; Lorusso, D.; Hoskins, P.; Freyer, G.; et al. PRIMA/ENGOT-OV26/GOG-3012 Investigators. Niraparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2391–2402. [Google Scholar] [CrossRef]
- Ray-Coquard, I.; Pautier, P.; Pignata, S.; Pérol, D.; González-Martín, A.; Berger, R.; Fujiwara, K.; Vergote, I.; Colombo, N.; Mäenpää, J.; et al. PAOLA-1 Investigators. Olaparib plus Bevacizumab as First-Line Maintenance in Ovarian Cancer. N. Engl. J. Med. 2019, 381, 2416–2428. [Google Scholar] [CrossRef]
- Xu, C.; Hong, Y. Rational design of biodegradable thermoplastic polyurethanes for tissue repair. Bioact. Mater. 2021, 15, 250–271. [Google Scholar] [CrossRef]
- Fukuhira, Y.; Ito, M.; Kaneko, H.; Sumi, Y.; Tanaka, M.; Yamamoto, S.; Shimomura, M. Prevention of postoperative adhesions by a novel honeycomb-patterned poly(lactide) film in a rat experimental model. J. Biomed. Mater. Res. B Appl. Biomater. 2008, 86, 353–359. [Google Scholar] [CrossRef]
- Fukuhira, Y.; Kaneko, H.; Yamaga, M.; Tanaka, M.; Yamamoto, S.; Shimomura, M. Effect of honeycomb-patterned structure on chondrocyte behavior in vitro. Colloids Surf. A 2008, 313–314, 520–525. [Google Scholar] [CrossRef]


| Category | ID | Description | p Value | padj | geneID | Count |
|---|---|---|---|---|---|---|
| BP | GO:0070268 | cornification | 4.04 × 10−15 | 1.18 × 10−11 | PKP1/KRT23/PERP/CSTA/PI3/KRT17/SPINK5/DSC3/KRT7/TCHH/KLK13/KRT24/KRT27/KRT5/KRT16/KRT14/KRT6A | 17 |
| BP | GO:0008544 | epidermis development | 7.88 × 10−15 | 1.18 × 10−11 | FOXC1/COL17A1/TP63/PKP1/PTHLH/FERMT1/KRT23/VDR/PERP/CSTA/PI3/EREG/KRT17/SPINK5/DSC3/KRT7/ZNF750/NOTCH1/TCHH/FOXQ1/KLK13/KRT24/KRT27/SFN/ALOX15B/KRT5/KRT16/KRT14/KRT6A | 29 |
| BP | GO:0030216 | keratinocyte differentiation | 1.31 × 10−14 | 1.31 × 10−11 | FOXC1/TP63/PKP1/KRT23/VDR/PERP/CSTA/PI3/EREG/KRT17/SPINK5/DSC3/KRT7/NOTCH1/TCHH/KLK13/KRT24/KRT27/SFN/ALOX15B/KRT5/KRT16/KRT14/KRT6A | 24 |
| BP | GO:0043588 | skin development | 3.47 × 10−14 | 2.59 × 10−11 | FOXC1/TP63/PKP1/FERMT1/KRT23/VDR/PERP/CSTA/PI3/EREG/KRT17/SPINK5/DSC3/KRT7/NOTCH1/TCHH/FOXQ1/KLK13/KRT24/KRT27/SFN/ALOX15B/KRT5/KRT16/KRT14/GJB3/KRT6A | 27 |
| BP | GO:0009913 | epidermal cell differentiation | 3.77 × 10−13 | 2.25 × 10−10 | FOXC1/TP63/PKP1/KRT23/VDR/PERP/CSTA/PI3/EREG/KRT17/SPINK5/DSC3/KRT7/NOTCH1/TCHH/KLK13/KRT24/KRT27/SFN/ALOX15B/KRT5/KRT16/KRT14/KRT6A | 24 |
| BP | GO:0031424 | keratinization | 3.52 × 10−11 | 1.75 × 10−8 | PKP1/KRT23/PERP/CSTA/PI3/KRT17/SPINK5/DSC3/KRT7/TCHH/KLK13/KRT24/KRT27/SFN/KRT5/KRT16/KRT14/KRT6A | 18 |
| BP | GO:0050663 | cytokine secretion | 2.41 × 10−9 | 9.01 × 10−7 | FERMT1/IL1A/GBP1/CD274/IL1B/CHI3L1/IL6/IL33/CASP1/NOTCH1/ZC3H12A/SAA1/ALOX15B/IL1RAP/CARD16 | 15 |
| BP | GO:0097529 | myeloid leukocyte migration | 2.66 × 10−9 | 9.01 × 10−7 | CXCL2/CHGA/SERPINE1/IL1A/CCL20/PGF/IL1B/IL6/MMP14/CXCL3/CXCL1/CXCL8/SAA1/CSF1 | 14 |
| BP | GO:1901342 | regulation of vasculature development | 2.72 × 10−9 | 9.01 × 10−7 | FOXC1/NGFR/SERPINE1/IL1A/PGF/IL1B/C3/ID1/SERPINF1/CHI3L1/SPINK5/IL6/NOTCH1/ZC3H12A/HEY1/CXCL8/MMRN2 | 17 |
| BP | GO:0045765 | regulation of angiogenesis | 4.79 × 10−9 | 1.43 × 10−6 | FOXC1/NGFR/SERPINE1/IL1A/PGF/IL1B/C3/ID1/SERPINF1/CHI3L1/SPINK5/IL6/NOTCH1/ZC3H12A/CXCL8/MMRN2 | 16 |
| BP | GO:0002687 | positive regulation of leukocyte migration | 6.28 × 10−9 | 1.70 × 10−6 | CXCL2/BDKRB1/SERPINE1/IL1A/CCL20/PGF/IL6/MMP14/CXCL3/CXCL1/CXCL8/CSF1 | 12 |
| BP | GO:0001525 | angiogenesis | 7.23 × 10−9 | 1.70 × 10−6 | FOXC1/NGFR/CALCRL/SERPINE1/IL1A/PGF/EREG/IL1B/C3/ID1/SERPINF1/CHI3L1/SPINK5/IL6/NOTCH1/MMP14/ZC3H12A/HEY1/CXCL8/MMRN2/NOTCH4 | 21 |
| BP | GO:0032496 | response to lipopolysaccharide | 7.41 × 10−9 | 1.70 × 10−6 | CD6/NGFR/CXCL2/BDKRB1/SERPINE1/CCL20/IL1B/IRAK2/IL6/CASP1/NOTCH1/CXCL3/CXCL1/ZC3H12A/CXCL8/THBD/CD55/CARD16 | 18 |
| BP | GO:0002237 | response to molecule of bacterial origin | 1.61 × 10−8 | 3.44 × 10−6 | CD6/NGFR/CXCL2/BDKRB1/SERPINE1/CCL20/IL1B/IRAK2/IL6/CASP1/NOTCH1/CXCL3/CXCL1/ZC3H12A/CXCL8/THBD/CD55/CARD16 | 18 |
| BP | GO:0002685 | regulation of leukocyte migration | 2.63 × 10−8 | 5.24 × 10−6 | CXCL2/BDKRB1/SERPINE1/IL1A/CCL20/PGF/IL6/IL33/MMP14/CXCL3/CXCL1/CXCL8/CSF1 | 13 |
| BP | GO:0050707 | regulation of cytokine secretion | 3.04 × 10−8 | 5.67 × 10−6 | FERMT1/IL1A/GBP1/CD274/IL1B/IL6/IL33/CASP1/ZC3H12A/SAA1/ALOX15B/IL1RAP/CARD16 | 13 |
| BP | GO:0050900 | leukocyte migration | 5.64 × 10−8 | 9.90 × 10−6 | CXCL2/CHGA/BDKRB1/SERPINE1/IL1A/CCL20/PGF/IL1B/IL6/IL33/SLC7A7/MMP14/CXCL3/CXCL1/INPP5D/CXCL8/SAA1/THBD/CSF1 | 19 |
| BP | GO:0060326 | cell chemotaxis | 9.33 × 10−8 | 1.55 × 10−5 | CXCL2/CHGA/SERPINE1/CCL20/PGF/IL1B/SAA2/IL6/NOTCH1/CXCL3/CXCL1/CXCL8/SAA1/CSF1/PLXNB3 | 15 |
| BP | GO:0032675 | regulation of interleukin-6 production | 2.27 × 10−7 | 3.57 × 10−5 | BPI/IL1A/EREG/IL1B/IL6/IL33/ZC3H12A/INPP5D/ADORA2B/IL1RAP | 10 |
| BP | GO:0010951 | negative regulation of endopeptidase activity | 3.25 × 10−7 | 4.86 × 10−5 | NGFR/TFPI2/SERPINE1/CSTA/PI3/C3/SERPINF1/SPINK5/IL6/SFN/SERPINA3/SERPINA1/CARD16/SERPINB5 | 14 |
| Category | ID | Description | p Value | padj | geneID | Count |
|---|---|---|---|---|---|---|
| MF | GO:0005126 | cytokine receptor binding | 1.07 × 10−5 | 0.000916 | CXCL2/IL1A/CCL20/TNFSF4/PGF/IL1B/IL6/INHBB/CXCL3/CXCL1/IL13/CXCL8/CSF1/LTA/ITGB3 | 15 |
| MF | GO:0045236 | CXCR chemokine receptor binding | 8.79 × 10−5 | 0.004524 | CXCL2/CXCL3/CXCL1/CXCL8 | 4 |
| MF | GO:0001664 | G-protein coupled receptor binding | 0.0005354 | 0.017232 | GNA15/CXCL2/RNF43/CCL20/C3/SHANK1/ADORA1/CXCL3/CXCL1/BDKRB2/CXCL8/SAA1 | 12 |
| MF | GO:0042379 | chemokine receptor binding | 0.0009311 | 0.023976 | CXCL2/CCL20/CXCL3/CXCL1/CXCL8 | 5 |
| MF | GO:0070851 | growth factor receptor binding | 0.0010385 | 0.025467 | AREG/IL1A/PGF/EREG/IL1B/FLRT1/IL6/ITGB3 | 8 |
| MF | GO:0030276 | clathrin binding | 0.002324 | 0.047874 | LRP1/TRPC6/SYT8/DOC2A/DNER | 5 |
| Category | ID | Description | p Value | padj | geneID | Count |
|---|---|---|---|---|---|---|
| BP | GO:0043616 | keratinocyte proliferation | 2.23 × 10−5 | 0.0015763 | SNAI2/TP63/FERMT1/VDR/EREG/SFN | 6 |
| BP | GO:0050678 | regulation of epithelial cell proliferation | 5.68 × 10−5 | 0.0028436 | SNAI2/NGFR/TP63/VDR/PGF/EREG/ID1/DLL4/SERPINF1/IL6/NOTCH1/SFN/PLXNB3/SERPINB5/ITGB3 | 15 |
| BP | GO:0014009 | glial cell proliferation | 0.0002361 | 0.007432 | AREG/IL33/NOTCH1/ASCL2/LTA | 5 |
| BP | GO:0050673 | epithelial cell proliferation | 2.17 × 10−6 | 0.0002763 | SNAI2/NGFR/TP63/FERMT1/AREG/VDR/PGF/EREG/ID1/DLL4/SERPINF1/IL6/LGR5/NOTCH1/MMP14/SFN/PLXNB3/SERPINB5/ITGB3 | 19 |
| BP | GO:0032946 | positive regulation of mononuclear cell proliferation | 0.0002448 | 0.0075068 | CD6/TNFSF4/CD274/IL1B/IL6/CD1D/IL13/CSF1/CD55 | 9 |
| BP | GO:0070665 | positive regulation of leukocyte proliferation | 0.000323 | 0.0091505 | CD6/TNFSF4/CD274/IL1B/IL6/CD1D/IL13/CSF1/CD55 | 9 |
| BP | GO:2000647 | negative regulation of stem cell proliferation | 0.0005807 | 0.0137109 | SNAI2/FERMT1/KCTD11 | 3 |
| BP | GO:0010839 | negative regulation of keratinocyte proliferation | 0.0007653 | 0.0167911 | SNAI2/VDR/SFN | 3 |
| BP | GO:0010837 | regulation of keratinocyte proliferation | 0.000794 | 0.017078 | SNAI2/TP63/VDR/SFN | 4 |
| BP | GO:0042102 | positive regulation of T cell proliferation | 0.0008037 | 0.017078 | CD6/TNFSF4/CD274/IL1B/IL6/CD1D/CD55 | 7 |
| BP | GO:0050671 | positive regulation of lymphocyte proliferation | 0.0011127 | 0.0213732 | CD6/TNFSF4/CD274/IL1B/IL6/CD1D/IL13/CD55 | 8 |
| BP | GO:0032944 | regulation of mononuclear cell proliferation | 0.0014005 | 0.0239278 | CD6/TNFSF4/CD274/IL1B/IL6/CD1D/INPP5D/IL13/CSF1/CD55 | 10 |
| BP | GO:0070663 | regulation of leukocyte proliferation | 0.0018681 | 0.028611 | CD6/TNFSF4/CD274/IL1B/IL6/CD1D/INPP5D/IL13/CSF1/CD55 | 10 |
| BP | GO:0050680 | negative regulation of epithelial cell proliferation | 0.0021854 | 0.0323065 | SNAI2/NGFR/VDR/EREG/DLL4/SERPINF1/SFN | 7 |
| BP | GO:0010812 | negative regulation of cell-substrate adhesion | 0.0014459 | 0.0244574 | SERPINE1/GBP1/NOTCH1/MMP14/MELTF | 5 |
| BP | GO:0030198 | extracellular matrix organization | 0.0021733 | 0.0322666 | TNC/ELN/FOXC1/FERMT1/SERPINE1/LRP1/SPINK5/NOTCH1/MMP14/MELTF/TMPRSS6/SERPINB5/ITGB3 | 13 |
| BP | GO:0045109 | intermediate filament organization | 0.0036129 | 0.0448321 | PKP1/KRT17/KRT14 | 3 |
| BP | GO:0045104 | intermediate filament cytoskeleton organization | 0.0004401 | 0.0116361 | PKP1/KRT17/KRT16/KRT14/KRT6A | 5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ohta, T.; Tanaka, M.; Taki, S.; Nakagawa, H.; Nagase, S. Honeycomb-like Structured Film, a Novel Therapeutic Device, Suppresses Tumor Growth in an In Vivo Ovarian Cancer Model. Cancers 2023, 15, 237. https://doi.org/10.3390/cancers15010237
Ohta T, Tanaka M, Taki S, Nakagawa H, Nagase S. Honeycomb-like Structured Film, a Novel Therapeutic Device, Suppresses Tumor Growth in an In Vivo Ovarian Cancer Model. Cancers. 2023; 15(1):237. https://doi.org/10.3390/cancers15010237
Chicago/Turabian StyleOhta, Tsuyoshi, Masaru Tanaka, Seitaro Taki, Hiroyuki Nakagawa, and Satoru Nagase. 2023. "Honeycomb-like Structured Film, a Novel Therapeutic Device, Suppresses Tumor Growth in an In Vivo Ovarian Cancer Model" Cancers 15, no. 1: 237. https://doi.org/10.3390/cancers15010237
APA StyleOhta, T., Tanaka, M., Taki, S., Nakagawa, H., & Nagase, S. (2023). Honeycomb-like Structured Film, a Novel Therapeutic Device, Suppresses Tumor Growth in an In Vivo Ovarian Cancer Model. Cancers, 15(1), 237. https://doi.org/10.3390/cancers15010237







