Evolutionary Analysis of TCGA Data Using Over- and Under- Mutated Genes Identify Key Molecular Pathways and Cellular Functions in Lung Cancer Subtypes
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Gene List Acquisition
2.2. Mutational Frequency
2.3. Expression Data
2.4. Identifying Conserved Pathways and Functions
3. Results
3.1. Cohort Demographics
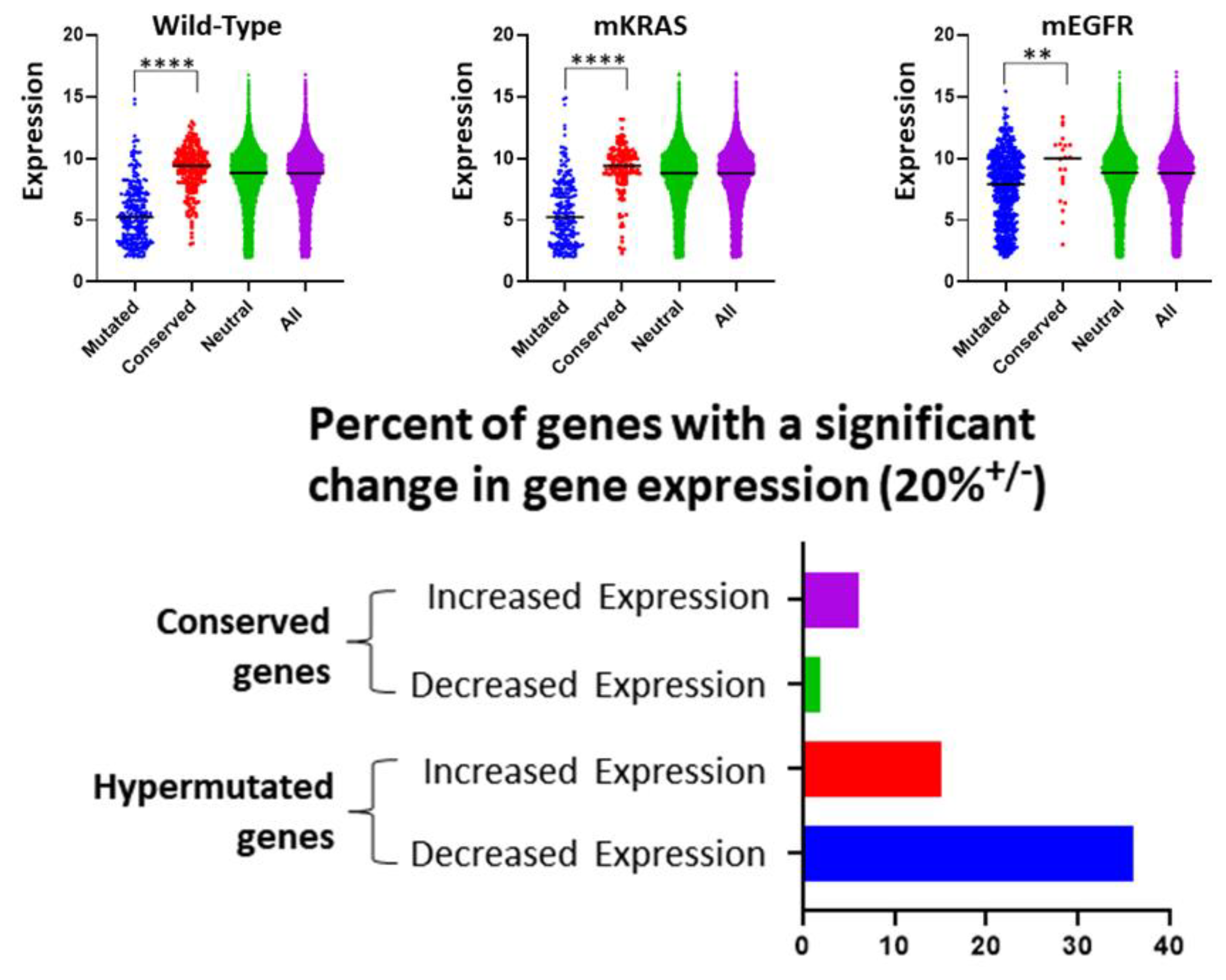
3.2. Gene Expression and Evolutionary Selection
3.3. Highly Mutated Genes under Directional Selection
3.4. Evolutionarily Conserved Genes as Evidence for Stabilizing Selection
3.5. Evolutionary Selection on Cellular Pathways
3.6. TP53 Interactome in Lung Cancer
3.7. EGFR-Mut and KRAS-Mut Signaling
3.8. Evolutionary Selection on Cellular Functions
3.9. Evolutionary Selection on Signaling Pathways
3.10. Evolutionary Selection on DNA Repair, Phenotypic Plasticity, and Epigenetic Modifications
3.11. Evolutionary Selection on Microenvironmental Interactions
3.12. Evolutionary Selection on Membrane Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gatenby, R.A.; Cunningham, J.J.; Brown, J.S. Evolutionary triage governs fitness in driver and passenger mutations and suggests targeting never mutations. Nat. Commun. 2014, 5, 5499. [Google Scholar] [CrossRef] [PubMed]
- Gatenby, R.A.; Brown, J. Mutations, evolution and the central role of a self-defined fitness function in the initiation and progression of cancer. Biochim. Biophys. Acta Rev. Cancer 2017, 1867, 162–166. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, M.C.; Cunningham, J.J.; Bui, M.M.; Gillies, R.J.; Brown, J.S.; Gatenby, R.A. Darwinian Dynamics of Intratumoral Heterogeneity: Not Solely Random Mutations but Also Variable Environmental Selection Forces. Cancer Res. 2016, 76, 3136–3144. [Google Scholar] [CrossRef] [PubMed]
- De Magalhaes, J.P. Every gene can (and possibly will) be associated with cancer. Trends Genet. 2022, 38, 216–217. [Google Scholar] [CrossRef]
- Mohammad, H.P.; Barbash, O.; Creasy, C.L. Targeting epigenetic modifications in cancer therapy: Erasing the roadmap to cancer. Nat. Med. 2019, 25, 403–418. [Google Scholar] [CrossRef]
- Gatenby, R.A.; Frieden, B.R. Application of information theory and extreme physical information to carcinogenesis. Cancer Res. 2002, 62, 3675–3684. [Google Scholar]
- Khadem, H.; Kebriaei, H.; Veisi, Z. Inactivation of tumor suppressor genes and cancer therapy: An evolutionary game theory approach. Math Biosci. 2017, 288, 84–93. [Google Scholar] [CrossRef]
- Wodarz, D.; Komarova, N.L. Dynamics of Cancer: Mathematical Foundations of Oncology; World Scientific: Hackensack, NJ, USA, 2014; 514p. [Google Scholar]
- Galor, O.; Michalopoulos, S. Evolution and the Growth Process: Natural Selection of Entrepreneurial Traits. J. Econ. Theory 2012, 147, 759–780. [Google Scholar] [CrossRef]
- Gatenby, R.A.; Vincent, T.L. An evolutionary model of carcinogenesis. Cancer Res. 2003, 63, 6212–6220. [Google Scholar]
- Gatenby, R.A.; Frieden, B.R. Information dynamics in carcinogenesis and tumor growth. Mutat. Res. 2004, 568, 259–273. [Google Scholar] [CrossRef]
- Temko, D.; Tomlinson, I.P.M.; Severini, S.; Schuster-Bockler, B.; Graham, T.A. The effects of mutational processes and selection on driver mutations across cancer types. Nat. Commun. 2018, 9, 1857. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Warrell, J.; Li, S.; McGillivray, P.D.; Meyerson, W.; Salichos, L.; Harmanci, A.; Martinez-Fundichely, A.; Chan, C.W.Y.; Nielsen, M.M.; et al. Passenger Mutations in More Than 2500 Cancer Genomes: Overall Molecular Functional Impact and Consequences. Cell 2020, 180, 915–927.e16. [Google Scholar] [CrossRef] [PubMed]
- Zapata, L.; Pich, O.; Serrano, L.; Kondrashov, F.A.; Ossowski, S.; Schaefer, M.H. Negative selection in tumor genome evolution acts on essential cellular functions and the immunopeptidome. Genome Biol. 2018, 19, 67. [Google Scholar] [CrossRef]
- Bailey, M.H.; Tokheim, C.; Porta-Pardo, E.; Sengupta, S.; Bertrand, D.; Weerasinghe, A.; Colaprico, A.; Wendl, M.C.; Kim, J.; Reardon, B.; et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell 2018, 173, 371–385.e18. [Google Scholar] [CrossRef] [PubMed]
- Martincorena, I.; Raine, K.M.; Gerstung, M.; Dawson, K.J.; Haase, K.; Van Loo, P.; Davies, H.; Stratton, M.R.; Campbell, P.J. Universal Patterns of Selection in Cancer and Somatic Tissues. Cell 2017, 171, 1029–1041.e1021. [Google Scholar] [CrossRef]
- Fearon, E.R.; Vogelstein, B. A genetic model for colorectal tumorigenesis. Cell 1990, 61, 759–767. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, L.B.; Stratton, M.R. Mutational signatures: The patterns of somatic mutations hidden in cancer genomes. Curr. Opin. Genet. Dev. 2014, 24, 52–60. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Campbell, P.J.; Stratton, M.R. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013, 3, 246–259. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Borresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014, 511, 543–550. [Google Scholar] [CrossRef]
- Gonzalez-Calderon, G.; Liu, R.; Carvajal, R.; Teer, J.K. A negative storage model for precise but compact storage of genetic variation data. Database 2020, 2020, baz158. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Qian, W.; Zhang, J. Genomic evidence for elevated mutation rates in highly expressed genes. EMBO Rep. 2012, 13, 1123–1129. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.S.; Stojanov, P.; Polak, P.; Kryukov, G.V.; Cibulskis, K.; Sivachenko, A.; Carter, S.L.; Stewart, C.; Mermel, C.H.; Roberts, S.A.; et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013, 499, 214–218. [Google Scholar] [CrossRef] [PubMed]
- Schott, B.H.; Assmann, A.; Schmierer, P.; Soch, J.; Erk, S.; Garbusow, M.; Mohnke, S.; Pohland, L.; Romanczuk-Seiferth, N.; Barman, A.; et al. Epistatic interaction of genetic depression risk variants in the human subgenual cingulate cortex during memory encoding. Transl. Psychiatry 2014, 4, e372. [Google Scholar] [CrossRef]
- Delling, M.; DeCaen, P.G.; Doerner, J.F.; Febvay, S.; Clapham, D.E. Primary cilia are specialized calcium signalling organelles. Nature 2013, 504, 311–314. [Google Scholar] [CrossRef]
- Agullo-Ortuno, M.T.; Garcia-Ruiz, I.; Diaz-Garcia, C.V.; Enguita, A.B.; Pardo-Marques, V.; Prieto-Garcia, E.; Ponce, S.; Iglesias, L.; Zugazagoitia, J.; Lopez-Martin, J.A.; et al. Blood mRNA expression of REV3L and TYMS as potential predictive biomarkers from platinum-based chemotherapy plus pemetrexed in non-small cell lung cancer patients. Cancer Chemother. Pharmacol. 2020, 85, 525–535. [Google Scholar] [CrossRef]
- Valdes-Mora, F.; Locke, W.J.; Bandres, E.; Gallego-Ortega, D.; Cejas, P.; Garcia-Cabezas, M.A.; Colino-Sanguino, Y.; Feliu, J.; Del Pulgar, T.G.; Lacal, J.C. Clinical relevance of the transcriptional signature regulated by CDC42 in colorectal cancer. Oncotarget 2017, 8, 26755–26770. [Google Scholar] [CrossRef]
- Thion, M.S.; Humbert, S. Cancer: From Wild-Type to Mutant Huntingtin. J. Huntingtons Dis. 2018, 7, 201–208. [Google Scholar] [CrossRef]
- Zhou, J.; Du, Y.R.; Qin, W.H.; Hu, Y.G.; Huang, Y.N.; Bao, L.; Han, D.; Mansouri, A.; Xu, G.L. RIM-BP3 is a manchette-associated protein essential for spermiogenesis. Development 2009, 136, 373–382. [Google Scholar] [CrossRef]
- Saudou, F.; Humbert, S. The Biology of Huntingtin. Neuron 2016, 89, 910–926. [Google Scholar] [CrossRef]
- McNulty, P.; Pilcher, R.; Ramesh, R.; Necuiniate, R.; Hughes, A.; Farewell, D.; Holmans, P.; Jones, L.; Registry Investigators of the European Huntington’s Disease Network. Reduced Cancer Incidence in Huntington’s Disease: Analysis in the Registry Study. J. Huntingtons Dis. 2018, 7, 209–222. [Google Scholar] [CrossRef] [PubMed]
- Jefri, M.; Bell, S.; Peng, H.; Hettige, N.; Maussion, G.; Soubannier, V.; Wu, H.; Silveira, H.; Theroux, J.F.; Moquin, L.; et al. Stimulation of L-type calcium channels increases tyrosine hydroxylase and dopamine in ventral midbrain cells induced from somatic cells. Stem Cells Transl. Med. 2020, 9, 697–712. [Google Scholar] [CrossRef] [PubMed]
- Dziurdzik, S.K.; Bean, B.D.M.; Davey, M.; Conibear, E. A VPS13D spastic ataxia mutation disrupts the conserved adaptor-binding site in yeast Vps13. Hum. Mol. Genet. 2020, 29, 635–648. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, H.A.; Wang, C.; Kanfer, G.; Shah, H.V.; Velayos-Baeza, A.; Dulovic-Mahlow, M.; Bruggemann, N.; Anding, A.; Baehrecke, E.H.; Maric, D.; et al. VPS13D promotes peroxisome biogenesis. J. Cell Biol. 2021, 220, e202001188. [Google Scholar] [CrossRef]
- Zhang, B.; Xie, Z.; Li, B. The clinicopathologic impacts and prognostic significance of GLUT1 expression in patients with lung cancer: A meta-analysis. Gene 2019, 689, 76–83. [Google Scholar] [CrossRef]
- Shepelev, M.V.; Korobko, I.V. The RHOV gene is overexpressed in human non-small cell lung cancer. Cancer Genet. 2013, 206, 393–397. [Google Scholar] [CrossRef]
- Tang, Y.; Zhang, Y.J.; Wu, Z.H. High GJB2 mRNA expression and its prognostic significance in lung adenocarcinoma: A study based on the TCGA database. Medicine 2020, 99, e19054. [Google Scholar] [CrossRef]
- Wang, T.; Liang, Y.; Thakur, A.; Zhang, S.; Yang, T.; Chen, T.; Gao, L.; Chen, M.; Ren, H. Diagnostic significance of S100A2 and S100A6 levels in sera of patients with non-small cell lung cancer. Tumour Biol. 2016, 37, 2299–2304. [Google Scholar] [CrossRef]
- Jiang, H.; Wang, H.; Wang, S.; Pei, Z.; Fu, Z.; Fang, C.; Wang, J.; Lu, Q.; Wang, E.; Li, J. Expression of ERCC1, TYMS, RRM1, TUBB3, non-muscle myosin II, myoglobin and MyoD1 in lung adenocarcinoma pleural effusions predicts survival in patients receiving platinum-based chemotherapy. Mol. Med. Rep. 2015, 11, 3523–3532. [Google Scholar] [CrossRef]
- Yang, F.; Shao, C.; Wei, K.; Jing, X.; Qin, Z.; Shi, Y.; Shu, Y.; Shen, H. miR-942 promotes tumor migration, invasion, and angiogenesis by regulating EMT via BARX2 in non-small-cell lung cancer. J. Cell Physiol. 2019, 234, 23596–23607. [Google Scholar] [CrossRef]
- Fan, C.; Chen, L.; Huang, Q.; Shen, T.; Welsh, E.A.; Teer, J.K.; Cai, J.; Cress, W.D.; Wu, J. Overexpression of major CDKN3 transcripts is associated with poor survival in lung adenocarcinoma. Br. J. Cancer 2015, 113, 1735–1743. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, J.; Xu, D.; Zhang, T.; Hu, W.; Feng, Z. Gain-of-function mutant p53 in cancer progression and therapy. J. Mol. Cell Biol. 2020, 12, 674–687. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, H.; Counter, C.M. Interrogating the protein interactomes of RAS isoforms identifies PIP5K1A as a KRAS-specific vulnerability. Nat. Commun. 2018, 9, 3646. [Google Scholar] [CrossRef] [PubMed]
- Irimia, M.; Fraga, M.F.; Sanchez-Cespedes, M.; Esteller, M. CpG island promoter hypermethylation of the Ras-effector gene NORE1A occurs in the context of a wild-type K-ras in lung cancer. Oncogene 2004, 23, 8695–8699. [Google Scholar] [CrossRef][Green Version]
- Uyama, T.; Ueda, N. Assay of NAT Activity. Methods Mol. Biol. 2016, 1412, 113–122. [Google Scholar] [CrossRef]
- Qin, T.; Xia, J.; Liu, S.; Wang, J.; Liu, H.; Zhang, Y.; Jia, Y.; Li, K. Clinical importance of VEGFC and PD-L1 co-expression in lung adenocarcinoma patients. Thorac. Cancer 2020, 11, 1139–1148. [Google Scholar] [CrossRef]
- Farwell, S.L.N.; Reylander, K.G.; Iovine, M.K.; Lowe-Krentz, L.J. Novel Heparin Receptor Transmembrane Protein 184a Regulates Angiogenesis in the Adult Zebrafish Caudal Fin. Front. Physiol. 2017, 8, 671. [Google Scholar] [CrossRef]
- Li, K.; Jiang, Y.; Xiang, X.; Gong, Q.; Zhou, C.; Zhang, L.; Ma, Q.; Zhuang, L. Long non-coding RNA SNHG6 promotes the growth and invasion of non-small cell lung cancer by downregulating miR-101-3p. Thorac. Cancer 2020, 11, 1180–1190. [Google Scholar] [CrossRef]
- Kessenbrock, K.; Plaks, V.; Werb, Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell 2010, 141, 52–67. [Google Scholar] [CrossRef]
- Momcilovic, M.; Jones, A.; Bailey, S.T.; Waldmann, C.M.; Li, R.; Lee, J.T.; Abdelhady, G.; Gomez, A.; Holloway, T.; Schmid, E.; et al. In vivo imaging of mitochondrial membrane potential in non-small-cell lung cancer. Nature 2019, 575, 380–384. [Google Scholar] [CrossRef]
- Blackiston, D.J.; McLaughlin, K.A.; Levin, M. Bioelectric controls of cell proliferation: Ion channels, membrane voltage and the cell cycle. Cell Cycle 2009, 8, 3527–3536. [Google Scholar] [CrossRef] [PubMed]
- Lu, A.; Shi, Y.; Liu, Y.; Lin, J.; Zhang, H.; Guo, Y.; Li, L.; Lin, Z.; Wu, J.; Ji, D.; et al. Integrative analyses identified ion channel genes GJB2 and SCNN1B as prognostic biomarkers and therapeutic targets for lung adenocarcinoma. Lung Cancer 2021, 158, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Lv, J.; Liu, Z. Identification of stage-specific biomarkers in lung adenocarcinoma based on RNA-seq data. Tumour Biol. 2015, 36, 6391–6399. [Google Scholar] [CrossRef] [PubMed]
- Debaugny, R.E.; Skok, J.A. CTCF and CTCFL in cancer. Curr. Opin. Genet. Dev. 2020, 61, 44–52. [Google Scholar] [CrossRef]
- Monroe, J.G.; Srikant, T.; Carbonell-Bejerano, P.; Becker, C.; Lensink, M.; Exposito-Alonso, M.; Klein, M.; Hildebrandt, J.; Neumann, M.; Kliebenstein, D.; et al. Mutation bias reflects natural selection in Arabidopsis thaliana. Nature 2022, 602, 101–105. [Google Scholar] [CrossRef]
- Fisk, J.N.; Mahal, A.R.; Dornburg, A.; Gaffney, S.G.; Aneja, S.; Contessa, J.N.; Rimm, D.; Yu, J.B.; Townsend, J.P. Premetastatic shifts of endogenous and exogenous mutational processes support consolidative therapy in EGFR-driven lung adenocarcinoma. Cancer Lett. 2022, 526, 346–351. [Google Scholar] [CrossRef]
- Chapman, A.M.; Sun, K.Y.; Ruestow, P.; Cowan, D.M.; Madl, A.K. Lung cancer mutation profile of EGFR, ALK, and KRAS: Meta-analysis and comparison of never and ever smokers. Lung Cancer 2016, 102, 122–134. [Google Scholar] [CrossRef]
- Wang, X.; Ricciuti, B.; Nguyen, T.; Li, X.; Rabin, M.S.; Awad, M.M.; Lin, X.; Johnson, B.E.; Christiani, D.C. Association between Smoking History and Tumor Mutation Burden in Advanced Non-Small Cell Lung Cancer. Cancer Res. 2021, 81, 2566–2573. [Google Scholar] [CrossRef]
- Hankey, W.; Zanghi, N.; Crow, M.M.; Dow, W.H.; Kratz, A.; Robinson, A.M. Using The Cancer Genome Atlas as an Inquiry Tool in the Undergraduate Classroom. Front Genet. 2020, 11, 573992. [Google Scholar] [CrossRef]

| Evolutionary Selection for Mutations or Conservation in Intracellular Pathways | |||||
|---|---|---|---|---|---|
| EGFR-Mut | KRAS-Mut | NEK | |||
| Conserved | Mutation | Conserved | Mutation | Conserved | Mutation |
| MAPK | |||||
| HRASLS | MAP3K13 | MAP3K6 | MAPK7 | MAP3K1 | FAM83C |
| EGF | MAP7D2 | MEGF11 | FAM83B | MAP4K5 | |
| MAP2K4 | VEGFC | INAVA | MEGF9 | ||
| MAP3K11 | |||||
| MAP3K9 | |||||
| RASA2 | |||||
| RASAL2 | |||||
| RASL10B | |||||
| MYC | |||||
| HECTD4 | |||||
| NOTCH | |||||
| NOTCH4 | NOTCH4 | ||||
| Rho receptors | |||||
| RHOV | RHOH | RHOV | RHOV | ||
| Rho/Rac guanine nucleotide exchange | |||||
| ARHGEF19 | ARHGEF40 | ARHGEF5 | ARHGEF6 | ARHGEF1 | |
| ARHGAP6 | ARHGAP36 | ARHGAP6 | ARGHEF39 | ||
| KIAA0355 (GARRE1) | ARHGEF39 | ||||
| Rab pathway | |||||
| RAB26 | RAB3GAP1 | RAB3B | |||
| RAB3GAP2 | RAB26 | ||||
| TBC1D2B | TBC1D3H | ||||
| TBC1D3C | |||||
| DENND4C | |||||
| DENND5A | |||||
| FAM71B GARIN3 | FAM71B GARIN3 | ||||
| ST5 | |||||
| Rac pathway | |||||
| RAC3 | |||||
| RAN pathway | |||||
| RANBP9 | |||||
| Protein tyrosine phosphatase receptors | |||||
| PTPRH | PTPRN | PTPRF | PTPRD | PTPRD | |
| PTPRT | PTPRT | ||||
| PTPRN | PTPRZ1 | ||||
| ERBB | |||||
| ERBB4 | ERBB4 | ||||
| Interleukin receptors | |||||
| IL36RN | ILRL1 | IL36RN | IL1R1 | ||
| IL23A | IL1RAPL2 | IL17C | IL27RA | ||
| IL37 | IL23A | ||||
| IL31RA | |||||
| IL22RA2 | |||||
| IL1RL2 | |||||
| IL41L | |||||
| IL36G | |||||
| Interferon | |||||
| IRF2BP1 | |||||
| IRF5 | |||||
| WNT/Catenin | |||||
| WNT3 | |||||
| CTNNA2 | CTNNA2 | ||||
| CTNND2 | CTNND2 | ||||
| FAM123B (AMER1) | FAM123C (AMER3) | ||||
| Lipid Receptors | |||||
| NLRP3 | NLRP3 | NLRP3 | |||
| NLRP14 | NLRP14 | NLRP14 | |||
| NLRP5 | NLRP5 | NLRP3 | |||
| LLRP1B | LRP1B | ||||
| NLRP10 | |||||
| LRP1B | |||||
| LRP1B | |||||
| HIPPO | |||||
| WWC2 | |||||
| NOTCH | |||||
| NOTCH4 | NOTCH4 | ||||
| Fibroblast Growth Factor | |||||
| FGF11 | FGF11 | FGF11 | |||
| FGF19 | |||||
| Insulin like growth factors | |||||
| IGF2BP3 | IGFL2 | IGFL2 | |||
| IGFBP3 | |||||
| Toll Like Receptors | |||||
| TLR4 | TLR4 | ||||
| Ephrin Receptors | |||||
| EPHX4 | EPHA3 | EPHA1 | EPHA3 | EPHA5 | EPHA3 |
| EPHB2 | EPHA6 | EPHB6 | EPHA6 | EPHA6 | |
| EPHA5 | EPHS5 | ||||
| EPHB6 | |||||
| EPHA3 | |||||
| EFNA4 | EFNA4 | ||||
| EFNA3 | EFNA3 | ||||
| Ryanodine Receptors | |||||
| RYR3 | RYR2 | RYR2 | RYR2 | ||
| RYR3 | RYR3 | ||||
| RYR1 | RYR1 | ||||
| Glutamate Receptors | |||||
| GRIN2A | GRIN2B | GRIN2A | |||
| GRIN2B | |||||
| GRID2 | GRID2 | ||||
| Semaphorin | |||||
| SEMA5A | SEMA5A | ||||
| SEMA5B | SEMA5B | ||||
| G-Coupled Protein | |||||
| GPR115 | GPR158 | GPR113 | GPR112 | GPR108 | GPR112 |
| GPR87 | GPR148 | GPR19 | GPR158 | ||
| GPR110 | GNL | GPR141 | |||
| GPR174 | |||||
| GPR139 | |||||
| GPR98 | |||||
| GPR158 | |||||
| Inositol | |||||
| ITPKA | ITPKA | ||||
| cAMP signaling | |||||
| ADCY2 | ADCY2 | ADCY2 | |||
| ADCY8 | ADCY8 | ||||
| ADCYAP | |||||
| Evolutionary Selection of Genes Associated with DNA Repair, Phenotypic Plasticity, and Epigenetic Modifications | |||||
|---|---|---|---|---|---|
| EGFR-Mut | KRAS-Mut | NEK | |||
| Conserved | Mutation | Conserved | Mutation | Conserved | Mutation |
| Histones | |||||
| HIST1H2AM | HIST1H2AM | ||||
| HIST1H2BD | HIST1H2BD | ||||
| HIST1H2BG | |||||
| Acetylation/Methylation | |||||
| CDYL2 | HDAC9 | HDAC9 | CDY1 | HDAC9 | |
| ACY3 | CDY1B | ||||
| BRD4 | CDY2A | ||||
| B4GALNT44 | CDY2B | ||||
| GCNT3 | CLOCK | ||||
| ST8SIA2 | HDAC5 | ||||
| HEXB | |||||
| GCNT3 | |||||
| HDAC5 | |||||
| KIAA0182 (GSE1) | |||||
| Cytochrome P450 family | |||||
| CYP2D6 | CYP11B2 | ||||
| CYP27B1 | CYP11B1 | ||||
| CYP27C1 | CYP26B1 | ||||
| CYP241 | |||||
| ABCB1 | |||||
| Homeobox/RNA Polymerase II | |||||
| BARX2 | BARX2 | BARX2 | |||
| ETV4 | ETV4 | ||||
| SIX4 | SIX1 | ||||
| SIX2 | |||||
| HOXC10 | HOXA13 | HOXB9 | HOXA13 | ||
| HOXB9 | HOXA1 | HOXC10 | HOXA1 | ||
| HOXC13 | HOXA3 | HOXB3 | HOXA3 | ||
| HOXC9 | HOXA10 | HOXA5 | |||
| HOXB13 | |||||
| HOXD4 | |||||
| HOXD3 | |||||
| HOXB8 | |||||
| HOXC11 | |||||
| HOXC8 | |||||
| HOXC6 | |||||
| HOXB7 | |||||
| FOXM1 | FOXG1 | FOXG1 | FOXF2 | ||
| FOXP3 | |||||
| FOXB1 | |||||
| ONECUT1 | ONECUT1 | ||||
| ONECUT2 | ONECUT2 | ||||
| OTX1 | |||||
| PAX7 | PAX4 | ||||
| PAX9 | |||||
| PITX2 | GREM1 | ||||
| E2F2 | |||||
| E2F8 | |||||
| ASXL1 | ASXL3 | ASXL3 | |||
| POU3F2 | POU3F4 | POU3F4 | |||
| CIITA | |||||
| NOMO2 | NOMO3 | ||||
| SATB2 | SATB2 | ||||
| TSHZ3 | TSHZ3 | ||||
| ZEB1 | ZEB1 | ||||
| ZFHX4 | ZFHX4 | ||||
| Multicellularity | |||||
| PLXNB3 | SEMA5A | SEMA5A | |||
| SEMA5B | SEMA4B | SEMA5B | |||
| SEMA6D | |||||
| FREM2 | SEMA3D | ||||
| ABCB5 | ABCB5 | ABCB5 | |||
| ABCB1 | |||||
| GLI2 | |||||
| GINS1 | GINS1 | ||||
| GINS2 | GINS2 | ||||
| E2F transcription factor 5 | |||||
| E2F8 | |||||
| E2F2 | |||||
| Cyclin-dependent genes | |||||
| CDKN3 | CDKN2A | CDK13 | CDKN2A | CDK5RAP1 | CDKN2A |
| CDKN3 | CDKN3 | ||||
| CDK6 | CDK5R2 | ||||
| CDKL2 | CDK5R2 | ||||
| Telomere | |||||
| TERT | |||||
| Zinc Finger proteins | |||||
| ZFHX3 | ZFPM2 | ZNF729 | ZNF479 | NF729 | ZNF479 |
| ZFHX4 | ZNF827 | ZNF536 | ZNF324 | ZNF536 | |
| ZNF839 | ZNF676 | ZNF84 | ZNF676 | ||
| ZNF687 | ZNF804B | ZNF726 | ZNF804B | ||
| ZNF845 | ZNF385D | ZNF749 | ZNF385D | ||
| ZC3H4 | ZNF208 | ZNF642 | ZNF208 | ||
| PDZD8 | ZNF257 | ZNF30 | ZNF257 | ||
| ZZEF1 | ZNF716 | ZNF57 | ZNF716 | ||
| ZNF521 | ZNF433 | ZNF521 | |||
| ZNF831 | ZNF655 | ZNF831 | |||
| ZNF98 | ZNF503 | ZNF98 | |||
| ZNF804A | ZNF846 | ZNF804A | |||
| ZNF711 | ZNF205 | ZNF648 | |||
| ZNF679 | ZNF26 | ZBTB1 | |||
| ZNF835 | ZFPM2 | ZBBX | |||
| NF479 | TSHZ3 | ZBTB1 | |||
| GLI3 | ZFP106 | ZFPM2 | |||
| ZEB1 | ZBTB1 | TSHZ3 | |||
| ZFPM2 | ZDHHC5 | ZEB1 | |||
| TSHZ3 | TSHZ3 | ZCCHC5 | |||
| ZIC4 | ZIC4 | ||||
| ZIC1 | ZIC1 | ||||
| ZSCAN4 | ZIM2 | ||||
| ZCCHC12 | TSHZ2 | ||||
| ZIC3 | ZFHX4 | ||||
| ZSCAN1 | |||||
| ZSCAN5B | |||||
| ZFHX4 | |||||
| DNA repair | |||||
| BRCA1 | |||||
| BRCA2 | |||||
| KIAA0101 (PCLAF) | KIAA0101 (PCLAF) | ||||
| REV3L | REV3L | ||||
| XRCC2 | |||||
| CHAF1B | |||||
| CLSPN | |||||
| EME1 | EME1 | ||||
| EXPO1 | |||||
| FOXM1 | |||||
| MCM10 | |||||
| TONSL | |||||
| UBE2T | UBE2T | UBE2A | |||
| UBE2C | |||||
| EXPO1 | |||||
| HROB | |||||
| AUNIP | |||||
| Extracellular Matrix | |||||
|---|---|---|---|---|---|
| EGFR-Mut | KRAS-Mut | NEK | |||
| Conserved | Mutation | Conserved | Mutation | Conserved | Mutation |
| Matrix Metalloproteinase | |||||
| MMP17 | MMP16 | MMP16 | |||
| MMP9 | MMP2 | ||||
| ADAMTS | |||||
| ADAMTS5 | ADAMTS8 | ADAMTS16 | ADAMTS16 | ||
| ADAMTS14 | ADAMTS2 | ADAMTS2 | |||
| ADAMTS20 | ADAMTS20 | ||||
| ADAMTS12 | ADAMTS12 | ||||
| ADAMTS18 | |||||
| Membrane anchored proteases | |||||
| TMPRSS4 | TMPRSS12 | TMPRSS4 | TMPRSS11E | ||
| TMPRSS11E | TMPRSS15 | TMPRSS15 | |||
| Bone Marrow Morphogenetic Proteins | |||||
| BMP8A | BMP1 | ||||
| FAM5C (BRINP3) | FAM5C (BRINP3) | FAM5C (BRINP3) | |||
| FAM5B (BRINP2) | |||||
| Protease inhibitors | |||||
| CST1 | CST1 | ||||
| CST2 | CST2 | ||||
| CST4 | |||||
| SPINK1 | SPINK1 | SPINK1 | |||
| SPINK13 | SPINK13 | ||||
| SPINK2 | |||||
| SERPINA7 | SERPINB4 | SERPINB3 | |||
| SERPINI1 | SERPINA5 | ||||
| Collagen | |||||
| COL1A1 | COL6A2 | COL6A2 | COL4A3BP | COL22A1 | |
| COL9A2 | COL7A1 | COL22A1 | COL19A1 | ||
| COL10A1 | COL23A1 | COL19A1 | COL25A1 | ||
| COL24A1 | COL25A1 | COL3A1 | |||
| COL3A1 | COL11A1 | ||||
| COL11A1 | COL5A2 | ||||
| COL21A1 | COL14A1 | ||||
| COL1A2 | COL6A3 | ||||
| COL6A3 | COL3A1 | ||||
| COL6A6 | |||||
| COL12A1 | |||||
| Tenascin | |||||
| ODZ1 | ODZ1 | ODZ1 (TENM3) | |||
| ODZ2 | ODZ3 | ODZ3 | |||
| TNR | TNR | TNR | |||
| TNN | TNN | ||||
| Other ECM components | |||||
| FLG | FLG | FLG | |||
| FLG2 | FLG2 | ||||
| Mucin | |||||
| MUC21 | MUC17 | MUC16 | MUC17 | ||
| MUC13 | MUC16 | MUC7 | |||
| MUC5B | |||||
| Laminin Beta Subunit | |||||
| LAMB4 | |||||
| Protocadherin | |||||
| PCDHB2 | PCDHB2 | PCDHGB5 | PCDHB2 | ||
| PCDH15 | PCDH15 | PCDH15 | |||
| PCDHGA2 | PCDHGA2 | PCDHGA2 | |||
| PCDHB7 | PCDHB7 | PCDHB7 | |||
| PCDHGB2 | PCDHGB2 | PCDHB11 | |||
| PCDHA4 | PCDHA4 | PCDHA2 | |||
| PCDHGB3 | PCDHGB3 | PCDHB4 | |||
| PCDHB8 | PCDHB11 | PCDH11X | |||
| PCDHA1 | PCDHA2 | PCDH10 | |||
| PCDHGA3 | PCDHB4 | PCDHA3 | |||
| PCDHGA7 | PCDH11X | PCDH178 | |||
| PCDHGA1 | PCDH10 | PCDHB12 | |||
| PCDHGB4 | PCDHA3 | PCDH18 | |||
| PCDHGC5 | PCDH178 | PCDHB14 | |||
| PCDHGC4 | PCDHB12 | PCDH11Y | |||
| PCDHGA10 | PCDHA6 | PCDH8 | |||
| PCDHGA5 | PCDHB3 | ||||
| PCDHGA9 | PCDH17 | ||||
| PCDHGA12 | |||||
| PCDHGA4 | |||||
| PCDHB6 | |||||
| PCDHGB | |||||
| PCDH19 | |||||
| PCDHGB5 | |||||
| PCDHGA8 | |||||
| PCDHGA11 | |||||
| PCDHGB6 | |||||
| PCDHGC3 | |||||
| PCDHGA6 | |||||
| PCDHGB1 | |||||
| Cadherins | |||||
| CDH3 | CDH10 | CDH10 | CDH10 | ||
| CDH17 | CDH6 | CDH18 | CDH6 | ||
| CDH13 | CDH9 | CDH18 | |||
| CDHR1 | CDH22 | CDH9 | |||
| CDHR2 | CDH7 | CDH22 | |||
| CDH18 | CDH8 | CDH7 | |||
| CDH23 | CDH2 | CDH12 | |||
| CDH11 | |||||
| Atypical cadherins | |||||
| FAT4 | FAT3 | FAT3 | |||
| FAT4 | FAT4 | ||||
| FAT1 | |||||
| Myosin | |||||
| MYO7A | MYO7B | MYO5C | MYO7B | ||
| MYOD1 | MYO18B | ||||
| MYO18A | |||||
| MYO1G | |||||
| Perlecan proteins | |||||
| HSPG2 | |||||
| Serine/threonine kinase | |||||
| STK32A | STK11 | STK11 | |||
| Intergrins | |||||
| INHA | |||||
| IBSP | |||||
| Evolutionary Selection for Mutations or Conservation of Genes Associated with the Cell Membrane | |||||
|---|---|---|---|---|---|
| EGFR-Mut | KRAS-Mut | NEK | |||
| Conserved | Mutation | Conserved | Mutation | Conserved | Mutation |
| Claudin | |||||
| CLDN3 | CLDN15 | CLDN1 | |||
| CLDN2 | |||||
| CLDN6 | |||||
| CLDN9 | |||||
| Gap junctions | |||||
| GJB2 | GJB2 | GJB2 | |||
| GJB6 | GJB6 | ||||
| GJB3 | |||||
| Spectrin family | |||||
| SPTBN5 | |||||
| SPTBN2 | |||||
| Cilia | |||||
| DNAH10 | DNAH11 | DNAH11 | |||
| DNAH9 | DNAH9 | ||||
| DNAH8 | |||||
| DNAH3 | |||||
| DNAH7 | |||||
| DNAH5 | |||||
| BBOF1 | |||||
| KIAA0753 | |||||
| IFT122 | |||||
| SPAG5 | SPAG11B | SPAG4 | |||
| SPAG9 | SPAG4 | ||||
| NEK2 | NEK2 | ||||
| C1orf174 ERICH3 | C1orf174 ERICH3 | C1orf174 ERICH3 | |||
| C16orf59 (TEDC2) | C16orf59 (TEDC2) | C16orf59 (TEDC2) | |||
| Ca++ Voltage gated channels | |||||
| CBARP | CACNA2D1 | CACNA2D2 | CACNA2D1 | CACNA2D2 | CACNA2D1 |
| CACNA1E | CACNA1E | ||||
| CACNA1C | CACNA1C | CACNA1C | |||
| CACNA2D3 | |||||
| K+ Voltage gated channels | |||||
| KCNQ5 | KCNK2 | KCNN4 | KCNK2 | KCNS1 | KCNK2 |
| KCNA5 | KCNA5 | KCNC3 | KCNA5 | ||
| KCNH1 | KCNH1 | KCNK10 | |||
| KCNH8 | KCNJ18 | KCNT2 | |||
| KCNH5 | KCNA4 | KCNK13 | |||
| KCNB2 | |||||
| KCNJ12 | |||||
| KCNJ3 | |||||
| KCNU1 | |||||
| KCNC2 | |||||
| KCNJ2 | |||||
| KCNS2 | |||||
| KCNQ2 | |||||
| K+ channel tetramerization domain-containing family | |||||
| KCTD19 | KCTD8 | ||||
| Hyperpolarization-activated cyclic nucleotide-gated potassium channel | |||||
| HCN1 | HCN1 | ||||
| Na+ channels | |||||
| SCN3A | SCN3A | ||||
| SCN2A | |||||
| Sodium leak Channel, non-selective | |||||
| NALCN | NALCN | ||||
| Acid sensing channel | |||||
| ASIC2 | |||||
| Glutamate-gated ion channel | |||||
| GRIN2A | GRIN2A | ||||
| GRIN2B | GRIN2B | ||||
| GRIN3A | |||||
| Cancer-testis genes | |||||
| XAGE1B | XAGE1B | XAGE1B | |||
| XAGE1D | XAGE1D | XAGE1D | |||
| MAGEH1 | MAGEC1 | MAGED4 | MAGEC1 | ||
| MAGED2 | MAGEC3 | MAGED4B | MAGEC3 | ||
| MAGED1 | MAGEC2 | MAGEA2 | MAGEC2 | ||
| MAGEB4 | MAGEB4 | ||||
| MAGEA10-MAGEA5 | MAGEB18 | ||||
| MAGEB16 | |||||
| MAGEA5 | |||||
| MAGEA6 | |||||
| MAGEA4 | |||||
| CXorf61 CT83 | CXorf61 CT83 | CXorf61 CT83 | |||
| Spermatogenesis | |||||
| FAM75D1 (SPATA31) | FAM75D1 (SPATA31) | ||||
| FAM75A4 (SPATA31A) | FAM75A4 (SPATA31A7) | FAM75A6 (SPATA31A6) | |||
| FAM75C2 (SPATA31C2) | FAM75C2 (SPATA31C2) | ||||
| FAM75A5 (SPATA31A5) | |||||
| C15orf2 (NPAP1) | C15orf2 (NPAP1) | C15orf2 (NPAP1) | |||
| CXorf59 (CFAP47) | CXorf59 (CFAP47) | CXorf59 (CFAP47) | |||
| Fucosyltransferases | |||||
| FUT2 | FUT4 | ||||
| FUT9 | FUT2 | ||||
| FUT3 | |||||
| FUT6 | |||||
| Solute Carriers | |||||
| SLC1A7 | SLC5A11 | SLC2A1 | SLC6A5 | SLC1A4 | SLC8A1 |
| SLC2A1 | SLC6A13 | SLC7A5 | SLC6A15 | SLC2A1 | SLC10A2 |
| SLC6A11 | SLC6A17 | SLC7A10 | SLC8A1 | SLC7A8 | SLC12A1 |
| SLC6A3 | SLC10A1 | SLC24A1 | SLC8A3 | SLC7A11 | SLC17A3 |
| SLC7A10 | SLC34A2 | SLC26A8 | SLC9A4 | SLC19A1 | SLC17A6 |
| SLC15A1 | SLCO1B1 | SLC17A6 SLC17A6 | SLC22A5 | SLC35F1 | |
| SLC16A9 | SLCO4A1 | SLC27A6 | SLC34A3 | SLC39A12 | |
| SLC29A4 | SLC30A10 | SLC44A2 | SLC44A5 | ||
| SLC52A1 | SLC39A12 | SLC44A4 | |||
| SLCO5A1 | |||||
| Transmembrane proteins | |||||
| TMEM184A | TMEM200A | TMEM184A | TMEM200A | TMEM180 | TMEM196 |
| TMEM63C | TMEM201 | TMEM132 | |||
| TMEM156 | |||||
| TMEM59L | |||||
| ATP-Binding Cassettes | |||||
| ABCA4 | ABCB5 | ABCB5 | ABCF1 | ABCB5 | |
| ABCC3 | ABCA13 | ABCB1 | |||
| ATP2A1 | ABCA13 | ||||
| ATP8B3 | |||||
| ATP2A2 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Freischel, A.R.; Teer, J.K.; Luddy, K.; Cunningham, J.; Artzy-Randrup, Y.; Epstein, T.; Tsai, K.Y.; Berglund, A.; Cleveland, J.L.; Gillies, R.J.; et al. Evolutionary Analysis of TCGA Data Using Over- and Under- Mutated Genes Identify Key Molecular Pathways and Cellular Functions in Lung Cancer Subtypes. Cancers 2023, 15, 18. https://doi.org/10.3390/cancers15010018
Freischel AR, Teer JK, Luddy K, Cunningham J, Artzy-Randrup Y, Epstein T, Tsai KY, Berglund A, Cleveland JL, Gillies RJ, et al. Evolutionary Analysis of TCGA Data Using Over- and Under- Mutated Genes Identify Key Molecular Pathways and Cellular Functions in Lung Cancer Subtypes. Cancers. 2023; 15(1):18. https://doi.org/10.3390/cancers15010018
Chicago/Turabian StyleFreischel, Audrey R., Jamie K. Teer, Kimberly Luddy, Jessica Cunningham, Yael Artzy-Randrup, Tamir Epstein, Kenneth Y. Tsai, Anders Berglund, John L. Cleveland, Robert J. Gillies, and et al. 2023. "Evolutionary Analysis of TCGA Data Using Over- and Under- Mutated Genes Identify Key Molecular Pathways and Cellular Functions in Lung Cancer Subtypes" Cancers 15, no. 1: 18. https://doi.org/10.3390/cancers15010018
APA StyleFreischel, A. R., Teer, J. K., Luddy, K., Cunningham, J., Artzy-Randrup, Y., Epstein, T., Tsai, K. Y., Berglund, A., Cleveland, J. L., Gillies, R. J., Brown, J. S., & Gatenby, R. A. (2023). Evolutionary Analysis of TCGA Data Using Over- and Under- Mutated Genes Identify Key Molecular Pathways and Cellular Functions in Lung Cancer Subtypes. Cancers, 15(1), 18. https://doi.org/10.3390/cancers15010018






