Multi-Transcriptomic Analysis Reveals the Heterogeneity and Tumor-Promoting Role of SPP1/CD44-Mediated Intratumoral Crosstalk in Gastric Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bulk RNA-Seq Dataset Retrieval and Analysis
2.2. Western Blot
2.3. Tissue Microarray (TMA), Clinical Samples, and IHC
2.4. Single-Cell RNA Sequencing (scRNA-seq) Data Acquisition, Processing, and Analysis
2.5. Cell—Cell Communication
2.6. Pathway Enrichment Analysis
2.7. Spatial Transcriptomic Data Collection, Quality Control, and Analysis
2.8. Statistical Analysis
3. Results
3.1. Expression Pattern of SPP1 in GC Specimens
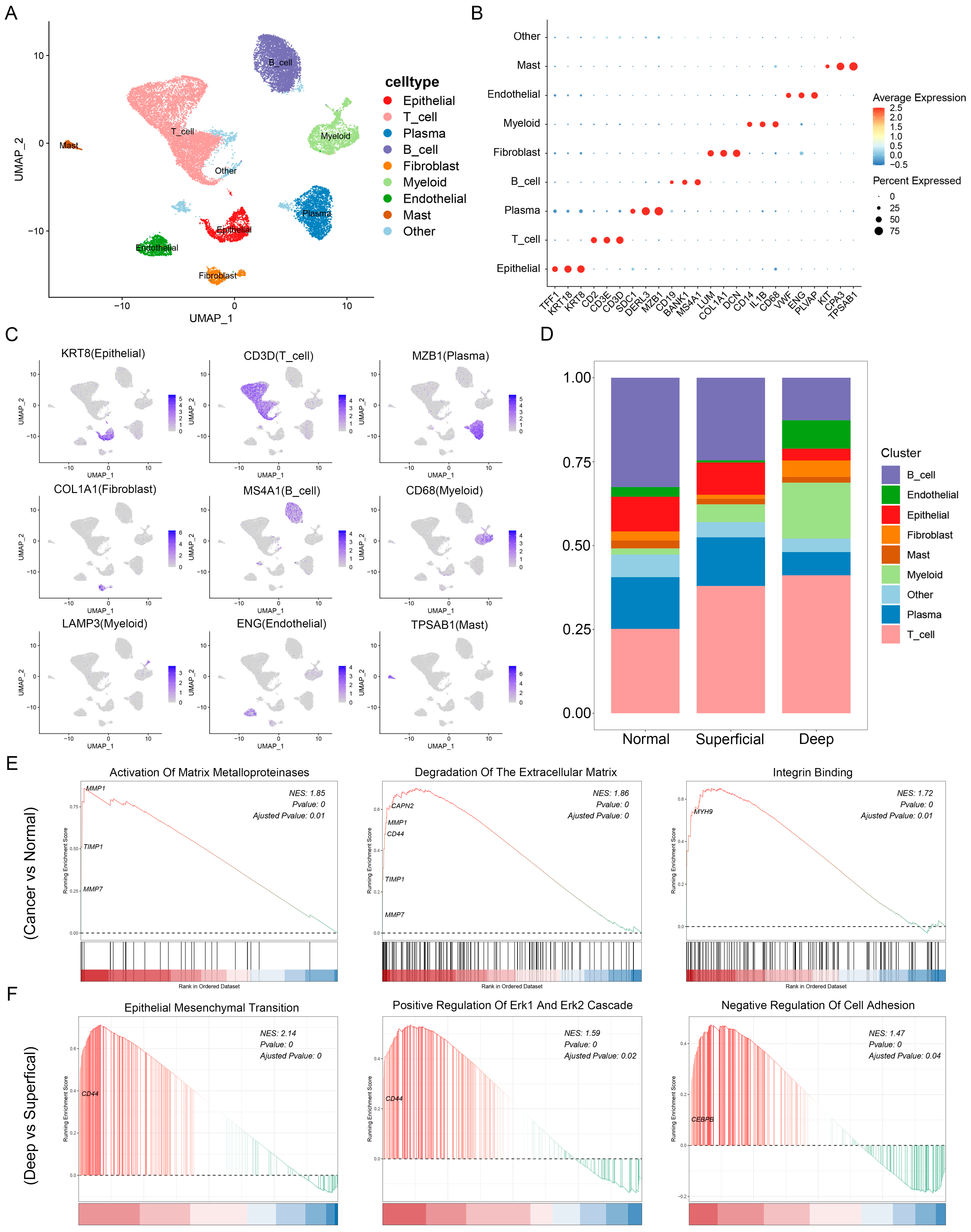
3.2. Single-Cell Landscape of GC
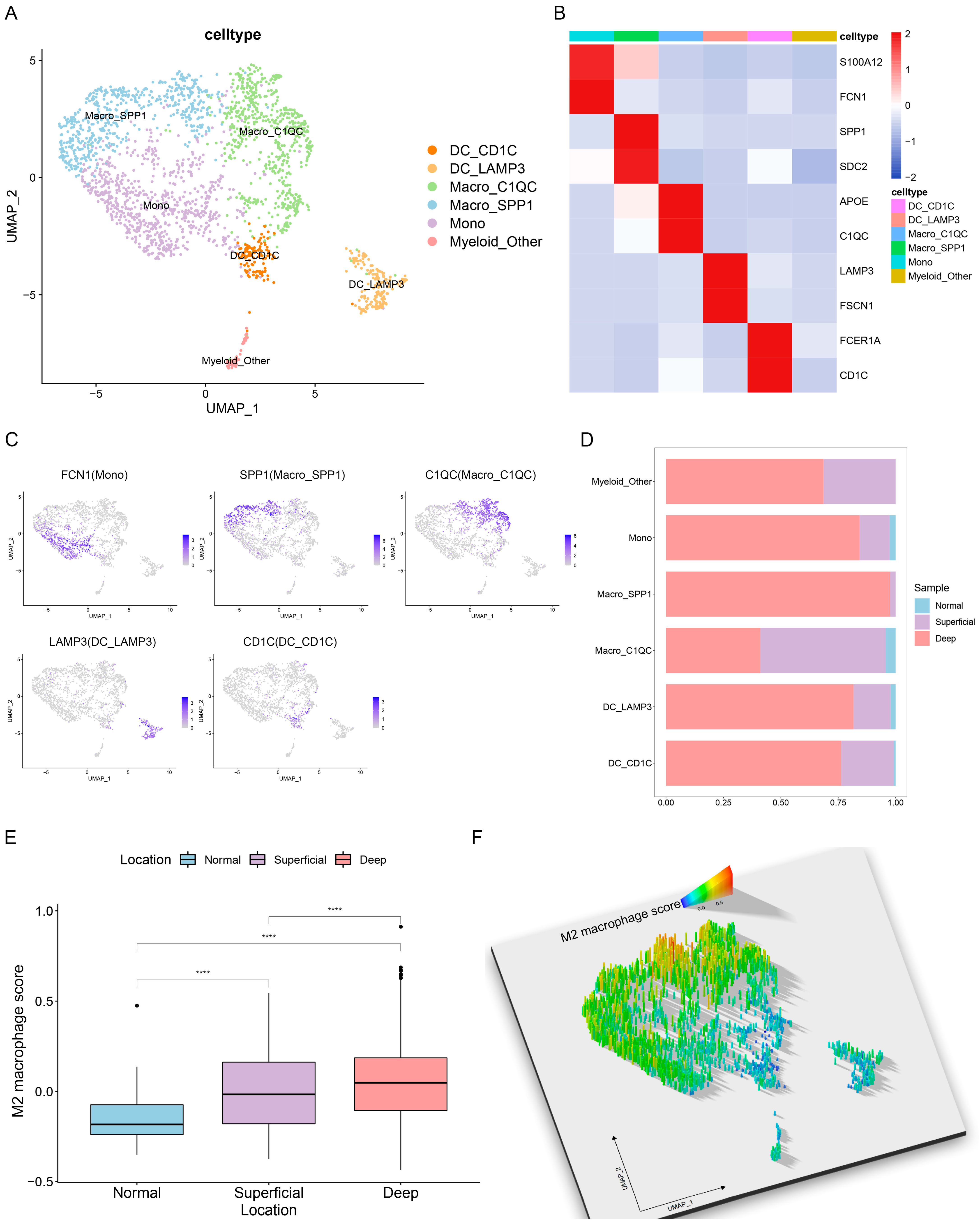
3.3. Single-Cell Characterization of Tumor-Infiltrating Myeloid Cells in GC
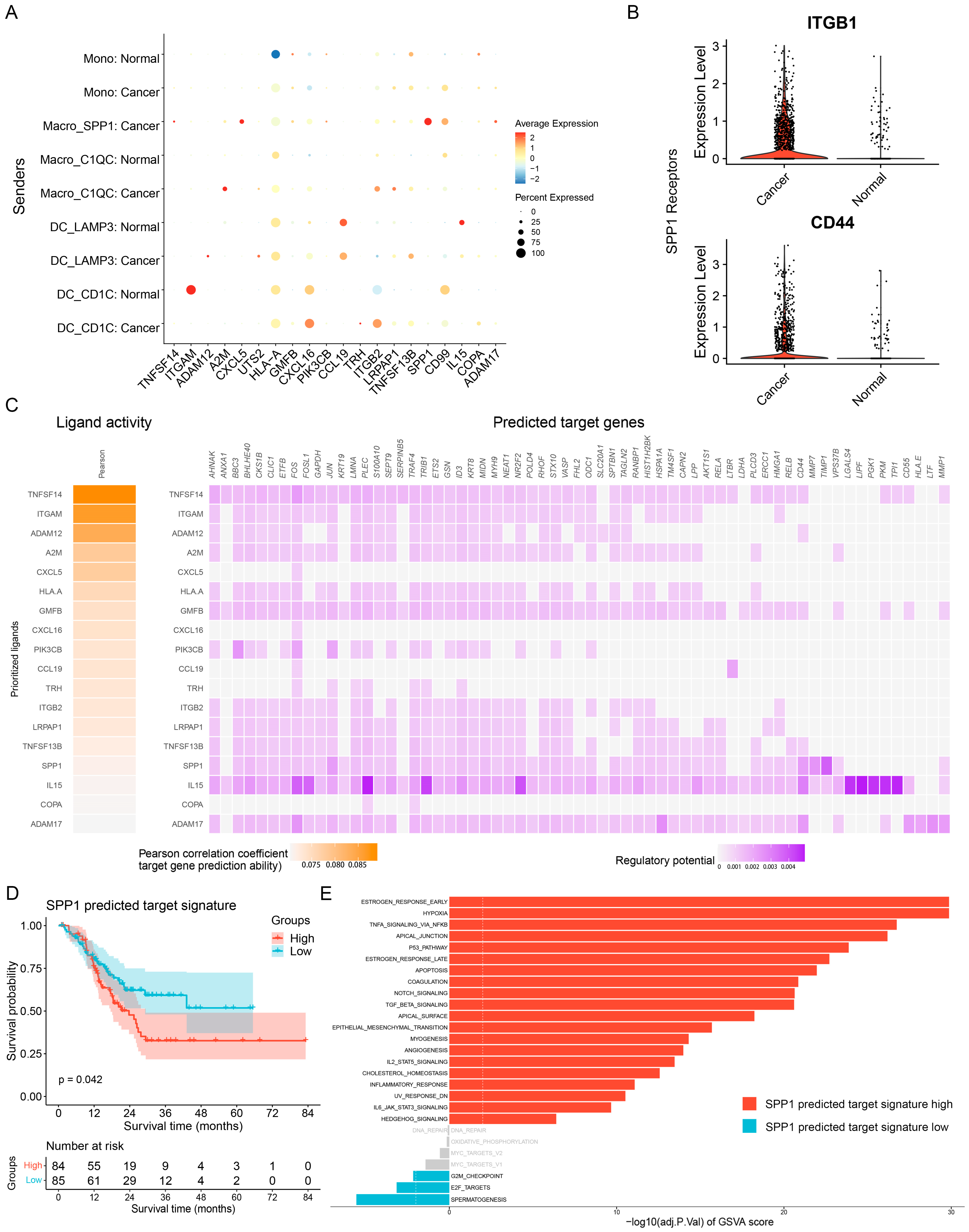
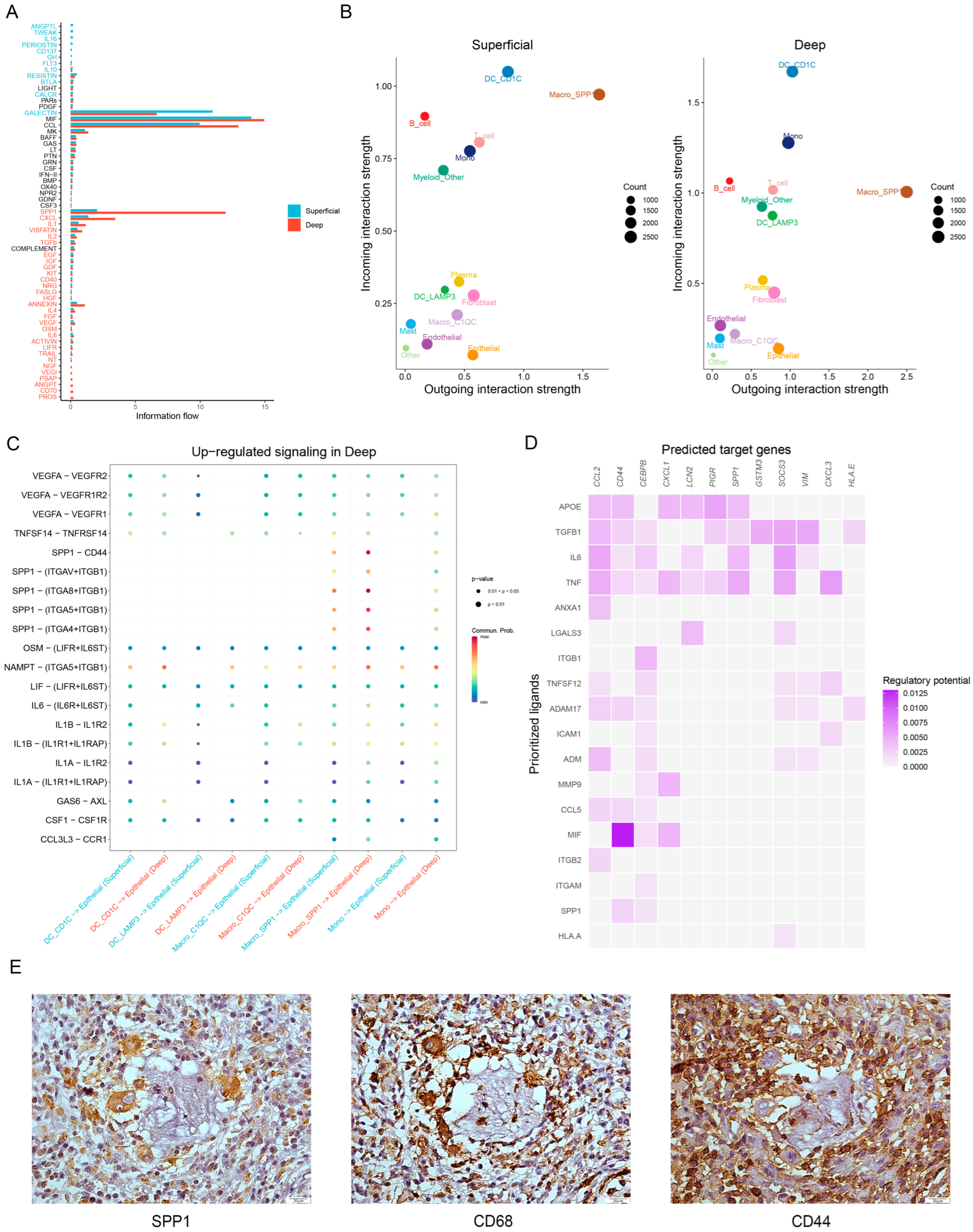
3.4. Ligand—Receptor Interactions in the TME
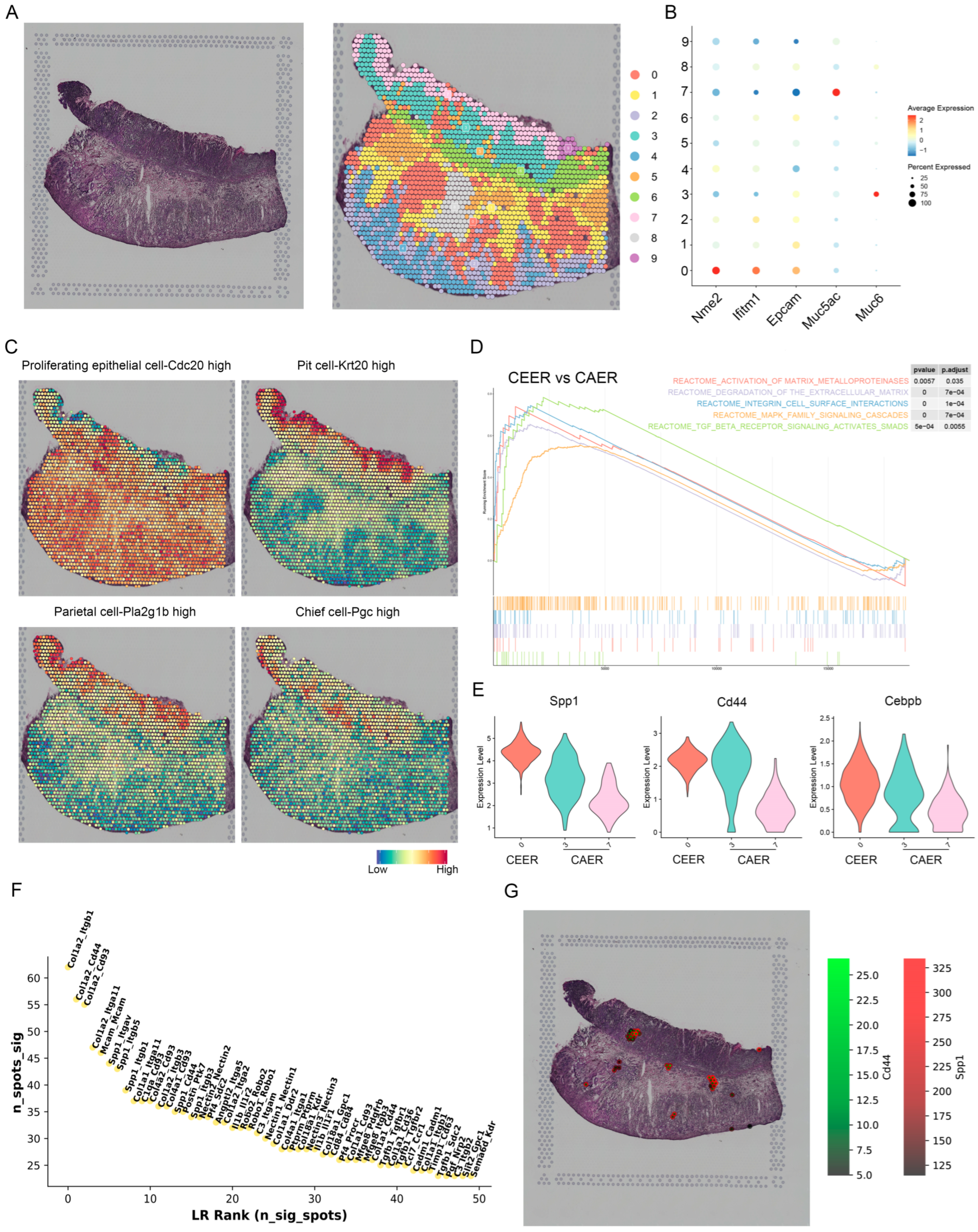
3.5. Spatial Expression Characteristics of SPP1 Revealed by Spatial Transcriptomics
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yeoh, K.G.; Tan, P. Mapping the genomic diaspora of gastric cancer. Nat. Rev. Cancer 2022, 22, 71–84. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Zhuo, H.; Wang, L.; Zheng, W.; Chen, X.; Hou, J.; Zhao, J.; Cai, J. Identification of the Combinatorial Effect of miRNA Family Regulatory Network in Different Growth Patterns of GC. Mol. Oncolytics 2020, 17, 531–546. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Zhuo, H.; Xu, M.; Wang, L.; Xu, H.; Peng, J.; Hou, J.; Lin, L.; Cai, J. Regulatory network of circRNA-miRNA-mRNA contributes to the histological classification and disease progression in gastric cancer. J. Transl. Med. 2018, 16, 216. [Google Scholar] [CrossRef]
- Cao, S.; Wang, J.R.; Ji, S.; Yang, P.; Dai, Y.; Guo, S.; Montierth, M.D.; Shen, J.P.; Zhao, X.; Chen, J. Estimation of tumor cell total mRNA expression in 15 cancer types predicts disease progression. Nat. Biotechnol. 2022, 40, 1624–1633. [Google Scholar] [CrossRef]
- Grünwald, B.T.; Devisme, A.; Andrieux, G.; Vyas, F.; Aliar, K.; McCloskey, C.W.; Macklin, A.; Jang, G.H.; Denroche, R.; Romero, J.M.; et al. Spatially confined sub-tumor microenvironments in pancreatic cancer. Cell 2021, 184, 5577–5592.e18. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Seehawer, M.; Polyak, K. Untangling the web of intratumour heterogeneity. Nat. Cell Biol. 2022, 24, 1192–1201. [Google Scholar] [CrossRef] [PubMed]
- Pittet, M.J.; Michielin, O.; Migliorini, D. Clinical relevance of tumour-associated macrophages. Nat. Rev. Clin. Oncol. 2022, 19, 402–421. [Google Scholar] [CrossRef]
- Casanova-Acebes, M.; Dalla, E.; Leader, A.M.; LeBerichel, J.; Nikolic, J.; Morales, B.M.; Brown, M.; Chang, C.; Troncoso, L.; Chen, S.T.; et al. Tissue-resident macrophages provide a pro-tumorigenic niche to early NSCLC cells. Nature 2021, 595, 578–584. [Google Scholar] [CrossRef]
- Ren, X.; Zhang, L.; Zhang, Y.; Li, Z.; Siemers, N.; Zhang, Z. Insights Gained from Single-Cell Analysis of Immune Cells in the Tumor Microenvironment. Annu. Rev. Immunol. 2021, 39, 583–609. [Google Scholar] [CrossRef]
- Qi, J.; Sun, H.; Zhang, Y.; Wang, Z.; Xun, Z.; Li, Z.; Ding, X.; Bao, R.; Hong, L.; Jia, W.; et al. Single-cell and spatial analysis reveal interaction of FAP(+) fibroblasts and SPP1(+) macrophages in colorectal cancer. Nat. Commun. 2022, 13, 1742. [Google Scholar] [CrossRef]
- Briones-Orta, M.A.; Avendaño-Vázquez, S.E.; Aparicio-Bautista, D.I.; Coombes, J.D.; Weber, G.F.; Syn, W.K. Osteopontin splice variants and polymorphisms in cancer progression and prognosis. Biochim. Biophys. Acta Rev. Cancer 2017, 1868, 93–108.A. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; LeSavage, B.L.; Hubka, K.M.; Ma, C.; Natarajan, S.; Eggold, J.T.; Xiao, Y.; Fuh, K.C.; Krishnan, V.; Enejder, A.; et al. Cancer-associated mesothelial cells promote ovarian cancer chemoresistance through paracrine osteopontin signaling. J. Clin. Investig. 2021, 131, e146186. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Marisetty, A.; Schrand, B.; Gabrusiewicz, K.; Hashimoto, Y.; Ott, M.; Grami, Z.; Kong, L.Y.; Ling, X.; Caruso, H.; et al. Osteopontin mediates glioblastoma-associated macrophage infiltration and is a potential therapeutic target. J. Clin. Investig. 2019, 129, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Deng, G.; Zeng, F.; Su, J.; Zhao, S.; Hu, R.; Zhu, W.; Hu, S.; Chen, X.; Yin, M. BET inhibitor suppresses melanoma progression via the noncanonical NF-κB/SPP1 pathway. Theranostics 2020, 10, 11428–11443. [Google Scholar] [CrossRef] [PubMed]
- Di Bartolomeo, M.; Pietrantonio, F.; Pellegrinelli, A.; Martinetti, A.; Mariani, L.; Daidone, M.G.; Bajetta, E.; Pelosi, G.; de Braud, F.; Floriani, I.; et al. Osteopontin, E-cadherin, and β-catenin expression as prognostic biomarkers in patients with radically resected gastric cancer. Gastric Cancer 2016, 19, 412–420. [Google Scholar] [CrossRef] [PubMed]
- Chivu-Economescu, M.; Necula, L.G.; Matei, L.; Dragu, D.; Bleotu, C.; Sorop, A.; Herlea, V.; Dima, S.; Popescu, I.; Diaconu, C.C. Collagen Family and Other Matrix Remodeling Proteins Identified by Bioinformatics Analysis as Hub Genes Involved in Gastric Cancer Progression and Prognosis. Int. J. Mol. Sci. 2022, 23, 3214. [Google Scholar] [CrossRef]
- Matsubara, E.; Komohara, Y.; Esumi, S.; Shinchi, Y.; Ishizuka, S.; Mito, R.; Pan, C.; Yano, H.; Kobayashi, D.; Fujiwara, Y.; et al. SPP1 Derived from Macrophages Is Associated with a Worse Clinical Course and Chemo-Resistance in Lung Adenocarcinoma. Cancers 2022, 14, 4374. [Google Scholar] [CrossRef]
- Lamort, A.S.; Giopanou, I.; Psallidas, I.; Stathopoulos, G.T. Osteopontin as a Link between Inflammation and Cancer: The Thorax in the Spotlight. Cells 2019, 8, 815. [Google Scholar] [CrossRef]
- Vivian, J.; Rao, A.A.; Nothaft, F.A.; Ketchum, C.; Armstrong, J.; Novak, A.; Pfeil, J.; Narkizian, J.; Deran, A.D.; Musselman-Brown, A.; et al. Toil enables reproducible, open source, big biomedical data analyses. Nat. Biotechnol. 2017, 35, 314–316. [Google Scholar] [CrossRef] [PubMed]
- Ming, S.C. Gastric carcinoma. A pathobiological classification. Cancer 1977, 39, 2475–2485. [Google Scholar] [CrossRef]
- Lin, A.; Qi, C.; Wei, T.; Li, M.; Cheng, Q.; Liu, Z.; Luo, P.; Zhang, J. CAMOIP: A web server for comprehensive analysis on multi-omics of immunotherapy in pan-cancer. Brief Bioinform. 2022, 23, bbac129. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.Y.; Lin, L.Y.; Hao, H.; Zhang, S.M.; Ma, F.; Hong, X.X.; Zhang, H.; Liu, Q.F.; Ye, G.D.; Sun, G.B.; et al. Yes-associated protein/TEA domain family member and hepatocyte nuclear factor 4-alpha (HNF4α) repress reciprocally to regulate hepatocarcinogenesis in rats and mice. Hepatology 2017, 65, 1206–1221. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.Y.; Ham, I.H.; Lee, S.H.; Ryu, D.; Son, S.Y.; Han, S.U.; Kim, T.M.; Hur, H. Spatially Distinct Reprogramming of the Tumor Microenvironment Based On Tumor Invasion in Diffuse-Type Gastric Cancers. Clin. Cancer Res. 2021, 27, 6529–6542. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Li, Z.; Gao, R.; Xing, B.; Gao, Y.; Yang, Y.; Qin, S.; Zhang, L.; Ouyang, H.; Du, P.; et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell 2021, 184, 792–809.e23. [Google Scholar] [CrossRef] [PubMed]
- Browaeys, R.; Saelens, W.; Saeys, Y. NicheNet: Modeling intercellular communication by linking ligands to target genes. Nat. Methods 2020, 17, 159–162. [Google Scholar] [CrossRef]
- Jin, S.; Guerrero-Juarez, C.F.; Zhang, L.; Chang, I.; Ramos, R.; Kuan, C.H.; Myung, P.; Plikus, M.V.; Nie, Q. Inference and analysis of cell-cell communication using CellChat. Nat. Commun. 2021, 12, 1088. [Google Scholar] [CrossRef]
- Pham, D.; Tan, X.; Xu, J.; Grice, L.F.; Lam, P.Y.; Raghubar, A.; Vukovic, J.; Ruitenberg, M.J.; Nguyen, Q. stLearn: Integrating spatial location, tissue morphology and gene expression to find cell types, cell-cell interactions and spatial trajectories within undissociated tissues. bioRxiv 2020. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Yamasaki, J.; Hirata, Y.; Otsuki, Y.; Suina, K.; Saito, Y.; Masuda, K.; Okazaki, S.; Ishimoto, T.; Saya, H.; Nagano, O. MEK inhibition suppresses metastatic progression of KRAS-mutated gastric cancer. Cancer Sci. 2022, 113, 916–925. [Google Scholar] [CrossRef]
- Fei, L.; Chen, H.; Ma, L.; Weigao, E.; Wang, R.; Fang, X.; Zhou, Z.; Sun, H.; Wang, J.; Jiang, M.; et al. Systematic identification of cell-fate regulatory programs using a single-cell atlas of mouse development. Nat. Genet. 2022, 54, 1051–1061. [Google Scholar] [CrossRef]
- Zhang, L.; Li, Z.; Skrzypczynska, K.M.; Fang, Q.; Zhang, W.; O’Brien, S.A.; He, Y.; Wang, L.; Zhang, Q.; Kim, A.; et al. Single-Cell Analyses Inform Mechanisms of Myeloid-Targeted Therapies in Colon Cancer. Cell 2020, 181, 442–459.e29. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Miao, J.; Wang, S.; Shen, R.; Zhang, S.; Tian, Y.; Li, M.; Zhu, D.; Yao, A.; Bao, W.; et al. Single-cell RNA-seq reveals the genesis and heterogeneity of tumor microenvironment in pancreatic undifferentiated carcinoma with osteoclast-like giant-cells. Mol. Cancer 2022, 21, 133. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Qu, J.; Qi, Y.; Duan, Y.; Huang, Y.W.; Zhou, Z.; Li, P.; Yao, J.; Huang, B.; Zhang, S.; et al. EZH2 engages TGFβ signaling to promote breast cancer bone metastasis via integrin β1-FAK activation. Nat. Commun. 2022, 13, 2543. [Google Scholar] [CrossRef] [PubMed]
- Nallasamy, P.; Nimmakayala, R.K.; Karmakar, S.; Leon, F.; Seshacharyulu, P.; Lakshmanan, I.; Rachagani, S.; Mallya, K.; Zhang, C.; Ly, Q.P.; et al. Pancreatic Tumor Microenvironment Factor Promotes Cancer Stemness via SPP1-CD44 Axis. Gastroenterology 2021, 161, 1998–2013.e7. [Google Scholar] [CrossRef]
- Du, S.; Yang, Z.; Lu, X.; Yousuf, S.; Zhao, M.; Li, W.; Miao, J.; Wang, X.; Yu, H.; Zhu, X.; et al. Anoikis resistant gastric cancer cells promote angiogenesis and peritoneal metastasis through C/EBPβ-mediated PDGFB autocrine and paracrine signaling. Oncogene 2021, 40, 5764–5779. [Google Scholar] [CrossRef]
- Wu, H.; Liu, B.; Chen, Z.; Li, G.; Zhang, Z. MSC-induced lncRNA HCP5 drove fatty acid oxidation through miR-3619-5p/AMPK/PGC1α/CEBPB axis to promote stemness and chemo-resistance of gastric cancer. Cell Death Dis. 2020, 11, 233. [Google Scholar] [CrossRef]
- Lee, J.; Goh, S.H.; Song, N.; Hwang, J.A.; Nam, S.; Choi, I.J.; Shin, A.; Kim, I.H.; Ju, M.H.; Jeong, J.S.; et al. Overexpression of IFITM1 has clinicopathologic effects on gastric cancer and is regulated by an epigenetic mechanism. Am. J. Pathol. 2012, 181, 43–52. [Google Scholar] [CrossRef]
- Qi, Y.; Wei, J.; Zhang, X. Requirement of transcription factor NME2 for the maintenance of the stemness of gastric cancer stem-like cells. Cell Death Dis. 2021, 12, 924. [Google Scholar] [CrossRef]
- Lu, X.Q.; Zhang, J.Q.; Zhang, S.X.; Qiao, J.; Qiu, M.T.; Liu, X.R.; Chen, X.X.; Gao, C.; Zhang, H.H. Identification of novel hub genes associated with gastric cancer using integrated bioinformatics analysis. BMC Cancer 2021, 21, 697. [Google Scholar] [CrossRef]
- Wu, K.; Lin, K.; Li, X.; Yuan, X.; Xu, P.; Ni, P.; Xu, D. Redefining Tumor-Associated Macrophage Subpopulations and Functions in the Tumor Microenvironment. Front. Immunol. 2020, 11, 1731. [Google Scholar] [CrossRef]
- Pasello, G.; Zago, G.; Lunardi, F.; Urso, L.; Kern, I.; Vlacic, G.; Grosso, F.; Mencoboni, M.; Ceresoli, G.L.; Schiavon, M.; et al. Malignant pleural mesothelioma immune microenvironment and checkpoint expression: Correlation with clinical-pathological features and intratumor heterogeneity over time. Ann. Oncol. 2018, 29, 1258–1265. [Google Scholar] [CrossRef] [PubMed]
- Tobin, J.W.D.; Keane, C.; Gunawardana, J.; Mollee, P.; Birch, S.; Hoang, T.; Lee, J.; Li, L.; Huang, L.; Murigneux, V.; et al. Progression of Disease Within 24 Months in Follicular Lymphoma Is Associated With Reduced Intratumoral Immune Infiltration. J. Clin. Oncol. 2019, 37, 3300–3309. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Paz, K.; Flynn, R.; Vulic, A.; Robinson, T.M.; Lineburg, K.E.; Alexander, K.A.; Meng, J.; Roy, S.; Panoskaltsis-Mortari, A.; et al. Pirfenidone ameliorates murine chronic GVHD through inhibition of macrophage infiltration and TGF-β production. Blood 2017, 129, 2570–2580. [Google Scholar] [CrossRef] [PubMed]
- Baghban, R.; Roshangar, L.; Jahanban-Esfahlan, R.; Seidi, K.; Ebrahimi-Kalan, A.; Jaymand, M.; Kolahian, S.; Javaheri, T.; Zare, P. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun. Signal. 2020, 18, 59. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, L.; Ju, X.; Wang, S.; Qie, J. Single-Cell Transcriptomic Analysis Reveals Macrophage-Tumor Crosstalk in Hepatocellular Carcinoma. Front. Immunol. 2022, 13, 955390. [Google Scholar] [CrossRef]
- Messex, J.K.; Byrd, C.J.; Thomas, M.U.; Liou, G.Y. Macrophages Cytokine Spp1 Increases Growth of Prostate Intraepithelial Neoplasia to Promote Prostate Tumor Progression. Int. J. Mol. Sci. 2022, 23, 4247. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, W.; Cheng, J.; Hong, Z.; Cai, W.; Zhuo, H.; Hou, J.; Lin, L.; Wei, X.; Wang, K.; Chen, X.; et al. Multi-Transcriptomic Analysis Reveals the Heterogeneity and Tumor-Promoting Role of SPP1/CD44-Mediated Intratumoral Crosstalk in Gastric Cancer. Cancers 2023, 15, 164. https://doi.org/10.3390/cancers15010164
Xie W, Cheng J, Hong Z, Cai W, Zhuo H, Hou J, Lin L, Wei X, Wang K, Chen X, et al. Multi-Transcriptomic Analysis Reveals the Heterogeneity and Tumor-Promoting Role of SPP1/CD44-Mediated Intratumoral Crosstalk in Gastric Cancer. Cancers. 2023; 15(1):164. https://doi.org/10.3390/cancers15010164
Chicago/Turabian StyleXie, Wen, Jia Cheng, Zhijun Hong, Wangyu Cai, Huiqin Zhuo, Jingjing Hou, Lingyun Lin, Xujin Wei, Kang Wang, Xin Chen, and et al. 2023. "Multi-Transcriptomic Analysis Reveals the Heterogeneity and Tumor-Promoting Role of SPP1/CD44-Mediated Intratumoral Crosstalk in Gastric Cancer" Cancers 15, no. 1: 164. https://doi.org/10.3390/cancers15010164
APA StyleXie, W., Cheng, J., Hong, Z., Cai, W., Zhuo, H., Hou, J., Lin, L., Wei, X., Wang, K., Chen, X., Song, Y., Wang, Z., & Cai, J. (2023). Multi-Transcriptomic Analysis Reveals the Heterogeneity and Tumor-Promoting Role of SPP1/CD44-Mediated Intratumoral Crosstalk in Gastric Cancer. Cancers, 15(1), 164. https://doi.org/10.3390/cancers15010164







