Glioblastoma Embryonic-like Stem Cells Exhibit Immune-Evasive Phenotype
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Single-Cell RNA-Seq Data Analysis
2.2. Cell Lines and Culture Conditions
2.3. Lentiviral Production and ic-GSCs Derivation
2.4. Immunostaining
2.5. Cell Sorting
2.6. RNA-Sequencing Profiling and Analysis
2.7. DNA Methylation Profiling and Analysis
2.8. Statistics
3. Results
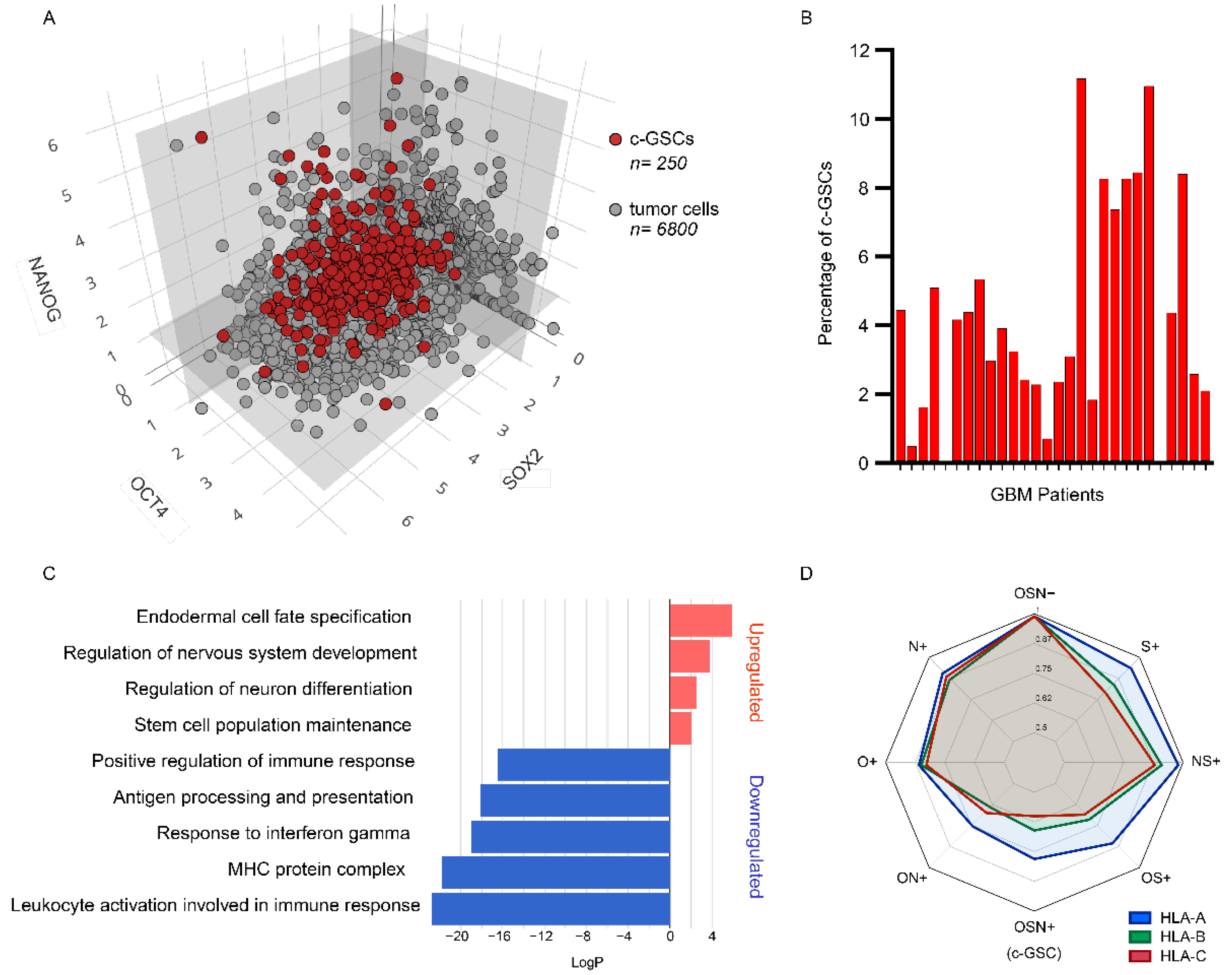
3.1. Identification and Characterization of Core-Glioma Stem Cells
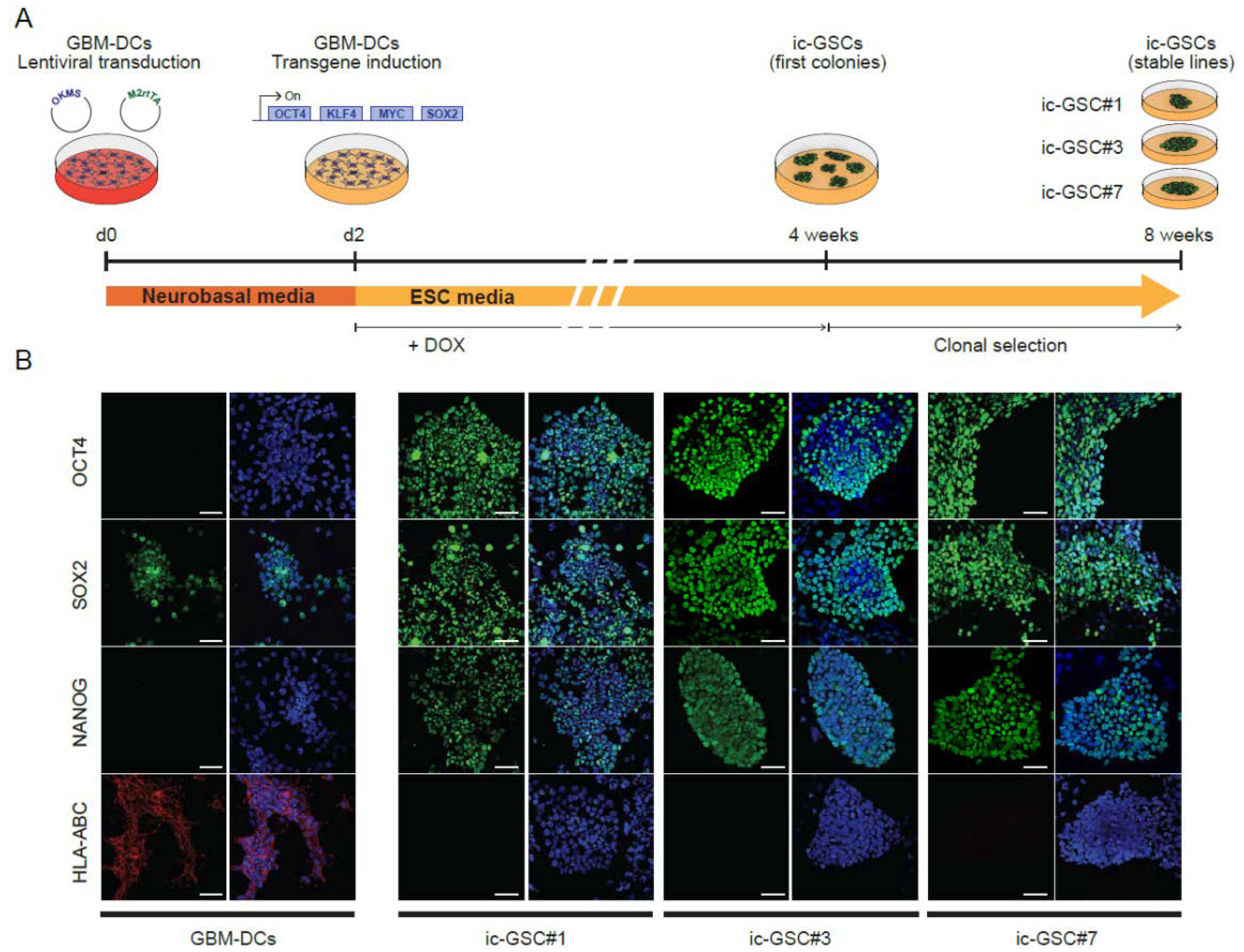
3.2. Reprogramming of Glioblastoma-Derived Cells Mimics the Core-Glioma Stem Cell Phenotype In Vitro
3.3. Gene Expression Analysis of Induced Core-Glioma Stem Cells
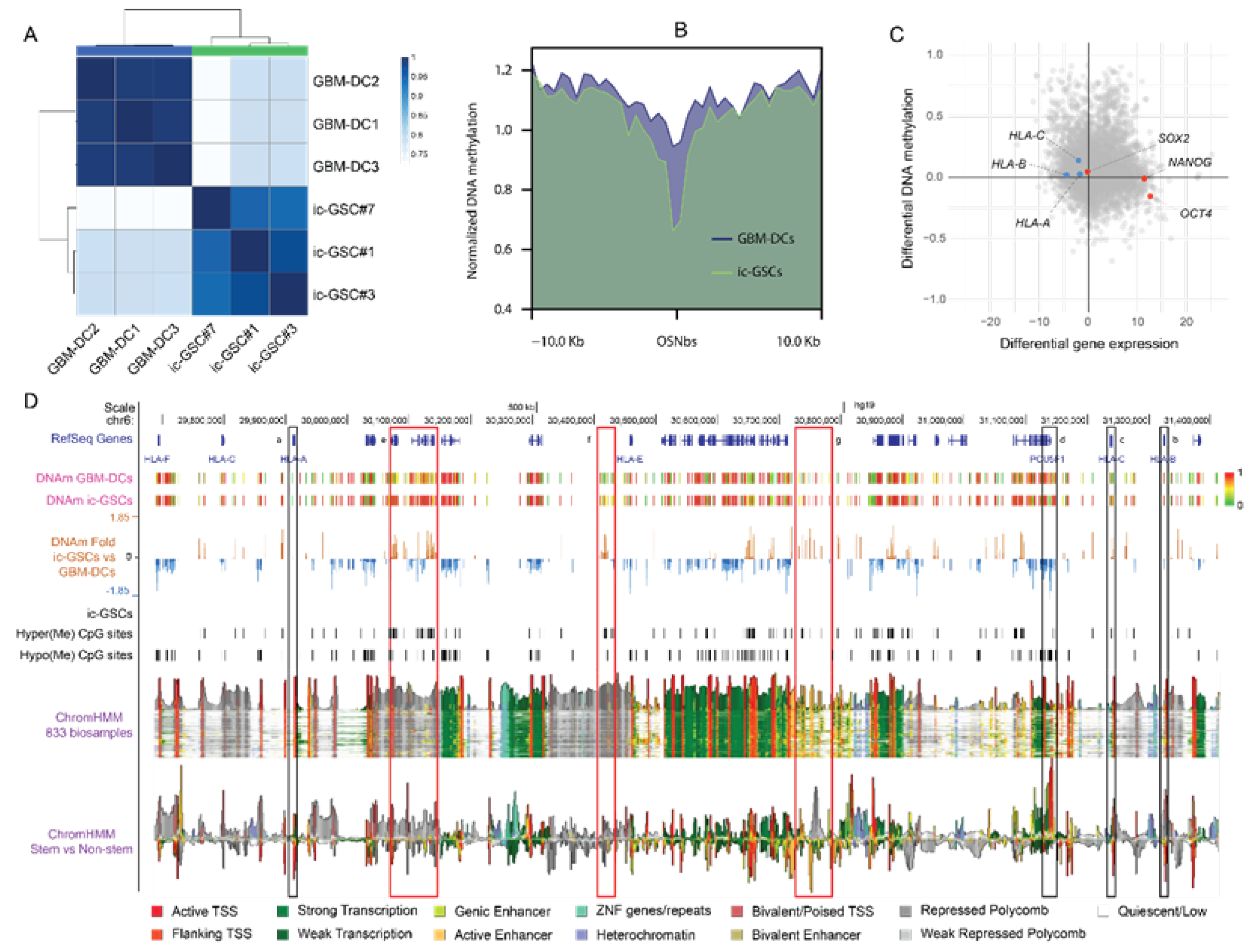
3.4. Epigenetic Changes behind the ic-GSC Transcriptome Reprogramming
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Gallego, O. Nonsurgical Treatment of Recurrent Glioblastoma. Curr. Oncol. 2015, 22, 273–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narita, Y.; Arakawa, Y.; Yamasaki, F.; Nishikawa, R.; Aoki, T.; Kanamori, M.; Nagane, M.; Kumabe, T.; Hirose, Y.; Ichikawa, T.; et al. A randomized, double-blind, phase III trial of personalized peptide vaccination for recurrent glioblastoma. Neuro-Oncology 2018, 21, 348–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gimple, R.C.; Bhargava, S.; Dixit, D.; Rich, J.N. Glioblastoma stem cells: Lessons from the tumor hierarchy in a lethal cancer. Genes Dev. 2019, 33, 591–609. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.-Q.; Chen, J.-T.; Zheng, M.-C.; Yang, L.-J.; Wang, J.-M.; Liu, Q.-L.; Chen, L.-F.; Ye, Z.-C.; Lin, J.-M.; Lin, Z.-X. Nestin+/CD31+ cells in the hypoxic perivascular niche regulate glioblastoma chemoresistance by upregulating JAG1 and DLL4. Neuro-Oncology 2021, 23, 905–919. [Google Scholar] [CrossRef] [PubMed]
- Alvarado, A.G.; Thiagarajan, P.S.; Mulkearns-Hubert, E.E.; Silver, D.J.; Hale, J.S.; Alban, T.; Turaga, S.; Jarrar, A.; Reizes, O.; Longworth, M.S.; et al. Glioblastoma Cancer Stem Cells Evade Innate Immune Suppression of Self-Renewal through Reduced TLR4 Expression. Cell Stem Cell 2017, 20, 450–461.e4. [Google Scholar] [CrossRef] [Green Version]
- Pollard, S.M.; Yoshikawa, K.; Clarke, I.D.; Danovi, D.; Stricker, S.; Russell, R.; Bayani, J.; Head, R.; Lee, M.; Bernstein, M.; et al. Glioma Stem Cell Lines Expanded in Adherent Culture Have Tumor-Specific Phenotypes and Are Suitable for Chemical and Genetic Screens. Cell Stem Cell 2009, 4, 568–580. [Google Scholar] [CrossRef] [Green Version]
- Reya, T.; Morrison, S.J.; Clarke, M.F.; Weissman, I.L. Stem cells, cancer, and cancer stem cells. Nature 2001, 414, 105–111. [Google Scholar] [CrossRef] [Green Version]
- Ben-Porath, I.; Thomson, M.W.; Carey, V.J.; Ge, R.; Bell, G.W.; Regev, A.; Weinberg, R.A. An embryonic stem cell–like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008, 40, 499–507. [Google Scholar] [CrossRef]
- Wong, D.J.; Liu, H.; Ridky, T.; Cassarino, D.; Segal, E.; Chang, H.Y. Module Map of Stem Cell Genes Guides Creation of Epithelial Cancer Stem Cells. Cell Stem Cell 2008, 2, 333–344. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Woo, A.; Chu, J.; Snow, J.W.; Fujiwara, Y.; Kim, C.G.; Cantor, A.B.; Orkin, S.H. A Myc Network Accounts for Similarities between Embryonic Stem and Cancer Cell Transcription Programs. Cell 2010, 143, 313–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradshaw, A.; Wickremesekera, A.; Brasch, H.D.; Chibnall, A.M.; Davis, P.F.; Tan, S.T.; Itinteang, T. Cancer Stem Cells in Glioblastoma Multiforme. Front. Surg. 2016, 3, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elsir, T.; Edqvist, P.; Carlson, J.; Ribom, D.; Bergqvist, M.; Ekman, S.; Popova, S.N.; Alafuzoff, I.; Ponten, F.; Nistér, M.; et al. A study of embryonic stem cell-related proteins in human astrocytomas: Identification of Nanog as a predictor of survival. Int. J. Cancer 2014, 134, 1123–1131. [Google Scholar] [CrossRef] [PubMed]
- Neftel, C.; Laffy, J.; Filbin, M.G.; Hara, T.; Shore, M.E.; Rahme, G.J.; Richman, A.R.; Silverbush, D.; Shaw, M.L.; Hebert, C.M.; et al. An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell 2019, 178, 835–849.e21. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Bergström, T.; Jiang, Y.; Johansson, P.; Marinescu, V.D.; Lindberg, N.; Segerman, A.; Wicher, G.; Niklasson, M.; Baskaran, S.; et al. The Human Glioblastoma Cell Culture Resource: Validated Cell Models Representing All Molecular Subtypes. EBioMedicine 2015, 2, 1351–1363. [Google Scholar] [CrossRef]
- Cacchiarelli, D.; Trapnell, C.; Ziller, M.J.; Soumillon, M.; Cesana, M.; Karnik, R.; Donaghey, J.; Smith, Z.D.; Ratanasirintrawoot, S.; Zhang, X.; et al. Integrative Analyses of Human Reprogramming Reveal Dynamic Nature of Induced Pluripotency. Cell 2015, 162, 412–424. [Google Scholar] [CrossRef] [Green Version]
- Hockemeyer, D.; Soldner, F.; Cook, E.G.; Gao, Q.; Mitalipova, M.; Jaenisch, R. A Drug-Inducible System for Direct Reprogramming of Human Somatic Cells to Pluripotency. Cell Stem Cell 2008, 3, 346–353. [Google Scholar] [CrossRef] [Green Version]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef] [Green Version]
- Batut, B.; Hiltemann, S.; Bagnacani, A.; Baker, D.; Bhardwaj, V.; Blank, C.; Bretaudeau, A.; Brillet-Guéguen, L.; Čech, M.; Chilton, J.; et al. Community-Driven Data Analysis Training for Biology. Cell Syst. 2018, 6, 752–758.e1. [Google Scholar] [CrossRef] [Green Version]
- Doyle, M.; Phipson, B.; Dashnow, H. 1: RNA-Seq Reads to Counts (Galaxy Training Materials). Available online: https://training.galaxyproject.org/training-material/topics/transcriptomics/tutorials/rna-seq-reads-to-counts/tutorial.html (accessed on 30 September 2021).
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dreos, R.; Ambrosini, G.; Groux, R.; Cavin Périer, R.; Bucher, P. The eukaryotic promoter database in its 30th year: Focus on non-vertebrate organisms. Nucleic Acids Res. 2017, 45, D51–D55. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karolchik, D.; Hinrichs, A.S.; Furey, T.S.; Roskin, K.M.; Sugnet, C.W.; Haussler, D.; Kent, W.J. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004, 32, D493–D496. [Google Scholar] [CrossRef] [PubMed]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The Human Genome Browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef] [Green Version]
- Tsankov, A.M.; Gu, H.; Akopian, V.; Ziller, M.J.; Donaghey, J.; Amit, I.; Gnirke, A.; Meissner, A. Transcription factor binding dynamics during human ES cell differentiation. Nature 2015, 518, 344–349. [Google Scholar] [CrossRef] [Green Version]
- Boix, C.A.; James, B.T.; Park, Y.P.; Meuleman, W.; Kellis, M. Regulatory genomic circuitry of human disease loci by integrative epigenomics. Nature 2021, 590, 300–307. [Google Scholar] [CrossRef]
- Sell, S. Stem cell origin of cancer and differentiation therapy. Crit. Rev. Oncol. 2004, 51, 1–28. [Google Scholar] [CrossRef]
- Wicha, M.S.; Liu, S.; Dontu, G. Cancer Stem Cells: An Old Idea—A Paradigm Shift. Cancer Res. 2006, 66, 1883–1890. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Kotliarova, S.; Kotliarov, Y.; Li, A.; Su, Q.; Donin, N.M.; Pastorino, S.; Purow, B.W.; Christopher, N.; Zhang, W.; et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 2006, 9, 391–403. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Yu, H.; Yuan, Y.; Yu, J.S.; Lou, Z.; Xue, Y.; Liu, Y. The necessity for standardization of glioma stem cell culture: A systematic review. Stem Cell Res. Ther. 2020, 11, 84. [Google Scholar] [CrossRef] [Green Version]
- Hazini, A.; Fisher, K.; Seymour, L. Deregulation of HLA-I in cancer and its central importance for immunotherapy. J. Immunother. Cancer. 2021, 9, e002899. [Google Scholar] [CrossRef] [PubMed]
- Wischhusen, J.; Friese, M.A.; Mittelbronn, M.; Meyermann, R.; Weller, M. HLA-E Protects Glioma Cells from NKG2D-Mediated Immune Responses In Vitro: Implications for Immune Escape In Vivo. J. Neuropathol. Exp. Neurol. 2005, 64, 523–528. [Google Scholar] [CrossRef] [PubMed]
- Feng, E.; Liang, T.; Wang, X.; Du, J.; Tang, K.; Wang, X.; Wang, F.; You, G. Correlation of alteration of HLA-F expression and clinical characterization in 593 brain glioma samples. J. Neuroinflamm. 2019, 16, 33. [Google Scholar] [CrossRef] [Green Version]
- Majc, B.; Novak, M.; Kopitar-Jerala, N.; Jewett, A.; Breznik, B. Immunotherapy of Glioblastoma: Current Strategies and Challenges in Tumor Model Development. Cells 2021, 10, 265. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wang, S.; Wang, Y.; Ma, W. Anti-PD-1 plus anti-VEGF therapy in multiple intracranial metastases of a hypermutated, IDH wild-type glioblastoma. Neuro-Oncology 2021, 23, 699–701. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Baroja, M.L.; Majumdar, A.; Chadwick, K.; Rouleau, A.; Gallacher, L.; Ferber, I.; Lebkowski, J.; Martin, T.; Madrenas, J.; et al. Human Embryonic Stem Cells Possess Immune-Privileged Properties. Stem Cells 2004, 22, 448–456. [Google Scholar] [CrossRef] [PubMed]
- Pick, M.; Ronen, D.; Yanuka, O.; Benvenisty, N. Reprogramming of the MHC-I and Its Regulation by NFκB in Human-Induced Pluripotent Stem Cells. Stem Cells 2012, 30, 2700–2708. [Google Scholar] [CrossRef]
- Gaunt, G.; Ramin, K. Immunological Tolerance of the Human Fetus. Am. J. Perinatol. 2001, 18, 299–312. [Google Scholar] [CrossRef]
- Chaligne, R.; Gaiti, F.; Silverbush, D.; Schiffman, J.S.; Weisman, H.R.; Kluegel, L.; Gritsch, S.; Deochand, S.D.; Castro, L.N.G.; Richman, A.R.; et al. Epigenetic encoding, heritability and plasticity of glioma transcriptional cell states. Nat. Genet. 2021, 53, 1469–1479. [Google Scholar] [CrossRef]
- Burr, M.; Sparbier, C.; Chan, K.L.; Chan, Y.-C.; Kersbergen, A.; Lam, E.Y.; Azidis-Yates, E.; Vassiliadis, D.; Bell, C.; Gilan, O.; et al. An Evolutionarily Conserved Function of Polycomb Silences the MHC Class I Antigen Presentation Pathway and Enables Immune Evasion in Cancer. Cancer Cell 2019, 36, 385–401.e8. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sesé, B.; Íñiguez-Muñoz, S.; Ensenyat-Mendez, M.; Llinàs-Arias, P.; Ramis, G.; Orozco, J.I.J.; Fernández de Mattos, S.; Villalonga, P.; Marzese, D.M. Glioblastoma Embryonic-like Stem Cells Exhibit Immune-Evasive Phenotype. Cancers 2022, 14, 2070. https://doi.org/10.3390/cancers14092070
Sesé B, Íñiguez-Muñoz S, Ensenyat-Mendez M, Llinàs-Arias P, Ramis G, Orozco JIJ, Fernández de Mattos S, Villalonga P, Marzese DM. Glioblastoma Embryonic-like Stem Cells Exhibit Immune-Evasive Phenotype. Cancers. 2022; 14(9):2070. https://doi.org/10.3390/cancers14092070
Chicago/Turabian StyleSesé, Borja, Sandra Íñiguez-Muñoz, Miquel Ensenyat-Mendez, Pere Llinàs-Arias, Guillem Ramis, Javier I. J. Orozco, Silvia Fernández de Mattos, Priam Villalonga, and Diego M. Marzese. 2022. "Glioblastoma Embryonic-like Stem Cells Exhibit Immune-Evasive Phenotype" Cancers 14, no. 9: 2070. https://doi.org/10.3390/cancers14092070
APA StyleSesé, B., Íñiguez-Muñoz, S., Ensenyat-Mendez, M., Llinàs-Arias, P., Ramis, G., Orozco, J. I. J., Fernández de Mattos, S., Villalonga, P., & Marzese, D. M. (2022). Glioblastoma Embryonic-like Stem Cells Exhibit Immune-Evasive Phenotype. Cancers, 14(9), 2070. https://doi.org/10.3390/cancers14092070





