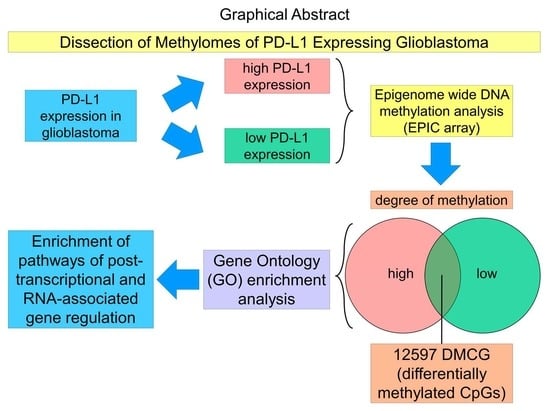
Methylome Profiling of PD-L1-Expressing Glioblastomas Shows Enrichment of Post-Transcriptional and RNA-Associated Gene Regulation
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Collection and Immunohistochemical Analysis
2.2. Molecular Genetic Analysis
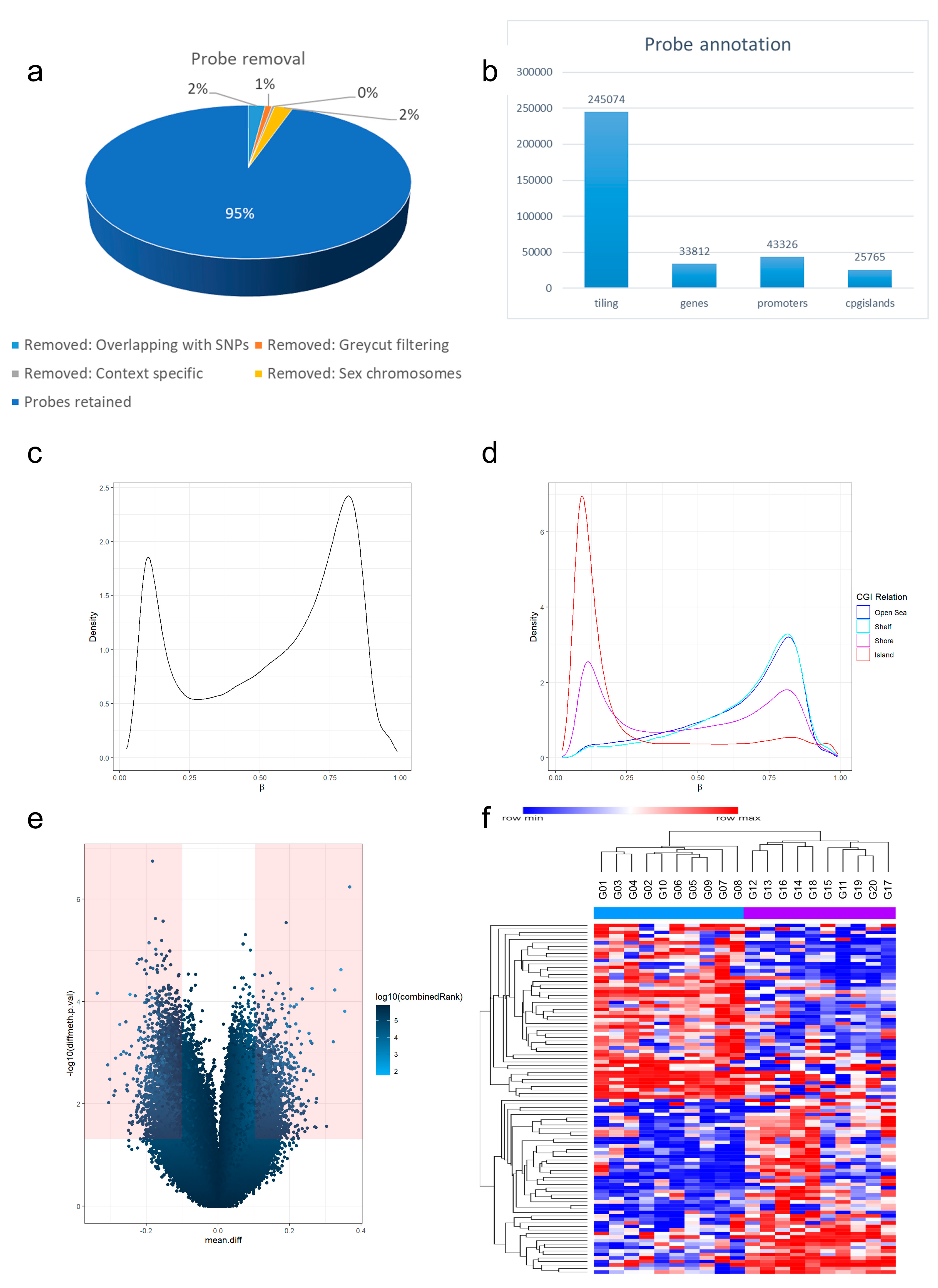
2.3. Computational Data Analysis
3. Results
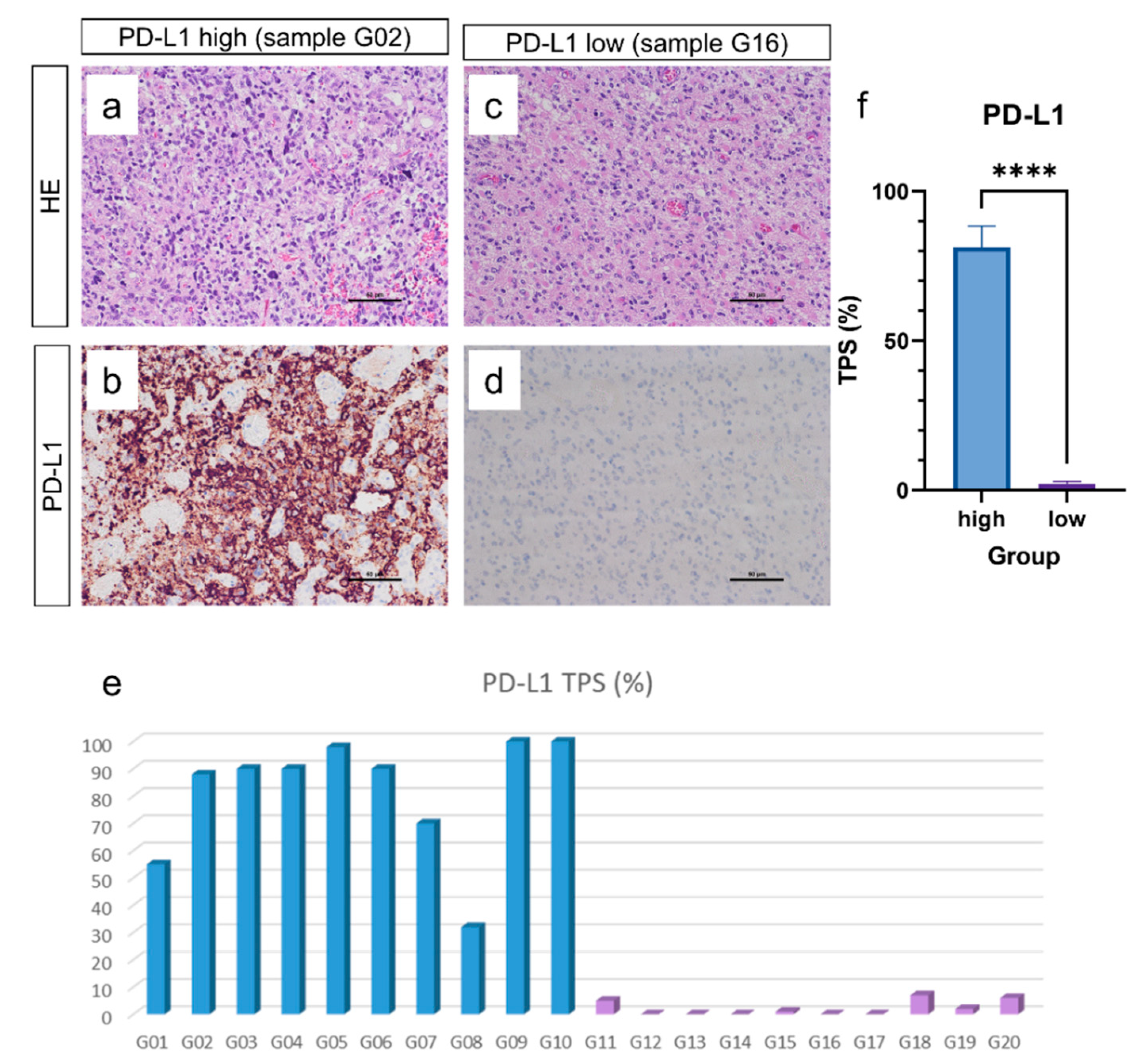
3.1. Glioblastoma IDH Wildtype CNS WHO Grade 4 Show Different Amount of PD-L1 Expression Quantified by Tumor Proportion Score (TPS)
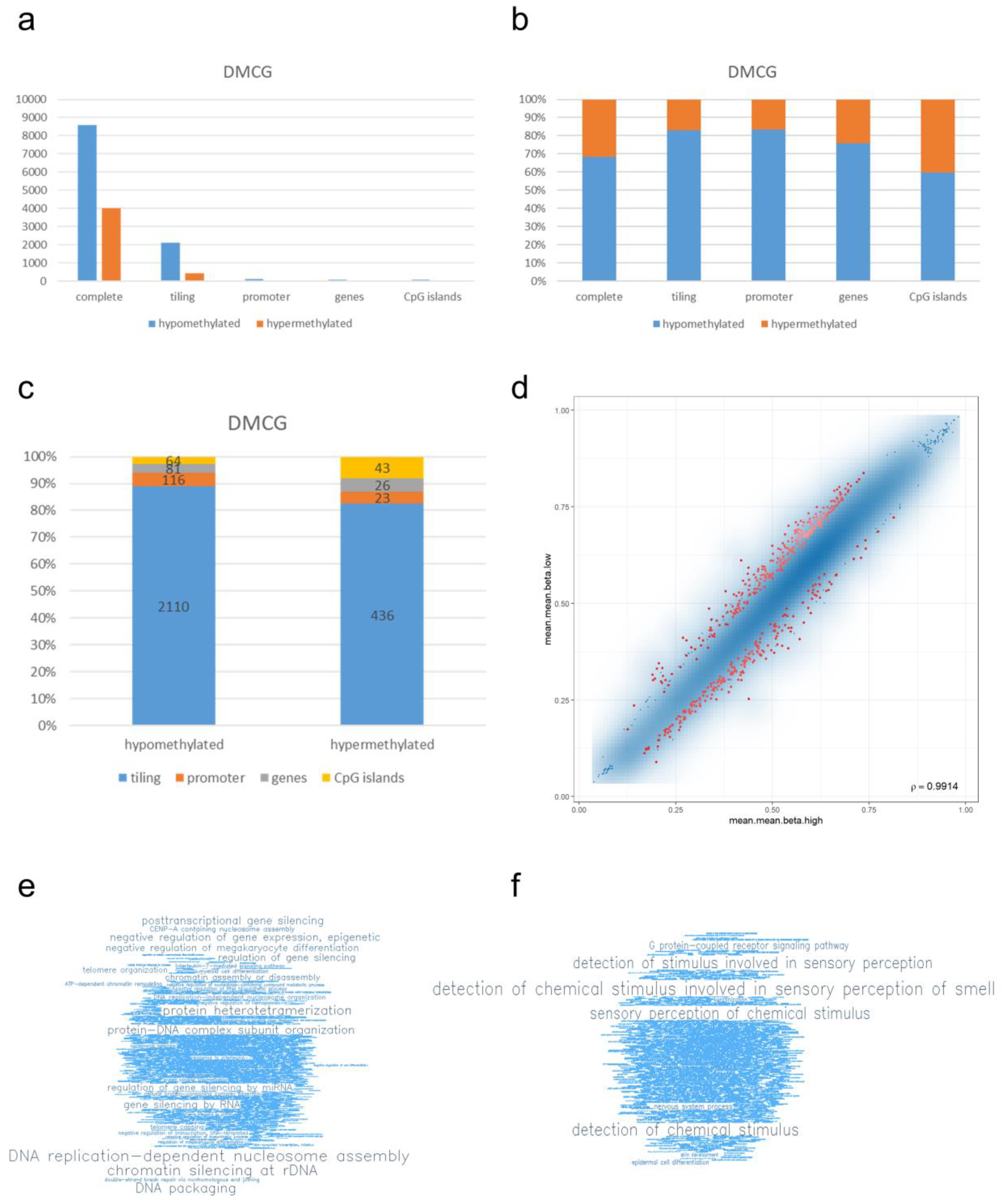
3.2. Differential Methylation Analysis Shows PD-L1 Correlation with Methylation Signatures in Glioblastoma IDH Mutant CNS WHO Grade 4
3.3. Enrichment Analysis of DMCGs Reveals Distinct Altered Pathways Correlating with PD-L1 Status
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 who classification of tumors of the central nervous system: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Hegi, M.E.; Liu, L.; Herman, J.G.; Stupp, R.; Wick, W.; Weller, M.; Mehta, M.P.; Gilbert, M.R. Correlation of o6-methylguanine methyltransferase (mgmt) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate mgmt activity. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2008, 26, 4189–4199. [Google Scholar] [CrossRef]
- Hegi, M.E.; Sciuscio, D.; Murat, A.; Levivier, M.; Stupp, R. Epigenetic deregulation of DNA repair and its potential for therapy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2009, 15, 5026–5031. [Google Scholar] [CrossRef]
- Kaina, B.; Christmann, M.; Naumann, S.; Roos, W.P. Mgmt: Key node in the battle against genotoxicity, carcinogenicity and apoptosis induced by alkylating agents. DNA Repair 2007, 6, 1079–1099. [Google Scholar] [PubMed]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. Mgmt gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Campesato, L.F.; Barroso-Sousa, R.; Jimenez, L.; Correa, B.R.; Sabbaga, J.; Hoff, P.M.; Reis, L.F.; Galante, P.A.; Camargo, A.A. Comprehensive cancer-gene panels can be used to estimate mutational load and predict clinical benefit to pd-1 blockade in clinical practice. Oncotarget 2015, 6, 34221–34227. [Google Scholar] [CrossRef]
- Gatalica, Z.; Snyder, C.; Maney, T.; Ghazalpour, A.; Holterman, D.A.; Xiao, N.; Overberg, P.; Rose, I.; Basu, G.D.; Vranic, S.; et al. Programmed cell death 1 (pd-1) and its ligand (pd-l1) in common cancers and their correlation with molecular cancer type. Cancer Epidemiol. Biomarkers Prev. 2014, 23, 2965–2970. [Google Scholar] [CrossRef]
- Ohaegbulam, K.C.; Assal, A.; Lazar-Molnar, E.; Yao, Y.; Zang, X. Human cancer immunotherapy with antibodies to the pd-1 and pd-l1 pathway. Trends Mol. Med. 2015, 21, 24–33. [Google Scholar] [CrossRef]
- Said, E.A.; Dupuy, F.P.; Trautmann, L.; Zhang, Y.; Shi, Y.; El-Far, M.; Hill, B.J.; Noto, A.; Ancuta, P.; Peretz, Y.; et al. Programmed death-1-induced interleukin-10 production by monocytes impairs cd4+ t cell activation during HIV infection. Nat. Med. 2010, 16, 452–459. [Google Scholar] [CrossRef]
- Sun, C.; Mezzadra, R.; Schumacher, T.N. Regulation and function of the pd-l1 checkpoint. Immunity 2018, 48, 434–452. [Google Scholar] [CrossRef]
- Honda, Y.; Otsuka, A.; Ono, S.; Yamamoto, Y.; Seidel, J.A.; Morita, S.; Hirata, M.; Kataoka, T.R.; Takenouchi, T.; Fujii, K.; et al. Infiltration of pd-1-positive cells in combination with tumor site pd-l1 expression is a positive prognostic factor in cutaneous angiosarcoma. Oncoimmunology 2017, 6, e1253657. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, K.; Ogawa, S. Genetic biomarkers for pd-1/pd-l1 blockade therapy. Oncoscience 2016, 3, 311–312. [Google Scholar] [CrossRef]
- Kataoka, K.; Shiraishi, Y.; Takeda, Y.; Sakata, S.; Matsumoto, M.; Nagano, S.; Maeda, T.; Nagata, Y.; Kitanaka, A.; Mizuno, S.; et al. Aberrant pd-l1 expression through 3′-utr disruption in multiple cancers. Nature 2016, 534, 402–406. [Google Scholar] [CrossRef] [PubMed]
- Isaacsson Velho, P.; Antonarakis, E.S. Pd-1/pd-l1 pathway inhibitors in advanced prostate cancer. Expert Rev. Clin. Pharmacol. 2018, 11, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Che, X.; Qu, J.; Hou, K.; Wen, T.; Li, Z.; Li, C.; Wang, S.; Xu, L.; Liu, Y.; et al. Exosomal pd-l1 retains immunosuppressive activity and is associated with gastric cancer prognosis. Ann. Surg. Oncol. 2019, 26, 3745–3755. [Google Scholar] [CrossRef] [PubMed]
- Reck, M.; Rodriguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csoszi, T.; Fulop, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S.; et al. Pembrolizumab versus chemotherapy for pd-l1-positive non-small-cell lung cancer. N. Engl. J. Med. 2016, 375, 1823–1833. [Google Scholar] [CrossRef]
- Li, X.; Wetherilt, C.S.; Krishnamurti, U.; Yang, J.; Ma, Y.; Styblo, T.M.; Meisel, J.L.; Peng, L.; Siddiqui, M.T.; Cohen, C.; et al. Stromal pd-l1 expression is associated with better disease-free survival in triple-negative breast cancer. Am. J. Clin. Pathol. 2016, 146, 496–502. [Google Scholar] [CrossRef]
- Fujita, Y.; Yagishita, S.; Hagiwara, K.; Yoshioka, Y.; Kosaka, N.; Takeshita, F.; Fujiwara, T.; Tsuta, K.; Nokihara, H.; Tamura, T.; et al. The clinical relevance of the mir-197/cks1b/stat3-mediated pd-l1 network in chemoresistant non-small-cell lung cancer. Mol. Ther. 2015, 23, 717–727. [Google Scholar] [CrossRef]
- Holzl, D.; Hutarew, G.; Zellinger, B.; Schlicker, H.U.; Schwartz, C.; Winkler, P.A.; Sotlar, K.; Kraus, T.F.J. Integrated analysis of programmed cell death ligand 1 expression reveals increased levels in high-grade glioma. J. Cancer Res. Clin. Oncol. 2021, 147, 2271–2280. [Google Scholar] [CrossRef]
- Nduom, E.K.; Wei, J.; Yaghi, N.K.; Huang, N.; Kong, L.Y.; Gabrusiewicz, K.; Ling, X.; Zhou, S.; Ivan, C.; Chen, J.Q.; et al. Pd-l1 expression and prognostic impact in glioblastoma. Neuro-Oncology 2016, 18, 195–205. [Google Scholar] [CrossRef]
- Heiland, D.H.; Haaker, G.; Delev, D.; Mercas, B.; Masalha, W.; Heynckes, S.; Gabelein, A.; Pfeifer, D.; Carro, M.S.; Weyerbrock, A.; et al. Comprehensive analysis of pd-l1 expression in glioblastoma multiforme. Oncotarget 2017, 8, 42214–42225. [Google Scholar] [CrossRef] [PubMed]
- Hao, C.; Chen, G.; Zhao, H.; Li, Y.; Chen, J.; Zhang, H.; Li, S.; Zhao, Y.; Chen, F.; Li, W.; et al. Pd-l1 expression in glioblastoma, the clinical and prognostic significance: A systematic literature review and meta-analysis. Front. Oncol. 2020, 10, 1015. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Huang, C.; Mok, T.S.; Zhuang, W.; Xu, H.; Miao, Q.; Fan, X.; Zhu, W.; Huang, Y.; Lin, X.; et al. Comparison of 22c3 pd-l1 expression between surgically resected specimens and paired tissue microarrays in non-small cell lung cancer. J. Thorac. Oncol. 2017, 12, 1536–1543. [Google Scholar] [CrossRef] [PubMed]
- Neuman, T.; London, M.; Kania-Almog, J.; Litvin, A.; Zohar, Y.; Fridel, L.; Sandbank, J.; Barshak, I.; Vainer, G.W. A harmonization study for the use of 22c3 pd-l1 immunohistochemical staining on Ventana’s platform. J. Thorac. Oncol. 2016, 11, 1863–1868. [Google Scholar] [CrossRef] [PubMed]
- Roge, R.; Vyberg, M.; Nielsen, S. Accurate pd-l1 protocols for non-small cell lung cancer can be developed for automated staining platforms with clone 22c3. Appl. Immunohistochem. Mol. Morphol. 2017, 25, 381–385. [Google Scholar] [CrossRef] [PubMed]
- Kraus, T.F.J.; Machegger, L.; Poppe, J.; Zellinger, B.; Dovjak, E.; Schlicker, H.U.; Schwartz, C.; Ladisich, B.; Spendel, M.; Kral, M.; et al. Diffuse midline glioma of the cervical spinal cord with h3 k27m genotype phenotypically mimicking anaplastic ganglioglioma: A case report and review of the literature. Brain Tumor Pathol. 2020, 37, 89–94. [Google Scholar] [CrossRef]
- Kraus, T.F.J.; Schwartz, C.; Machegger, L.; Zellinger, B.; Holzl, D.; Schlicker, H.U.; Poppe, J.; Ladisich, B.; Spendel, M.; Kral, M.; et al. A patient with two gliomas with independent oligodendroglioma and glioblastoma biology proved by DNA-methylation profiling: A case report and review of the literature. Brain Tumor Pathol. 2022, 39, 111–119. [Google Scholar] [CrossRef]
- Holzl, D.; Hutarew, G.; Zellinger, B.; Alinger-Scharinger, B.; Schlicker, H.U.; Schwartz, C.; Sotlar, K.; Kraus, T.F.J. Egfr amplification is a phenomenon of idh wildtype and tert mutated high-grade glioma: An integrated analysis using fluorescence in situ hybridization and DNA methylome profiling. Biomedicines 2022, 10, 794. [Google Scholar] [CrossRef]
- Capper, D.; Jones, D.T.W.; Sill, M.; Hovestadt, V.; Schrimpf, D.; Sturm, D.; Koelsche, C.; Sahm, F.; Chavez, L.; Reuss, D.E.; et al. DNA methylation-based classification of central nervous system tumours. Nature 2018, 555, 469–474. [Google Scholar] [CrossRef]
- Muller, F.; Scherer, M.; Assenov, Y.; Lutsik, P.; Walter, J.; Lengauer, T.; Bock, C. Rnbeads 2.0: Comprehensive analysis of DNA methylation data. Genome Biol. 2019, 20, 55. [Google Scholar] [CrossRef]
- De Jager, P.L.; Srivastava, G.; Lunnon, K.; Burgess, J.; Schalkwyk, L.C.; Yu, L.; Eaton, M.L.; Keenan, B.T.; Ernst, J.; McCabe, C.; et al. Alzheimer’s disease: Early alterations in brain DNA methylation at ank1, bin1, rhbdf2 and other loci. Nat. Neurosci. 2014, 17, 1156–1163. [Google Scholar] [CrossRef] [PubMed]
- Lunnon, K.; Smith, R.; Hannon, E.; De Jager, P.L.; Srivastava, G.; Volta, M.; Troakes, C.; Al-Sarraj, S.; Burrage, J.; Macdonald, R.; et al. Methylomic profiling implicates cortical deregulation of ank1 in Alzheimer’s disease. Nat. Neurosci. 2014, 17, 1164–1170. [Google Scholar] [CrossRef] [PubMed]
- Ilie, M.; Khambata-Ford, S.; Copie-Bergman, C.; Huang, L.; Juco, J.; Hofman, V.; Hofman, P. Use of the 22c3 anti-pd-l1 antibody to determine pd-l1 expression in multiple automated immunohistochemistry platforms. PLoS ONE 2017, 12, e0183023. [Google Scholar]
- Yi, M.; Niu, M.; Xu, L.; Luo, S.; Wu, K. Regulation of PD-L1 expression in the tumor microenvironment. J. Hematol. Oncol. 2021, 14, 10. [Google Scholar] [CrossRef]
- Reardon, D.A.; Kim, T.M.; Frenel, J.-S.; Simonelli, M.; Lopez, J.; Subramaniam, D.S.; Siu, L.L.; Wang, H.; Krishnan, S.; Stein, K.; et al. Treatment with Pembrolizumab in Programmed Death Ligand 1–Positive Recurrent Glioblastoma: Results from the Multicohort Phase 1 KEYNOTE-028 Trial. Cancer 2021, 127, 1620–1629. [Google Scholar] [CrossRef]
- Jiang, F.; Lang, X.; Chen, N.; Jin, L.; Liu, L.; Wei, X.; Pan, J.; Yu, F.; Blake, A.; Xiao, S. A novel hnrnph1::Erg rearrangement in aggressive acute myeloid leukemia. Genes Chromosomes Cancer 2022, 61, 503–508. [Google Scholar] [CrossRef]
- Liu, M.; Yang, L.; Liu, X.; Nie, Z.; Zhang, X.; Lu, Y.; Pan, Y.; Wang, X.; Luo, J. Hnrnph1 is a novel regulator of cellular proliferation and disease progression in chronic myeloid leukemia. Front. Oncol. 2021, 11, 682859. [Google Scholar] [CrossRef]
- Aoki, T.; Miyamoto, T.; Yoshida, S.; Yamamoto, A.; Yamauchi, T.; Yoshimoto, G.; Mori, Y.; Kamezaki, K.; Iwasaki, H.; Takenaka, K.; et al. Additional acquisition of t(1;21) (p32;q22) in a patient relapsing with acute myelogenous leukemia with nup98-hoxa9. Int. J. Hematol. 2008, 88, 571–574. [Google Scholar] [CrossRef]
- Miyamoto, R.; Kanai, A.; Okuda, H.; Komata, Y.; Takahashi, S.; Matsui, H.; Inaba, T.; Yokoyama, A. Hoxa9 promotes myc-mediated leukemogenesis by maintaining gene expression for multiple anti-apoptotic pathways. Elife 2021, 10, e64148. [Google Scholar] [CrossRef]
- Choong, L.Y.; Lim, S.; Chong, P.K.; Wong, C.Y.; Shah, N.; Lim, Y.P. Proteome-wide profiling of the mcf10at breast cancer progression model. PLoS ONE 2010, 5, e11030. [Google Scholar] [CrossRef]
- Li, Y.; Wei, Z.; Huang, S.; Yang, B. Mrna expression and DNA methylation analysis of the inhibitory mechanism of h2o2 on the proliferation of a549 cells. Oncol. Lett. 2020, 20, 288. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, J.; Liu, Z.; Zhang, Y. High expression of mir-196b predicts poor prognosis in patients with ovarian cancer. OncoTargets Ther. 2020, 13, 9797–9806. [Google Scholar] [CrossRef] [PubMed]
- Davey, M.G.; Davies, M.; Lowery, A.J.; Miller, N.; Kerin, M.J. The role of microrna as clinical biomarkers for breast cancer surgery and treatment. Int. J. Mol. Sci. 2021, 22, 8290. [Google Scholar] [CrossRef]
- Liuksiala, T.; Teittinen, K.J.; Granberg, K.; Heinaniemi, M.; Annala, M.; Maki, M.; Nykter, M.; Lohi, O. Overexpression of snord114-3 marks acute promyelocytic leukemia. Leukemia 2014, 28, 233–236. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Wen, J.; Huang, Z.; Chen, X.P.; Zhang, B.X.; Chu, L. Small nucleolar rnas: Insight into their function in cancer. Front. Oncol. 2019, 9, 587. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.C.; Ni, J.J.; Cui, W.Y.; Wang, B.Y.; Zhuo, W. Emerging roles of lncrna in cancer and therapeutic opportunities. Am. J. Cancer Res. 2019, 9, 1354–1366. [Google Scholar]
- Li, Y.; Li, W.; Liang, B.; Li, L.; Wang, L.; Huang, H.; Guo, S.; Wang, Y.; He, Y.; Chen, L.; et al. Identification of cancer risk lncrnas and cancer risk pathways regulated by cancer risk lncrnas based on genome sequencing data in human cancers. Sci. Rep. 2016, 6, 39294. [Google Scholar] [CrossRef]
- Ma, R.; Yan, W.; Zhang, G.; Lv, H.; Liu, Z.; Fang, F.; Zhang, W.; Zhang, J.; Tao, T.; You, Y.; et al. Upregulation of miR-196b confers a poor prognosis in glioblastoma patients via inducing a proliferative phenotype. PLoS ONE 2012, 7, e38096. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hutarew, G.; Hölzl, D.; Schiefer, T.; Langwieder, C.K.; Alinger-Scharinger, B.; Schlicker, H.U.; Schwartz, C.; Sotlar, K.; Kraus, T.F.J. Methylome Profiling of PD-L1-Expressing Glioblastomas Shows Enrichment of Post-Transcriptional and RNA-Associated Gene Regulation. Cancers 2022, 14, 5375. https://doi.org/10.3390/cancers14215375
Hutarew G, Hölzl D, Schiefer T, Langwieder CK, Alinger-Scharinger B, Schlicker HU, Schwartz C, Sotlar K, Kraus TFJ. Methylome Profiling of PD-L1-Expressing Glioblastomas Shows Enrichment of Post-Transcriptional and RNA-Associated Gene Regulation. Cancers. 2022; 14(21):5375. https://doi.org/10.3390/cancers14215375
Chicago/Turabian StyleHutarew, Georg, Dorothee Hölzl, Tanja Schiefer, Celina K. Langwieder, Beate Alinger-Scharinger, Hans U. Schlicker, Christoph Schwartz, Karl Sotlar, and Theo F. J. Kraus. 2022. "Methylome Profiling of PD-L1-Expressing Glioblastomas Shows Enrichment of Post-Transcriptional and RNA-Associated Gene Regulation" Cancers 14, no. 21: 5375. https://doi.org/10.3390/cancers14215375
APA StyleHutarew, G., Hölzl, D., Schiefer, T., Langwieder, C. K., Alinger-Scharinger, B., Schlicker, H. U., Schwartz, C., Sotlar, K., & Kraus, T. F. J. (2022). Methylome Profiling of PD-L1-Expressing Glioblastomas Shows Enrichment of Post-Transcriptional and RNA-Associated Gene Regulation. Cancers, 14(21), 5375. https://doi.org/10.3390/cancers14215375