SFN Enhanced the Radiosensitivity of Cervical Cancer Cells via Activating LATS2 and Blocking Rad51/MDC1 Recruitment to DNA Damage Site
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Antibodies
2.2. Cell Culture and Reagents
2.3. Ionizing Radiation
2.4. Plasmids and Cell Transfection
2.5. RT-PCR and Illumina Hiseq xten/Nova seq 6000 Sequencing
2.6. Clonogenic Assays
2.7. Cell Immunofluorescence Staining
2.8. Western Blot
2.9. In Vitro Proliferation Assays
2.10. Cell Apoptosis Analysis
2.11. In Vivo Analysis
2.12. Statistical Analysis
3. Results
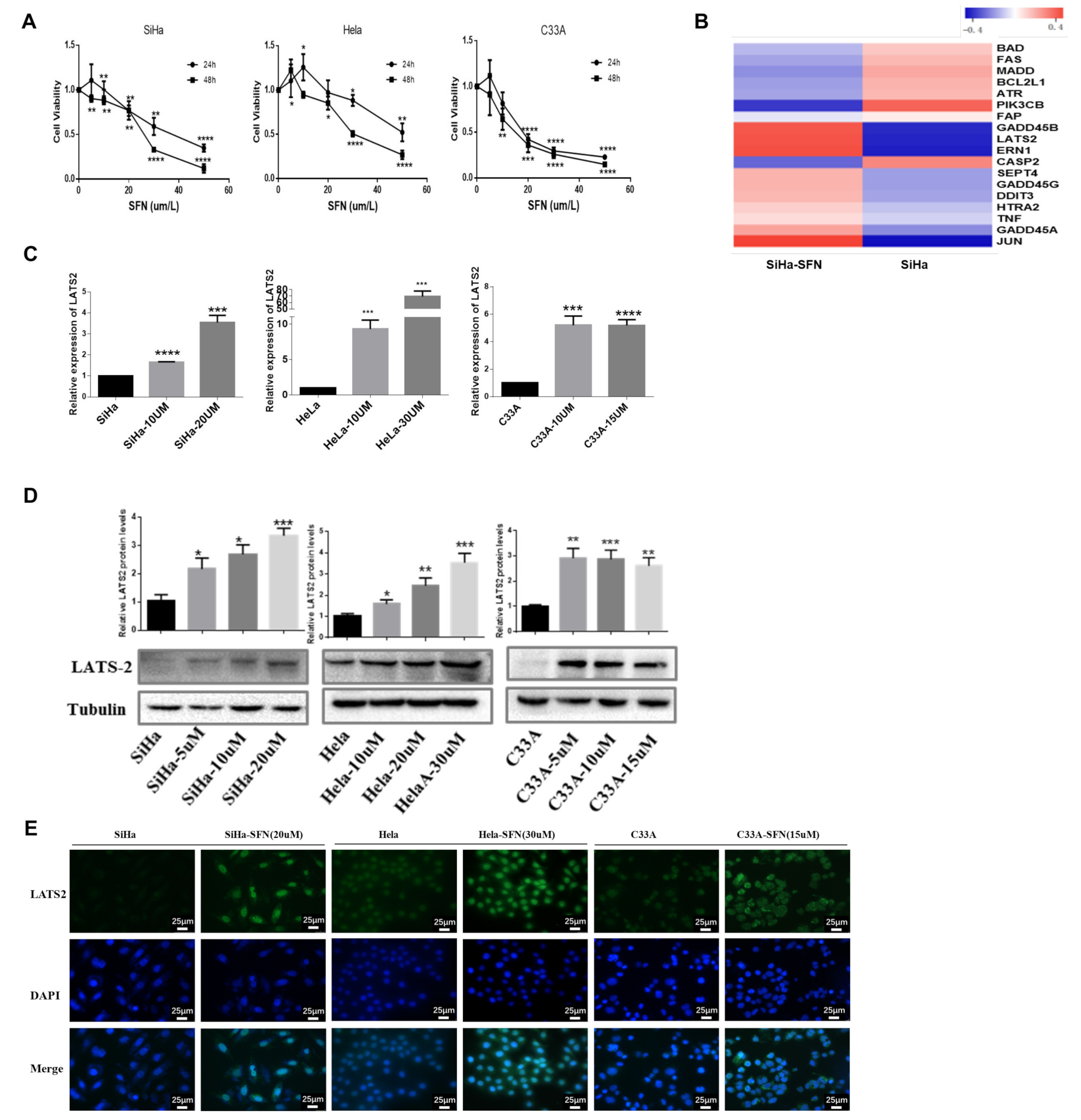
3.1. SFN Inhibits the Viability of Cervical Cancer Cells and Increases the Expression of LATS2
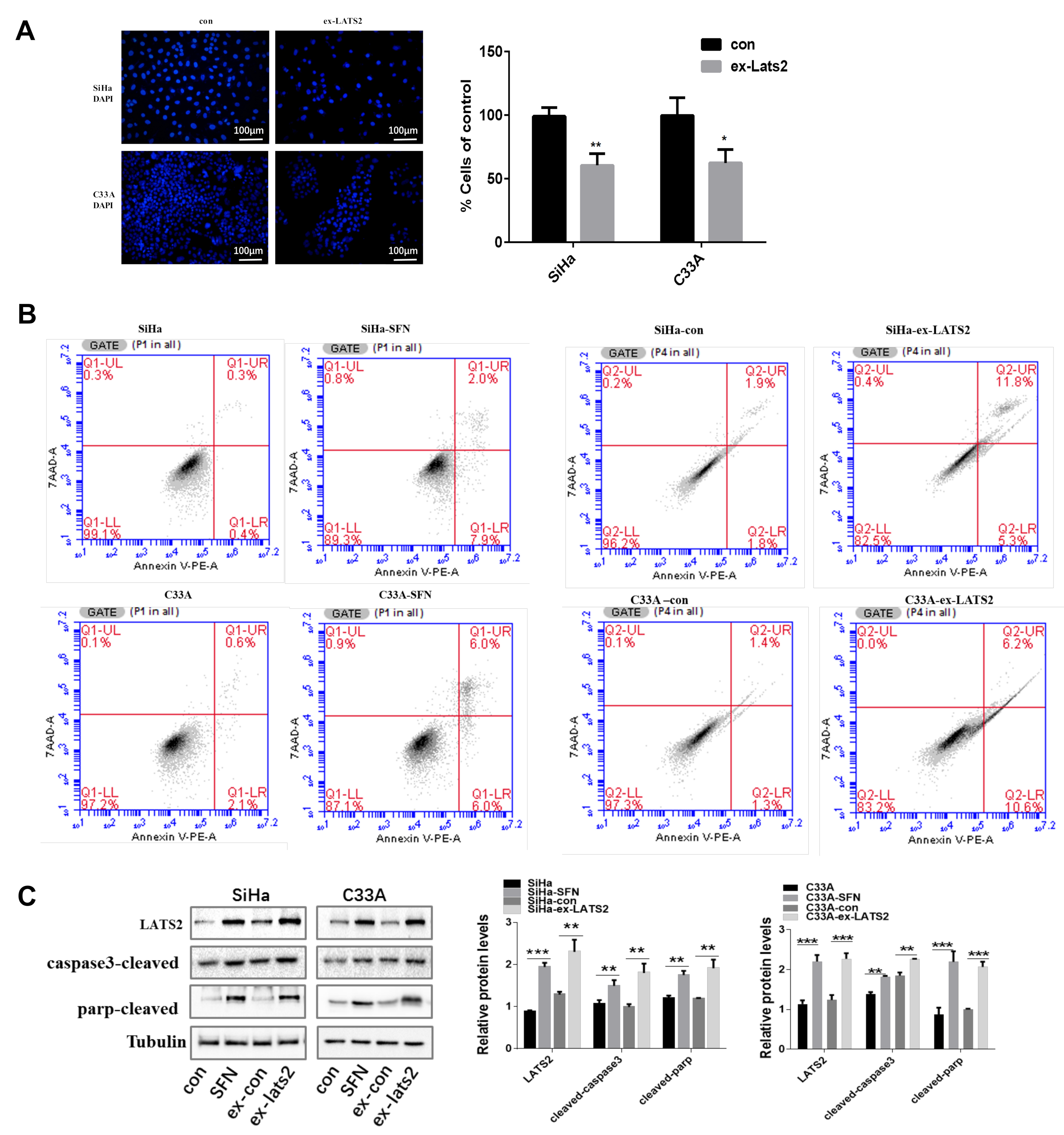
3.2. Overexpression of LATS2 Suppresses Cervical Cancer Cell Tumorigenicity In Vitro
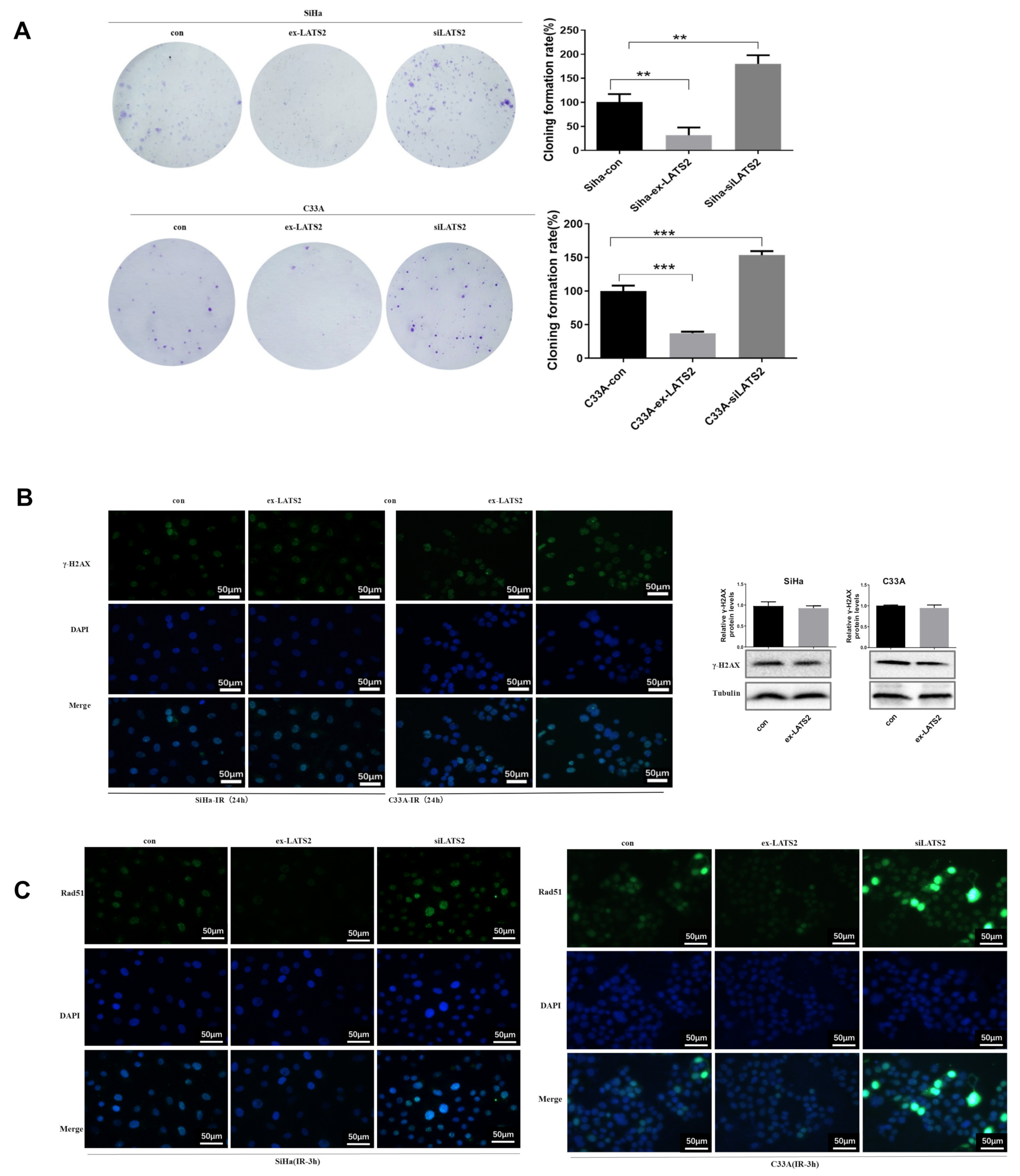
3.3. SFN Enhances Radiosensitivity In Vitro by Inhibiting DNA Damage Repair
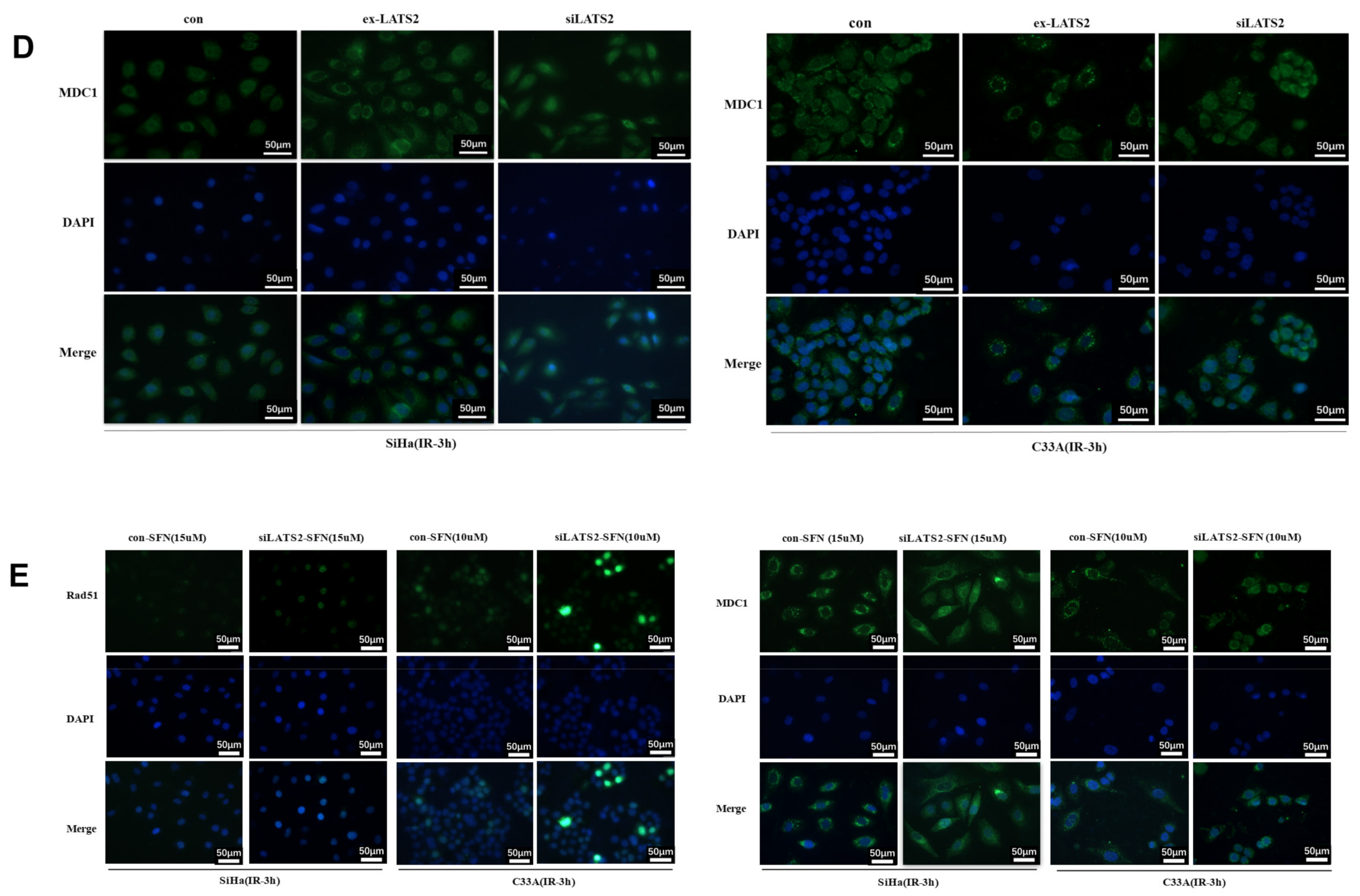
3.4. LATS2 Has Made a Contribution Role in SFN-Facilitated Reduced DNA Damage Repair
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| SFN | sulforaphane |
| HR | homologous recombination |
| IR | ionizing radiation |
| DSB | DNA double-strand breaks |
| DDR | DNA damage repair |
| CCRT | concurrent chemoradiotherapy |
| NHEJ | non-homologous end joining |
| LATS2 | large tumor suppressors 2 |
| MDC1 | DNA damage checkpoint protein 1 |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, 359–386. [Google Scholar] [CrossRef] [PubMed]
- Scarth, J.A.; Patterson, M.R.; Morgan, E.L.; Macdonald, A. The human papillomavirus oncoproteins: A review of the host pathways targeted on the road to transformation. J. Gen. Virol. 2021, 102, 001540. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.A.; Jhingran, A.; Oaknin, A.; Denny, L. Cervical cancer. Lancet 2019, 393, 69–182. [Google Scholar] [CrossRef]
- Falcaro, M.; Castañon, A.; Ndlela, B.; Checchi, M.; Soldan, K.; Lopez-Bernal, J.; Elliss-Brookes, L.; Sasieni, P. The effects of the national HPV vaccination programme in England, UK, on cervical cancer and grade 3 cervical intraepithelial neoplasia incidence: A register-based observational study. Lancet 2021, 398, 2084–2092. [Google Scholar] [CrossRef]
- Lei, J.; Ploner, A.; Elfström, K.M.; Wang, J.; Roth, A.; Fang, F.; Sundström, K.; Dillner, J.; Sparén, P. HPV Vaccination and the Risk of Invasive Cervical Cancer. N. Engl. J. Med. 2020, 383, 1340–1348. [Google Scholar] [CrossRef]
- Gauri, A.; Messiah, S.E.; Bouzoubaa, L.A.; Moore, K.J.; Koru-Sengul, T. Cervical cancer sociodemographic and diagnostic disparities in Florida: A population-based study (1981–2013) by stage at presentation. Ethn. Health 2020, 25, 995–1003. [Google Scholar] [CrossRef]
- Abu-Rustum, N.R.; Yashar, C.M.; Bean, S.; Bradley, K.; Campos, S.M.; Chon, H.S.; Chu, C.; Cohn, D.; Crispens, M.A.; Damast, S.; et al. NCCN Guidelines Insights: Cervical Cancer, Version 1.2020. J. Natl. Compr. Cancer Netw. JNCCN 2020, 18, 660–666. [Google Scholar] [CrossRef]
- Sedlis, A.; Bundy, B.N.; Rotman, M.Z.; Lentz, S.S.; Muderspach, L.I.; Zaino, R.J. A randomized trial of pelvic radiation therapy versus no further therapy in selected patients with stage IB carcinoma of the cervix after radical hysterectomy and pelvic lymphadenectomy: A Gynecologic Oncology Group Study. Gynecol. Oncol. 1999, 73, 177–183. [Google Scholar] [CrossRef]
- Liu, Y.; Murray-Stewart, T.; Casero, R.A., Jr.; Kagiampakis, I.; Jin, L.; Zhang, J.; Wang, H.; Che, Q.; Tong, H.; Ke, J.; et al. Targeting hexokinase 2 inhibition promotes radiosensitization in HPV16 E7-induced cervical cancer and suppresses tumor growth. Int. J. Oncol. 2017, 50, 2011–2023. [Google Scholar] [CrossRef] [Green Version]
- Maier, P.; Hartmann, L.; Wenz, F.; Herskind, C. Cellular Pathways in Response to Ionizing Radiation and Their Targetability for Tumor Radiosensitization. Int. J. Mol. Sci. 2016, 17, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeWeerdt, S. Food: The omnivore’s labyrinth. Nature 2011, 471, 22–24. [Google Scholar] [CrossRef] [PubMed]
- Vanduchova, A.; Anzenbacher, P.; Anzenbacherova, E. Isothiocyanate from Broccoli, Sulforaphane, and Its Properties. J. Med. Food 2019, 22, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Russo, M.; Spagnuolo, C.; Russo, G.L.; Skalicka-Woźniak, K.; Daglia, M.; Sobarzo-Sánchez, E.; Nabavi, S.F.; Nabavi, S.M. Nrf2 targeting by sulforaphane: A potential therapy for cancer treatment. Crit. Rev. Food Sci. Nutr. 2018, 58, 1391–1405. [Google Scholar] [CrossRef]
- Meng, Q.; Xia, Y. C-Jun, at the crossroad of the signaling network. Protein Cell 2011, 2, 889–898. [Google Scholar] [CrossRef] [Green Version]
- Šuštić, T.; Van Wageningen, S.; Bosdriesz, E.; Reid, R.; Dittmar, J.; Lieftink, C.; Beijersbergen, R.L.; Wessels, L.F.A.; Rothstein, R.; Bernards, R. A role for the unfolded protein response stress sensor ERN1 in regulating the response to MEK inhibitors in KRAS mutant colon cancers. Genome Med. 2018, 10, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Taguchi, Y.; Horiuchi, Y.; Kano, F.; Murata, M. Novel prosurvival function of Yip1A in human cervical cancer cells: Constitutive activation of the IRE1 and PERK pathways of the unfolded protein response. Cell Death Dis. 2017, 8, 2718. [Google Scholar] [CrossRef] [Green Version]
- Munoz, J.P.; Carrillo-Beltran, D.; Aedo-Aguilera, V.; Calaf, G.M.; Leon, O.; Maldonado, E.; Tapia, J.C.; Boccardo, E.; Ozbun, M.A.; Aguayo, F. Tobacco Exposure Enhances Human Papillomavirus 16 Oncogene Expression via EGFR/PI3K/Akt/c-Jun Signaling Pathway in Cervical Cancer Cells. Front. Microbiol. 2018, 9, 3022. [Google Scholar] [CrossRef]
- Morgan, E.L.; Scarth, J.A.; Patterson, M.R.; Wasson, C.W.; Hemingway, G.C.; Barba-Moreno, D.; Macdonald, A. E6-mediated activation of JNK drives EGFR signalling to promote proliferation and viral oncoprotein expression in cervical cancer. Cell Death Differ. 2021, 28, 1669–1687. [Google Scholar] [CrossRef]
- Bonner, W.M.; Redon, C.E.; Dickey, J.S.; Nakamura, A.J.; Sedelnikova, O.A.; Solier, S.; Pommier, Y. GammaH2AX and cancer. Nat. Rev. Cancer 2008, 8, 957–967. [Google Scholar] [CrossRef]
- Wang, Y.; Farmer, M.; Izaguirre, E.W.; Schwartz, D.L.; Somer, B.; Tillmanns, T.; Ballo, M.T. Association of Definitive Pelvic Radiation Therapy With Survival Among Patients With Newly Diagnosed Metastatic Cervical Cancer. JAMA Oncol. 2018, 4, 1288–1291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kocher, S.; Rieckmann, T.; Rohaly, G.; Mansour, W.Y.; Dikomey, E.; Dornreiter, I.; Dahm-Daphi, J. Radiation-induced double-strand breaks require ATM but not Artemis for homologous recombination during S-phase. Nucleic Acids Res. 2012, 40, 8336–8347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hao, S.; Song, H.; Zhang, W.; Seldomridge, A.; Jung, J.; Giles, A.J.; Hutchinson, M.K.; Cao, X.; Colwell, N.; Lita, A.; et al. Protein phosphatase 2A inhibition enhances radiation sensitivity and reduces tumor growth in chordoma. Neuro-Oncology 2018, 20, 799–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ran, Q.; Xiang, Y.; Stephen, P.; Wu, C.; Li, T. CRIF1-CDK2 Interface Inhibitors: An Unprecedented Strategy for Modulation of Cell Radiosensitivity. J. Am. Chem. Soc. 2019, 141, 1420–1424. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Lyu, Y.; Tang, X.; Zhang, Y.; Chen, J.; Zheng, D.; Liang, Y. Lupeol enhances radiosensitivity of human hepatocellular carcinoma cell line SMMC-7721 in vitro and in vivo. Int. J. Radiat. Biol. 2015, 91, 202–208. [Google Scholar] [CrossRef] [PubMed]
- Lewinska, A.; Adamczyk-Grochala, J.; Deregowska, A.; Wnuk, M. Sulforaphane-Induced Cell Cycle Arrest and Senescence are accompanied by DNA Hypomethylation and Changes in microRNA Profile in Breast Cancer Cells. Theranostics 2017, 7, 3461–3477. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.M.; Tsai, C.C.; Hsu, Y.C. Sulforaphane, a Dietary Isothiocyanate, Induces G(2)/M Arrest in Cervical Cancer Cells through CyclinB1 Downregulation and GADD45beta/CDC2 Association. Int. J. Mol. Sci. 2018, 53, 539–550. [Google Scholar]
- Okonkwo, A.; Mitra, J.; Johnson, G.S.; Li, L.; Dashwood, W.M.; Hegde, M.L.; Yue, C.; Dashwood, R.H.; Rajendran, P. Heterocyclic Analogs of Sulforaphane Trigger DNA Damage and Impede DNA Repair in Colon Cancer Cells: Interplay of HATs and HDACs. Mol. Nutr. Food Res. 2018, 62, 1800228. [Google Scholar] [CrossRef]
- Yu, D.; Sekine-Suzuki, E.; Xue, L.; Fujimori, A.; Kubota, N.; Okayasu, R. Chemopreventive agent sulforaphane enhances radiosensitivity in human tumor cells. Int. J. Cancer 2009, 125, 1205–1211. [Google Scholar] [CrossRef]
- Sekine-Suzuki, E.; Yu, D.; Kubota, N.; Okayasu, R.; Anzai, K. Sulforaphane induces DNA double strand breaks predominantly repaired by homologous recombination pathway in human cancer cells. Biochem. Biophys. Res. Commun. 2008, 377, 341–345. [Google Scholar] [CrossRef]
- McPherson, J.P.; Tamblyn, L.; Elia, A.; Migon, E.; Shehabeldin, A.; Matysiak-Zablocki, E.; Lemmers, B.; Salmena, L.; Hakem, A.; Fish, J.; et al. Lats2/Kpm is required for embryonic development, proliferation control and genomic integrity. EMBO J. 2004, 23, 3677–3688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, B.; Tumaneng, K.; Guan, K.L. The Hippo pathway in organ size control, tissue regeneration and stem cell self-renewal. Nat. Cell Biol. 2011, 13, 877–883. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Mao, D.; Hua, G.; Lv, X.; Chen, X.; Angeletti, P.C.; Dong, J. Remmenga SW, Rodabaugh KJ, Zhou J et al: The Hippo/YAP pathway interacts with EGFR signaling and HPV oncoproteins to regulate cervical cancer progression. EMBO Mol. Med. 2015, 7, 1426–1449. [Google Scholar] [CrossRef] [PubMed]
- Morgan, E.L.; Patterson, M.R.; Ryder, E.L.; Lee, S.Y.; Wasson, C.W.; Harper, K.L.; Li, Y.; Griffin, S.; Blair, G.E.; Whitehouse, A.; et al. MicroRNA-18a targeting of the STK4/MST1 tumour suppressor is necessary for transformation in HPV positive cervical cancer. PLoS Pathog. 2020, 16, 1008624. [Google Scholar] [CrossRef]
- Ma, B.; Cheng, H.; Gao, R.; Mu, C.; Chen, L.; Wu, S.; Chen, Q.; Zhu, Y. Zyxin-Siah2-Lats2 axis mediates cooperation between Hippo and TGF-beta signalling pathways. Nat. Commun. 2016, 7, 11123. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Yi, J.; Zhang, K.; Bai, F.; Feng, B.; Wang, R.; Chu, X.; Chen, L.; Song, H. Downregulation of MiR-31 stimulates expression of LATS2 via the hippo pathway and promotes epithelial-mesenchymal transition in esophageal squamous cell carcinoma. J. Exp. Clin. Cancer Res. CR 2017, 36, 161. [Google Scholar] [CrossRef] [Green Version]
- Mitamura, T.; Watari, H.; Wang, L.; Kanno, H.; Kitagawa, M.; Hassan, M.K.; Kimura, T.; Tanino, M.; Nishihara, H.; Tanaka, S.; et al. microRNA 31 functions as an endometrial cancer oncogene by suppressing Hippo tumor suppressor pathway. Mol. Cancer 2014, 13, 97. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, Y.; Miyoshi, Y.; Takahata, C.; Irahara, N.; Taguchi, T.; Tamaki, Y.; Noguchi, S. Down-regulation of LATS1 and LATS2 mRNA expression by promoter hypermethylation and its association with biologically aggressive phenotype in human breast cancers. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2005, 11, 1380–1385. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Wu, S.; Barrera, J.; Matthews, K.; Pan, D. The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila Homolog of YAP. Cell 2005, 122, 421–434. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, H.; Yabuta, N.; Okada, N.; Torigata, K.; Aylon, Y.; Oren, M.; Nojima, H. Lats2 phosphorylates p21/CDKN1A after UV irradiation and regulates apoptosis. J. Cell Sci. 2013, 126, 4358–4368. [Google Scholar] [CrossRef] [Green Version]
- Zhao, B.; Wei, X.; Li, W.; Udan, R.S.; Yang, Q.; Kim, J.; Xie, J.; Ikenoue, T.; Yu, J.; Li, L.; et al. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007, 21, 2747–2761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Pei, J.; Xia, H.; Ke, H.; Wang, H.; Tao, W. Lats2, a putative tumor suppressor, inhibits G1/S transition. Oncogene 2003, 22, 4398–4405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yabuta, N.; Okada, N.; Ito, A.; Hosomi, T.; Nishihara, S.; Sasayama, Y.; Fujimori, A.; Okuzaki, D.; Zhao, H.; Ikawa, M.; et al. Lats2 is an essential mitotic regulator required for the coordination of cell division. J. Biol. Chem. 2007, 282, 19259–19271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, C.W.; Chang, Y.L.; Chang, Y.C.; Lin, J.C.; Chen, C.C.; Pan, S.H.; Wu, C.T.; Chen, H.Y.; Yang, S.C.; Hong, T.M.; et al. MicroRNA-135b promotes lung cancer metastasis by regulating multiple targets in the Hippo pathway and LZTS1. Nat. Commun. 2013, 4, 1877. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Zuo, W.; Zeng, Q.; Qian, Y.; Li, Y.; Liu, C.; Wang, J.; Zhong, S.; Bu, Y.; Hu, G. Loss of NFBD1/MDC1 disrupts homologous recombination repair and sensitizes nasopharyngeal carcinoma cells to PARP inhibitors. J. Biomed. Sci. 2019, 26, 14. [Google Scholar] [CrossRef] [Green Version]
- Miller, R.E.; Leary, A.; Scott, C.L.; Serra, V.; Lord, C.J.; Bowtell, D.; Chang, D.K.; Garsed, D.W.; Jonkers, J.; Ledermann, J.A.; et al. ESMO recommendations on predictive biomarker testing for homologous recombination deficiency and PARP inhibitor benefit in ovarian cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2020, 31, 1606–1622. [Google Scholar] [CrossRef]
- Cruz, C.; Castroviejo-Bermejo, M.; Gutiérrez-Enríquez, S.; Llop-Guevara, A.; Ibrahim, Y.H.; Gris-Oliver, A.; Bonache, S.; Morancho, B.; Bruna, A.; Rueda, O.M.; et al. RAD51 foci as a functional biomarker of homologous recombination repair and PARP inhibitor resistance in germline BRCA-mutated breast cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2018, 29, 1203–1210. [Google Scholar] [CrossRef]
- Zhang, J.; Ma, Z.; Treszezamsky, A.; Powell, S.N. MDC1 interacts with Rad51 and facilitates homologous recombination. Nat. Struct. Mol. Biol. 2005, 12, 902–909. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Wang, Y.; Liu, X.; Yang, Y.; Wu, S.; Liu, Y. SFN Enhanced the Radiosensitivity of Cervical Cancer Cells via Activating LATS2 and Blocking Rad51/MDC1 Recruitment to DNA Damage Site. Cancers 2022, 14, 1872. https://doi.org/10.3390/cancers14081872
Wang S, Wang Y, Liu X, Yang Y, Wu S, Liu Y. SFN Enhanced the Radiosensitivity of Cervical Cancer Cells via Activating LATS2 and Blocking Rad51/MDC1 Recruitment to DNA Damage Site. Cancers. 2022; 14(8):1872. https://doi.org/10.3390/cancers14081872
Chicago/Turabian StyleWang, Shiyu, Yanan Wang, Xiangnan Liu, Yongbin Yang, Sufang Wu, and Yuan Liu. 2022. "SFN Enhanced the Radiosensitivity of Cervical Cancer Cells via Activating LATS2 and Blocking Rad51/MDC1 Recruitment to DNA Damage Site" Cancers 14, no. 8: 1872. https://doi.org/10.3390/cancers14081872
APA StyleWang, S., Wang, Y., Liu, X., Yang, Y., Wu, S., & Liu, Y. (2022). SFN Enhanced the Radiosensitivity of Cervical Cancer Cells via Activating LATS2 and Blocking Rad51/MDC1 Recruitment to DNA Damage Site. Cancers, 14(8), 1872. https://doi.org/10.3390/cancers14081872




