Krüppel-like Factor 9 (KLF9) Suppresses Hepatocellular Carcinoma (HCC)-Promoting Oxidative Stress and Inflammation in Mice Fed High-Fat Diet
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals and Diet
2.2. Histology and Immunohistochemistry (IHC)
2.3. Serum Leptin and Adiponectin
2.4. Triglyceride Assay
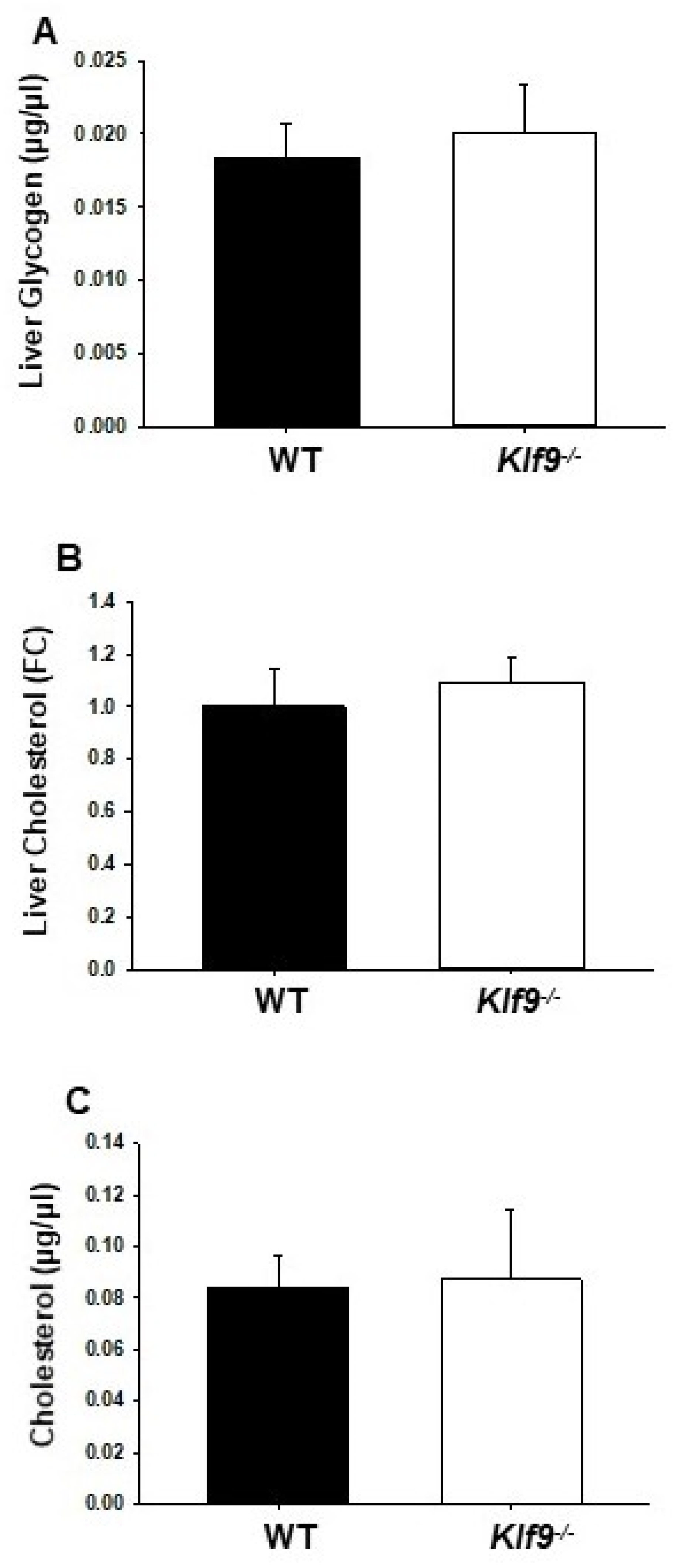
2.5. Glycogen and Cholesterol Assays
2.6. Oxidative Stress
2.7. RNA Isolation and Quantitative RT-PCR (qPCR)
2.8. Statistical Analysis
3. Results
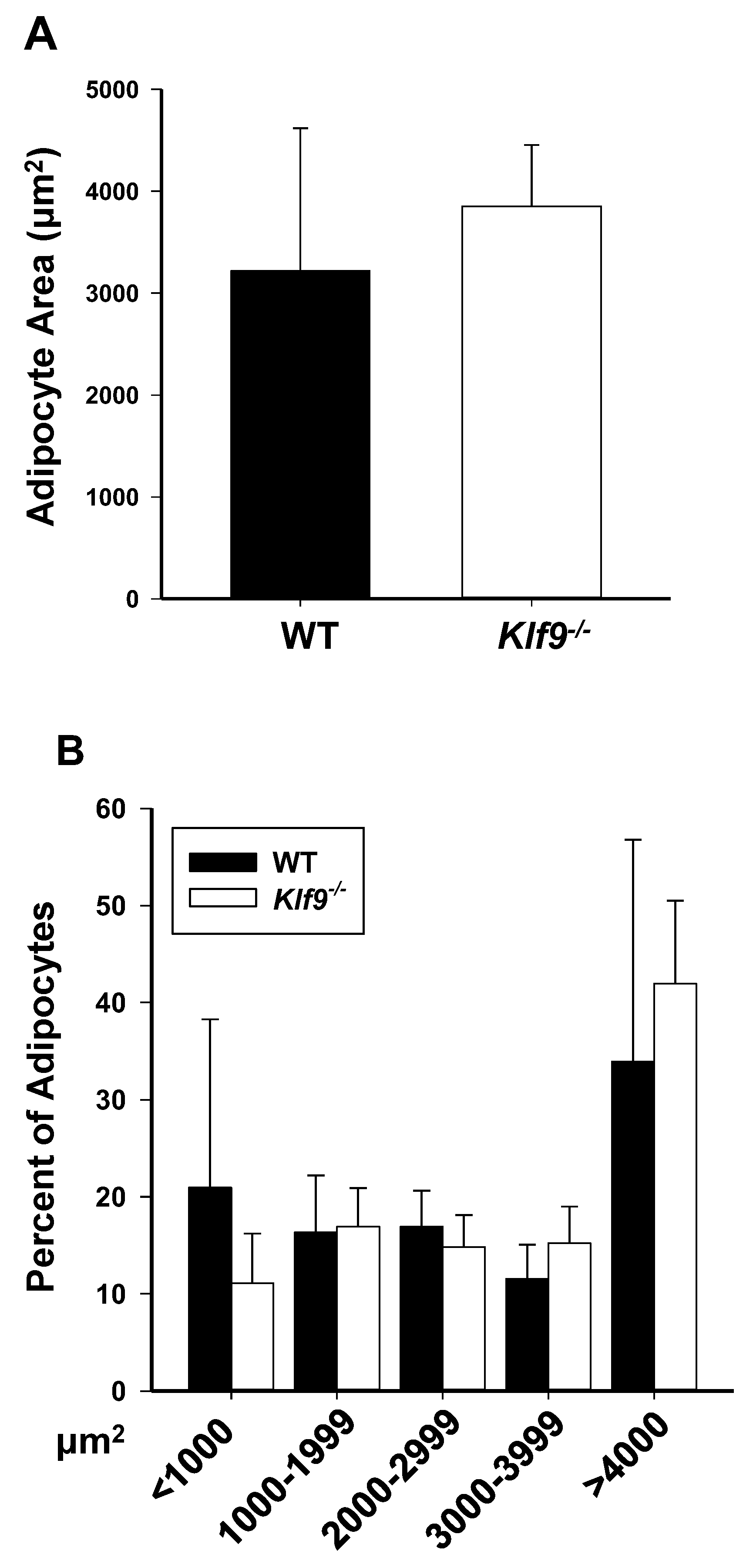
3.1. KO of Klf9 Had No Effect on Weight Gain, Fat Deposition, or Adipocyte Size in Mice Fed HFD
3.2. Increased Liver Weights in HFD-fed Klf9 KO Mice
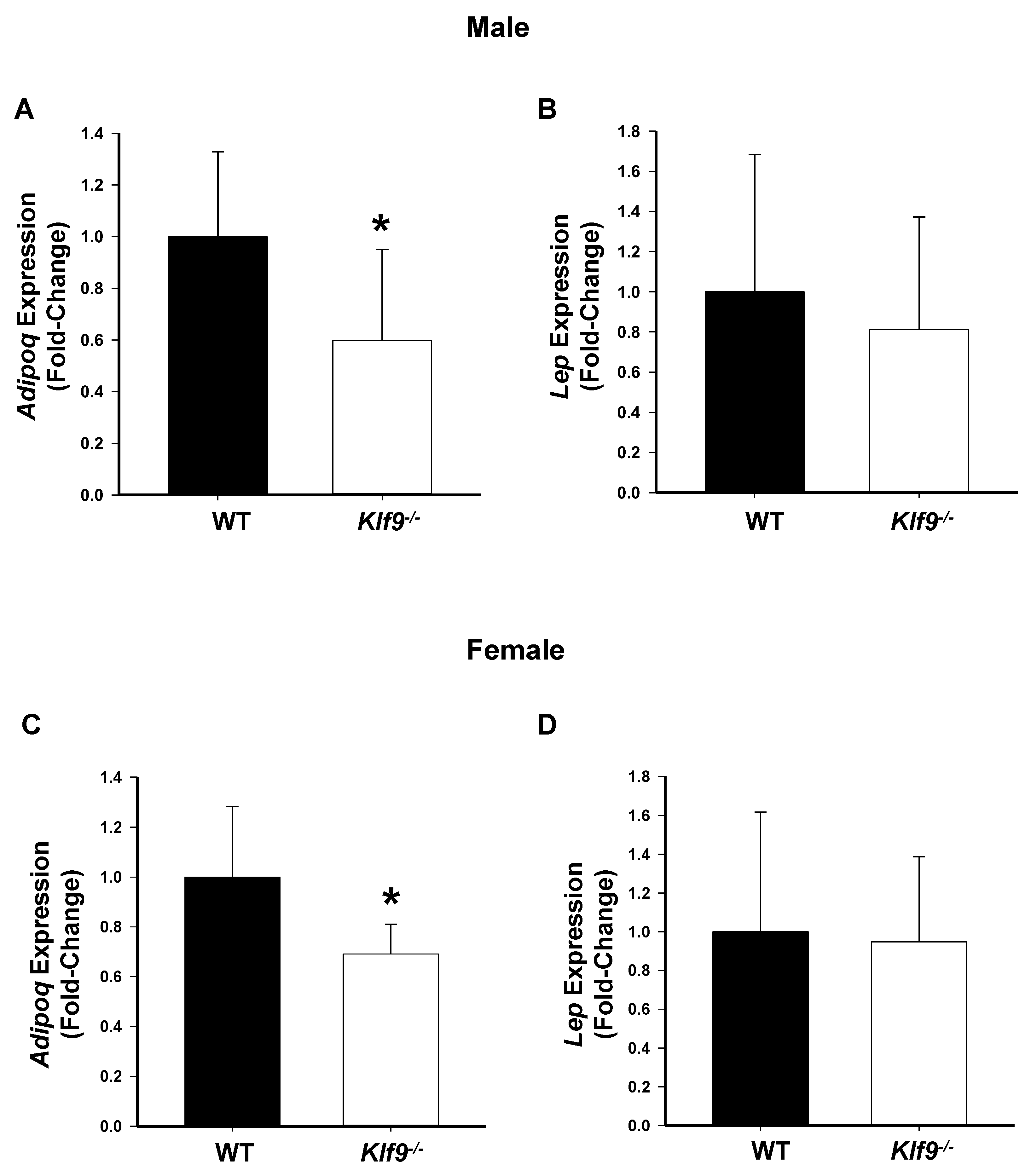
3.3. Effect of Klf9 KO on Circulating Leptin and Adiponectin Levels
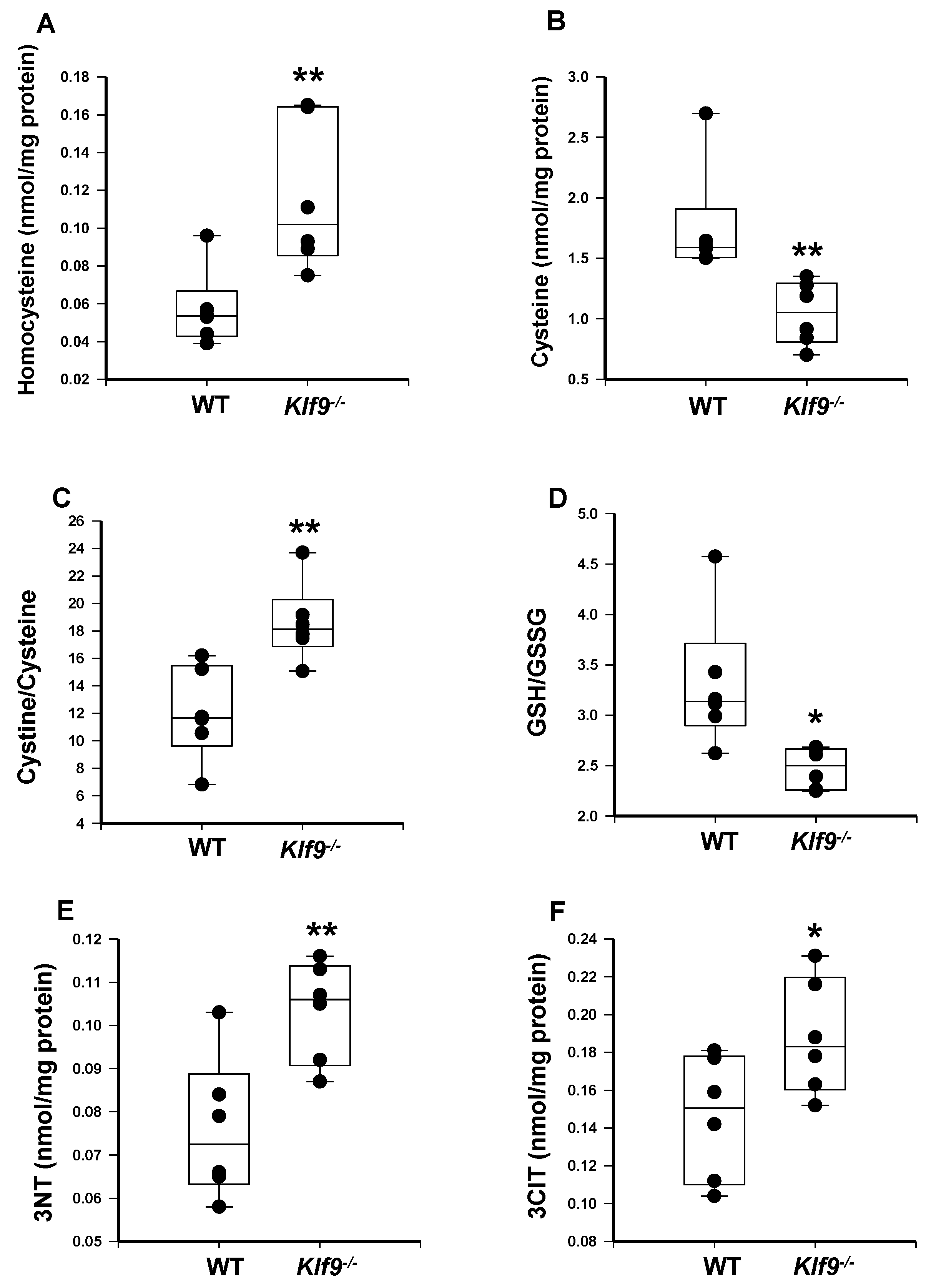
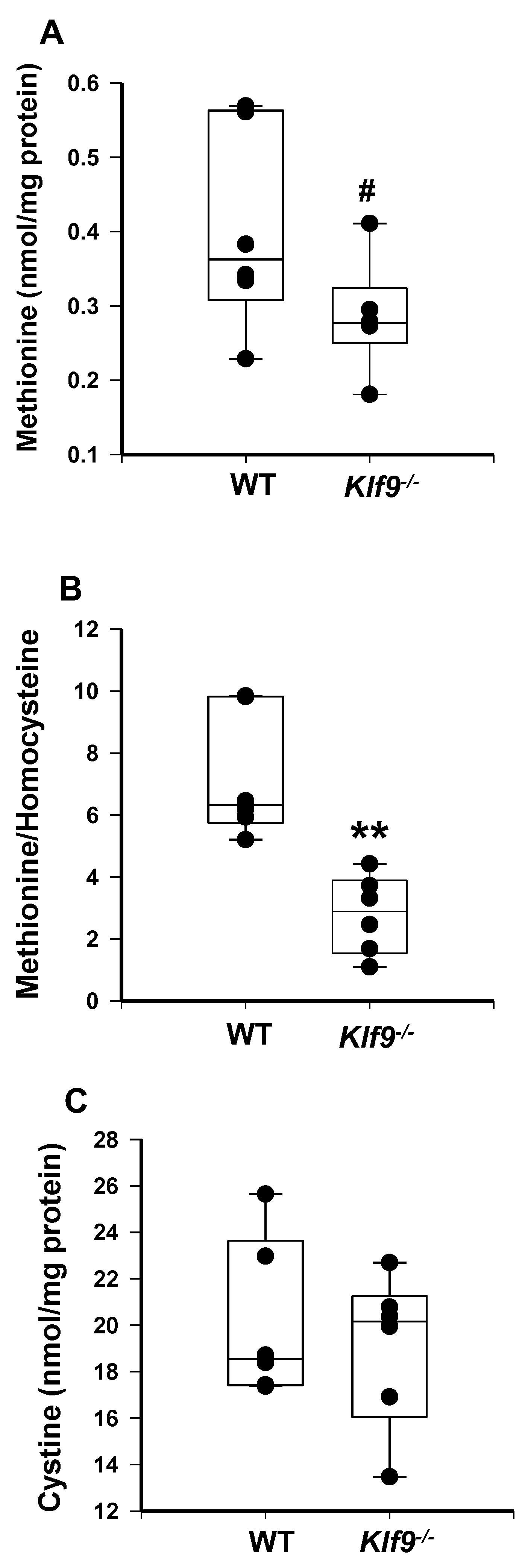
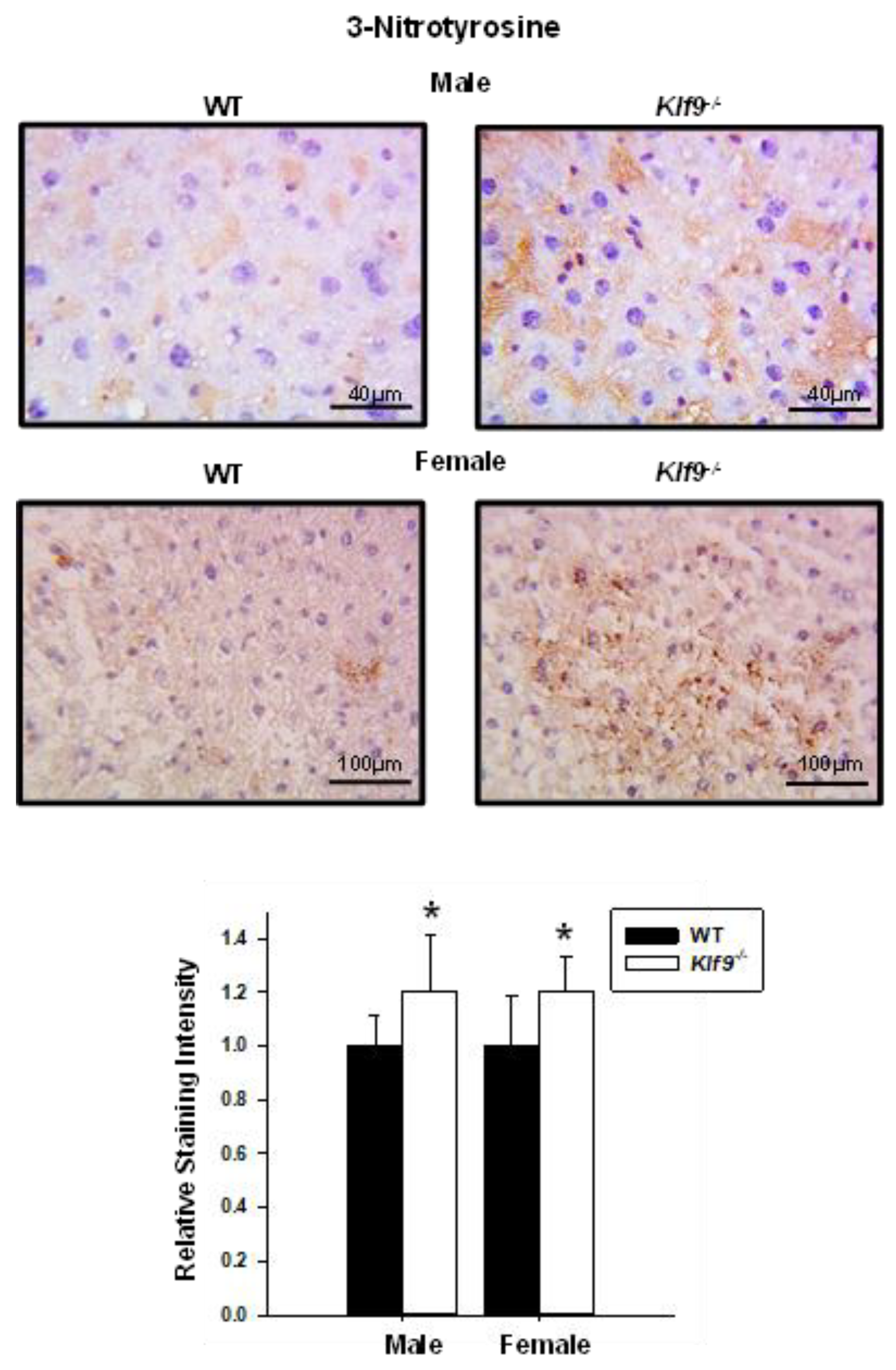
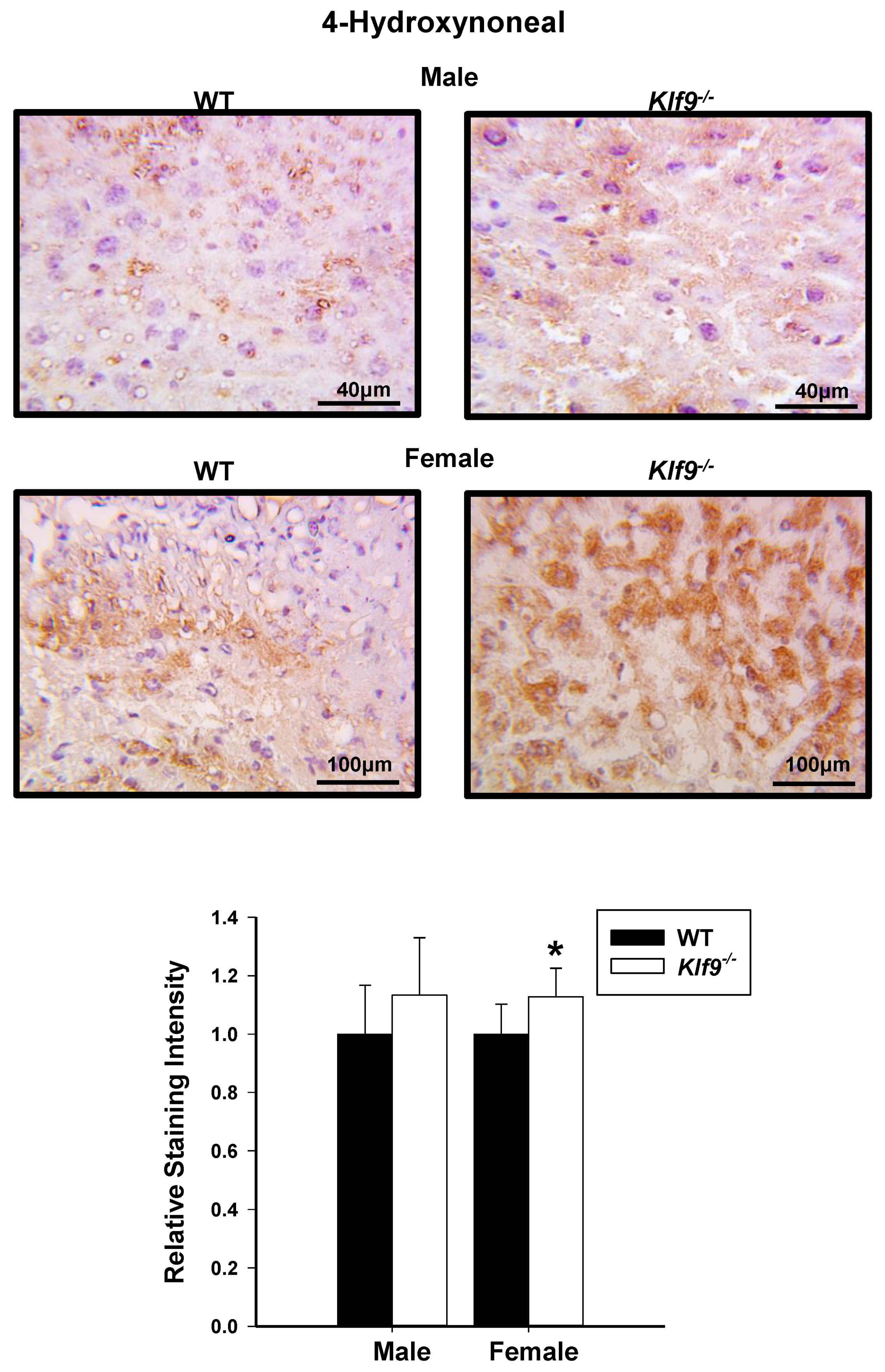
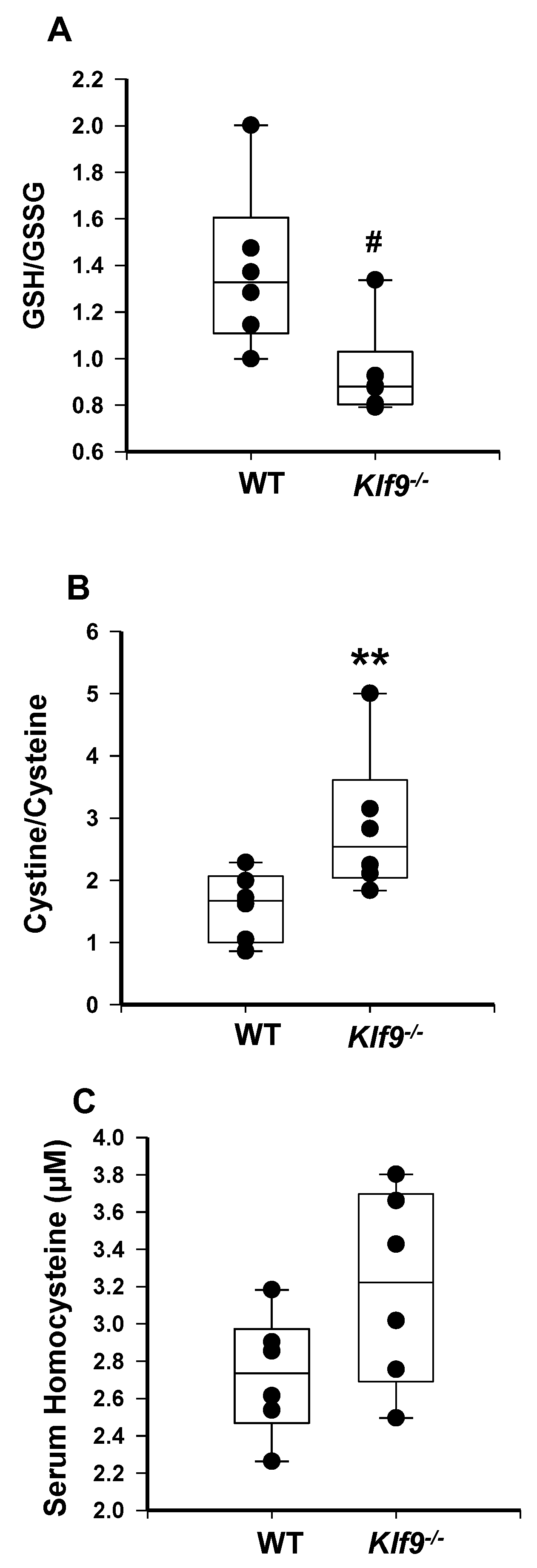
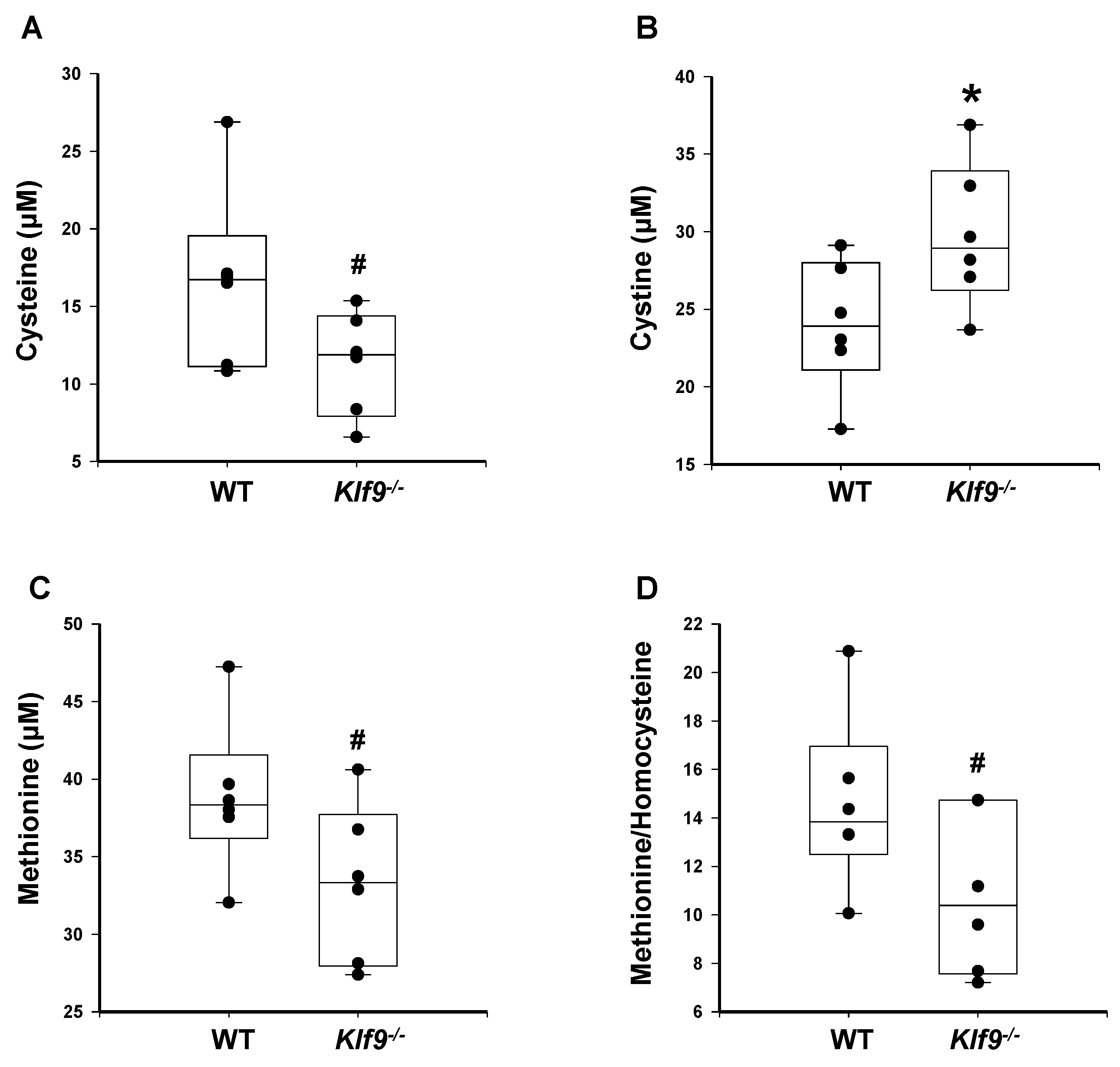
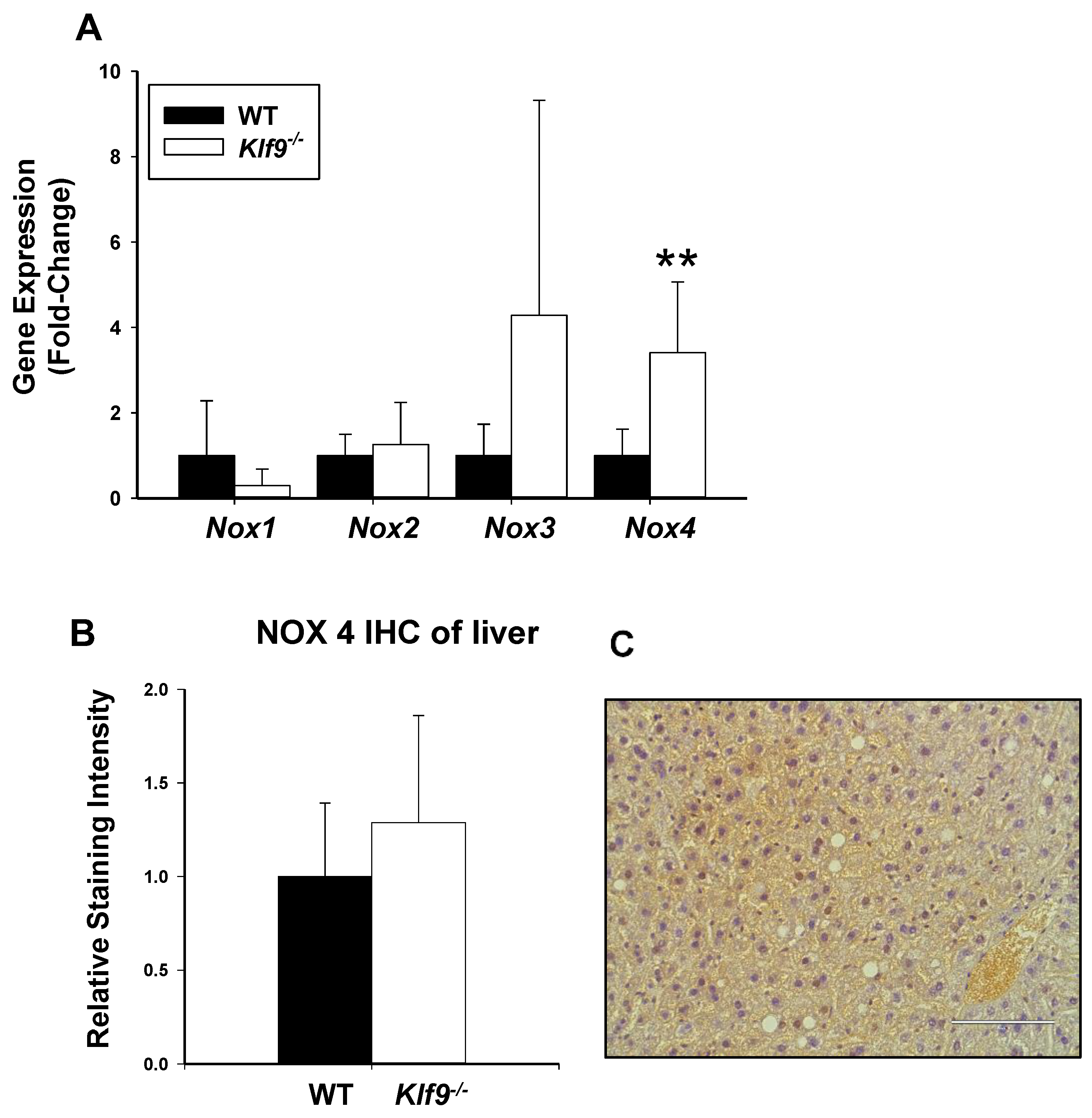
3.4. Increased Oxidative and Nitrosative Stress in Livers of Klf9 KO Mice
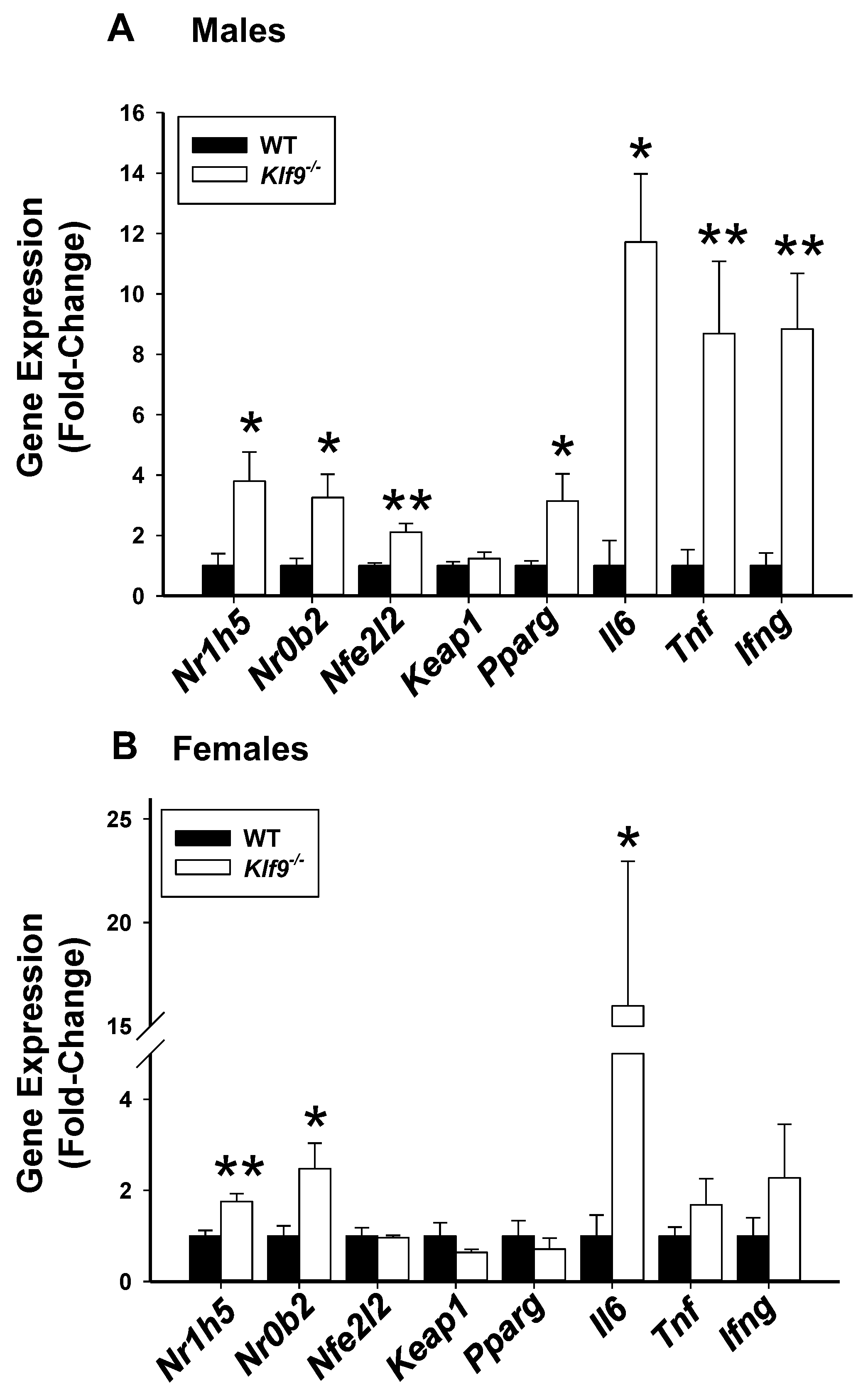
3.5. Hepatic Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lifshitz, F.; Lifshitz, J.Z. Globesity: The root causes of the obesity epidemic in the USA and now worldwide. Pediatr. Endocrinol. Rev. 2014, 12, 17–34. [Google Scholar] [PubMed]
- Simmen, F.A.; Simmen, R.C. The maternal womb: A novel target for cancer prevention in the era of the obesity pandemic? Eur. J. Cancer Prev. 2011, 20, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Liu, S.; Yang, M. Hepatocellular Carcinoma and Obesity, Type 2 Diabetes Mellitus, Cardiovascular Disease: Causing Factors, Molecular Links, and Treatment Options. Front. Endocrinol. 2021, 23, 808526. [Google Scholar] [CrossRef] [PubMed]
- Zelber-Sagi, S.; Noureddin, M.; Shibolet, O. Lifestyle and Hepatocellular Carcinoma What Is the Evidence and Prevention Recommendations. Cancers 2021, 14, 103. [Google Scholar] [CrossRef]
- Evseeva, M.N.; Balashova, M.S.; Kulebyakin, K.Y.; Rubtsov, Y.P. Adipocyte Biology from the Perspective of In Vivo Research: Review of Key Transcription Factors. Int. J. Mol. Sci. 2021, 23, 322. [Google Scholar] [CrossRef]
- Okada, Y.; Kubo, M.; Ohmiya, H.; Takahashi, A.; Kumasaka, N.; Hosono, N.; Maeda, S.; Wen, W.; Dorajoo, R.; Go, M.J.; et al. Common variants at CDKAL1 and KLF9 are associated with body mass index in east Asian populations. Nat. Genet. 2012, 44, 302–306. [Google Scholar] [CrossRef]
- Pei, H.; Yao, Y.; Yang, Y.; Liao, K.; Wu, J.R. Krüppel-like factor KLF9 regulates PPARγ transactivation at the middle stage of adipogenesis. Cell Death Differ. 2011, 18, 315–327. [Google Scholar] [CrossRef]
- Kimura, H.; Fujimori, K. Activation of early phase of adipogenesis through Krüppel-like factor KLF9-mediated, enhanced expression of CCAAT/enhancer-binding protein β in 3T3-L1 cells. Gene 2014, 534, 169–176. [Google Scholar] [CrossRef]
- Fan, H.; Zhang, Y.; Zhang, J.; Yao, Q.; Song, Y.; Shen, Q.; Lin, J.; Gao, Y.; Wang, X.; Zhang, L.; et al. Cold-Inducible Klf9 Regulates Thermogenesis of Brown and Beige Fat. Diabetes 2020, 69, 2603–2618. [Google Scholar] [CrossRef]
- Betteridge, D.J. What is oxidative stress? Metabolism 2000, 49, 3–8. [Google Scholar] [CrossRef]
- Haklar, G.; Sayin-Ozveri, E.; Yüksel, M.; Aktan, A.O.; Yalçin, A.S. Different kinds of reactive oxygen and nitrogen species were detected in colon and breast tumors. Cancer Lett. 2001, 165, 219–224. [Google Scholar] [CrossRef]
- Marra, M.; Sordelli, I.M.; Lombardi, A.; Lamberti, M.; Tarantino, L.; Giudice, A.; Stiuso, P.; Abbruzzese, A.; Sperlongano, R.; Accardo, M.; et al. Molecular targets and oxidative stress biomarkers in hepatocellular carcinoma: An overview. J. Transl. Med. 2011, 10, 171. [Google Scholar] [CrossRef] [PubMed]
- Reuter, S.; Gupta, S.C.; Chaturvedi, M.M.; Aggarwal, B.B. Oxidative stress, inflammation, and cancer: How are they linked? Free Radic. Biol. Med. 2010, 49, 1603–1616. [Google Scholar] [CrossRef] [PubMed]
- Skalicky, J.; Muzakova, V.; Kandar, R.; Meloun, M.; Rousar, T.; Palicka, V. Evaluation of oxidative stress and inflammation in obese adults with metabolic syndrome. Clin. Chem. Lab. Med. 2008, 46, 499–505. [Google Scholar] [CrossRef]
- Furukawa, S.; Fujita, T.; Shimabukuro, M.; Iwaki, M.; Yamada, Y.; Nakajima, Y.; Nakayama, O.; Makishima, M.; Matsuda, M.; Shimomura, I. Increased oxidative stress in obesity and its impact on metabolic syndrome. J. Clin. Investig. 2004, 114, 1752–1761. [Google Scholar] [CrossRef]
- Matsuzawa-Nagata, N.; Takamura, T.; Ando, H.; Nakamura, S.; Kurita, S.; Misu, H.; Ota, T.; Yokoyama, M.; Honda, M.; Miyamoto, K.; et al. Increased oxidative stress precedes the onset of high-fat diet-induced insulin resistance and obesity. Metabolism 2008, 57, 1071–1077. [Google Scholar] [CrossRef]
- Imataka, H.; Sogawa, K.; Yasumoto, K.; Kikuchi, Y.; Sasano, K.; Kobayashi, A.; Hayami, M.; Fujii-Kuriyama, Y. Two regulatory proteins that bind to the basic transcription element (BTE), a GC box sequence in the promoter region of the rat P-4501A1 gene. EMBO J. 1992, 11, 3663–3671. [Google Scholar] [CrossRef]
- Imataka, H.; Mizuno, A.; Fujii-Kuriyama, Y.; Hayami, M. Activation of the human immunodeficiency virus type 1 long terminal repeat by BTEB, a GC box-binding transcription factor. AIDS Res. Hum. Retrovir. 1993, 9, 825–831. [Google Scholar] [CrossRef]
- Foti, D.; Stroup, D.; Chiang, J.Y. Basic transcription element binding protein (BTEB) transactivates the cholesterol 7 alpha-hydroxylase gene (CYP7A). Biochem. Biophys. Res. Commun. 1998, 253, 109–113. [Google Scholar] [CrossRef]
- Koh, K.H.; Pan, X.; Zhang, W.; McLachlan, A.; Urrutia, R.; Jeong, H. Krüppel-like factor 9 promotes hepatic cytochrome P450 2D6 expression during pregnancy in CYP2D6-humanized mice. Mol. Pharmacol. 2014, 86, 727–735. [Google Scholar] [CrossRef]
- Cui, A.; Fan, H.; Zhang, Y.; Zhang, Y.; Niu, D.; Liu, S.; Liu, Q.; Ma, W.; Shen, Z.; Shen, L.; et al. Dexamethasone-induced Krüppel-like factor 9 expression promotes hepatic gluconeogenesis and hyperglycemia. J. Clin. Investig. 2019, 129, 2266–2278. [Google Scholar] [CrossRef]
- Tien, E.S.; Matsui, K.; Moore, R.; Negishi, M. The nuclear receptor constitutively active/androstane receptor regulates type 1 deiodinase and thyroid hormone activity in the regenerating mouse liver. J. Pharmacol. Exp. Ther. 2007, 320, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Cvoro, A.; Devito, L.; Milton, F.A.; Noli, L.; Zhang, A.; Filippi, C.; Sakai, K.; Suh, J.H.; Sieglaff, D.H.; Dhawan, A.; et al. A thyroid hormone receptor/KLF9 axis in human hepatocytes and pluripotent stem cells. Stem Cells. 2015, 33, 416–428. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.Z.; Cheng, Y.; He, H.; Liu, H.Y.; Liu, Y.F. The fate of Krüppel-like factor 9-positive hepatic carcinoma cells may be determined by the programmed cell death protein 5. Int. J. Oncol. 2014, 44, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Wang, B.; Liu, Y.; Zhang, L.; Ma, A.; Yang, Z.; Ji, Y.; Liu, Y. Transcription factor KLF9 suppresses the growth of hepatocellular carcinoma cells in vivo and positively regulates p53 expression. Cancer Lett. 2014, 355, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Kowalik, M.A.; Puliga, E.; Cabras, L.; Sulas, P.; Petrelli, A.; Perra, A.; Ledda-Columbano, G.M.; Morandi, A.; Merlin, S.; Orrù, C.; et al. Thyroid hormone inhibits hepatocellular carcinoma progression via induction of differentiation and metabolic reprogramming. J. Hepatol. 2020, 72, 1159–1169. [Google Scholar] [CrossRef]
- Zucker, S.N.; Fink, E.E.; Bagati, A.; Mannava, S.; Bianchi-Smiraglia, A.; Bogner, P.N.; Wawrzyniak, J.A.; Foley, C.; Leonova, K.I.; Grimm, M.J.; et al. Nrf2 amplifies oxidative stress via induction of Klf9. Mol. Cell 2014, 53, 916–928. [Google Scholar] [CrossRef]
- Bettaieb, A.; Jiang, J.X.; Sasaki, Y.; Chao, T.I.; Kiss, Z.; Chen, X.; Tian, J.; Katsuyama, M.; Yabe-Nishimura, C.; Xi, Y.; et al. Hepatocyte Nicotinamide Adenine Dinucleotide Phosphate Reduced Oxidase 4 Regulates Stress Signaling, Fibrosis, and Insulin Sensitivity During Development of Steatohepatitis in Mice. Gastroenterology 2015, 149, 468–480.e10. [Google Scholar] [CrossRef]
- Matsumoto, M.; Zhang, J.; Zhang, X.; Liu, J.; Jiang, J.X.; Yamaguchi, K.; Taruno, A.; Katsuyama, M.; Iwata, K.; Ibi, M.; et al. The NOX1 isoform of NADPH oxidase is involved in dysfunction of liver sinusoids in nonalcoholic fatty liver disease. Free Radic. Biol. Med. 2018, 115, 412–420. [Google Scholar] [CrossRef]
- Velarde, M.C.; Geng, Y.; Eason, R.R.; Simmen, F.A.; Simmen, R.C. Null mutation of Kruppel-like factor9/basic transcription element binding protein-1 alters peri-implantation uterine development in mice. Biol. Reprod. 2005, 73, 472–481. [Google Scholar] [CrossRef][Green Version]
- Melnyk, S.; Pogribna, M.; Pogribny, I.; Hine, R.J.; James, S.J. A new HPLC method for the simultaneous determination of oxidized and reduced plasma aminothiols using coulometric electrochemical detection. J. Nutr. Biochem. 1999, 10, 490–497. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef] [PubMed]
- Trujillo, M.E.; Scherer, P.E. Adipose tissue-derived factors: Impact on health and disease. Endocr. Rev. 2006, 27, 762–778. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, C.; Surmacz, E. Leptin and cancer. J. Cell. Physiol. 2006, 207, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Kelesidis, I.; Kelesidis, T.; Mantzoros, C.S. Adiponectin and cancer: A systematic review. Br. J. Cancer 2006, 94, 1221–1225. [Google Scholar] [CrossRef] [PubMed]
- Simmons, C.D.; Pabona, J.M.; Zeng, Z.; Velarde, M.C.; Gaddy, D.; Simmen, F.A.; Simmen, R.C. Response of adult mouse uterus to early disruption of estrogen receptor-alpha signaling is influenced by Krüppel-like factor 9. J. Endocrinol. 2010, 205, 147–157. [Google Scholar] [CrossRef][Green Version]
- Simmons, C.D.; Pabona, J.M.; Heard, M.E.; Friedman, T.M.; Spataro, M.T.; Godley, A.L.; Simmen, F.A.; Burnett, A.F.; Simmen, R.C. Krüppel-like factor 9 loss-of-expression in human endometrial carcinoma links altered expression of growth-regulatory genes with aberrant proliferative response to estrogen. Biol. Reprod. 2011, 85, 378–385. [Google Scholar] [CrossRef]
- Brown, A.R.; Simmen, R.C.; Raj, V.R.; Van, T.T.; MacLeod, S.L.; Simmen, F.A. Krüppel-like factor 9 (KLF9) prevents colorectal cancer through inhibition of interferon-related signaling. Carcinogenesis 2015, 36, 946–955. [Google Scholar] [CrossRef]
- Bai, X.; Jiang, X.; Liu, Y.; Wang, Y.; Jiang, X.; Song, G.; Qiu, H.; Zhang, Q. Krüppel-like factor 9 upregulates E-cadherin transcription and represses breast cancer invasion and metastasis. Am. J. Cancer Res. 2021, 11, 3660–3673. [Google Scholar]
- Ying, M.; Sang, Y.; Li, Y.; Guerrero-Cazares, H.; Quinones-Hinojosa, A.; Vescovi, A.L.; Eberhart, C.G.; Xia, S.; Laterra, J. Krüppel-like family of transcription factor 9, a differentiation-associated transcription factor, suppresses Notch1 signaling and inhibits glioblastoma-initiating stem cells. Stem Cells 2011, 29, 20–31. [Google Scholar] [CrossRef]
- Wang, K.; Liu, S.; Dou, Z.; Zhang, S.; Yang, X. Loss of Krüppel-like factor 9 facilitates stemness in ovarian cancer ascites-derived multicellular spheroids via Notch1/slug signaling. Cancer Sci. 2021, 112, 4220–4233. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Yang, L.; Li, X. MicroRNA-652 promotes cell proliferation and osteosarcoma invasion by directly targeting KLF9. Exp. Ther. Med. 2020, 20, 2953–2960. [Google Scholar] [CrossRef]
- Collins, S.; Martin, T.L.; Surwit, R.S.; Robidoux, J. Genetic vulnerability to diet-induced obesity in the C57BL/6J mouse: Physiological and molecular characteristics. Physiol. Behav. 2004, 81, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Arciero, P.J.; Goran, M.I.; Poehlman, E.T. Resting metabolic rate is lower in women than in men. J. Appl. Physiol. 1993, 75, 2514–2520. [Google Scholar] [CrossRef]
- Pettersson, U.S.; Waldén, T.B.; Carlsson, P.O.; Jansson, L.; Phillipson, M. Female mice are protected against high-fat diet induced metabolic syndrome and increase the regulatory T cell population in adipose tissue. PLoS ONE 2012, 7, e46057. [Google Scholar] [CrossRef]
- Yang, D.; Lv, Z.; Zhang, H.; Liu, B.; Jiang, H.; Tan, X.; Lu, J.; Baiyun, R.; Zhang, Z. Activation of the Nrf2 Signaling Pathway Involving KLF9 Plays a Critical Role in Allicin Resisting Against Arsenic Trioxide-Induced Hepatotoxicity in Rats. Biol. Trace Elem. Res. 2017, 176, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Festi, D.; Colecchia, A.; Sacco, T.; Bondi, M.; Roda, E.; Marchesini, G. Hepatic steatosis in obese patients: Clinical aspects and prognostic significance. Obes. Rev. 2004, 5, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; Chait, A. Hypertriglyceridemia secondary to obesity and diabetes. Biochim. Biophys. Acta 2012, 1821, 819–825. [Google Scholar] [CrossRef] [PubMed]
- Hu, E.; Liang, P.; Spiegelman, B.M. AdipoQ is a novel adipose-specific gene dysregulated in obesity. J. Biol. Chem. 1996, 271, 10697–10703. [Google Scholar] [CrossRef]
- Kelesidis, T.; Kelesidis, I.; Chou, S.; Mantzoros, C.S. Narrative review: The role of leptin in human physiology: Emerging clinical applications. Ann. Intern. Med. 2010, 152, 93–100. [Google Scholar] [CrossRef]
- Heard, M.E.; Pabona, J.M.; Clayberger, C.; Krensky, A.M.; Simmen, F.A.; Simmen, R.C. The reproductive phenotype of mice null for transcription factor Krüppel-like factor 13 suggests compensatory function of family member Krüppel-like factor 9 in the peri-implantation uterus. Biol. Reprod. 2012, 87, 115. [Google Scholar] [CrossRef] [PubMed]
- Basu, P.; Lung, T.K.; Lemsaddek, W.; Sargent, T.G.; Williams, D.C., Jr.; Basu, M.; Redmond, L.C.; Lingrel, J.B.; Haar, J.L.; Lloyd, J.A. EKLF and KLF2 have compensatory roles in embryonic beta-globin gene expression and primitive erythropoiesis. Blood 2007, 110, 3417–3425. [Google Scholar] [CrossRef]
- McConnell, B.B.; Yang, V.W. Mammalian Krüppel-like factors in health and diseases. Physiol. Rev. 2010, 90, 1337–1381. [Google Scholar] [CrossRef] [PubMed]
- Hergenreider, E.; Heydt, S.; Tréguer, K.; Boettger, T.; Horrevoets, A.J.; Zeiher, A.M.; Scheffer, M.P.; Frangakis, A.S.; Yin, X.; Mayr, M.; et al. Atheroprotective communication between endothelial cells and smooth muscle cells through miRNAs. Nat. Cell Biol. 2012, 14, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Natarelli, L.; Schober, A. Janus-faced role of Krüppel-like factor 2-dependent regulation of microRNAs in endothelial proliferation. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 1605–1606. [Google Scholar] [CrossRef][Green Version]
- Abdelmalek, M.F.; Diehl, A.M. Nonalcoholic fatty liver disease as a complication of insulin resistance. Med. Clin. N. Am. 2007, 91, 1125–1149. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.G.; Yang, C.W.; Chung, C.H.; Ho, C.L.; Chen, W.L.; Chien, W.C. The association between metabolic risk factors, nonalcoholic fatty liver disease, and the incidence of liver cancer: A nationwide population-based cohort study. Hepatol. Int. 2022. [Google Scholar] [CrossRef]
- Bialkowska, A.B.; Yang, V.W.; Mallipattu, S.K. Krüppel-like factors in mammalian stem cells and development. Development 2017, 144, 737–754. [Google Scholar] [CrossRef]
- Bonett, R.M.; Hu, F.; Bagamasbad, P.; Denver, R.J. Stressor and glucocorticoid-dependent induction of the immediate early gene kruppel-like factor 9: Implications for neural development and plasticity. Endocrinology 2009, 150, 1757–1765. [Google Scholar] [CrossRef] [PubMed]
- Vacca, M.; D’Amore, S.; Graziano, G.; D’Orazio, A.; Cariello, M.; Massafra, V.; Salvatore, L.; Martelli, N.; Murzilli, S.; Lo Sasso, G.; et al. Clustering nuclear receptors in liver regeneration identifies candidate modulators of hepatocyte proliferation and hepatocarcinoma. PLoS ONE 2014, 9, e1044492014. [Google Scholar] [CrossRef] [PubMed]
- Batusic, D.S.; von Bargen, A.; Blaschke, S.; Dudas, J.; Ramadori, G. Different physiology of interferon-α/-γ in models of liver regeneration in the rat. Histochem. Cell Biol. 2011, 136, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Norris, C.A.; He, M.; Kang, L.I.; Ding, M.Q.; Radder, J.E.; Haynes, M.M.; Yang, Y.; Paranjpe, S.; Bowen, W.C.; Orr, A.; et al. Correction: Synthesis of IL-6 by Hepatocytes Is a Normal Response to Common Hepatic Stimuli. PLoS ONE 2019, 14, e0224498. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Arras, D.; Rose-John, S. IL-6 pathway in the liver: From physiopathology to therapy. J. Hepatol. 2016, 64, 1403–1415. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Yang, H.; Learned, R.M.; Tian, H.; Ling, L. Non-cell-autonomous activation of IL-6/STAT3 signaling mediates FGF19-driven hepatocarcinogenesis. Nat. Commun. 2017, 8, 15433. [Google Scholar] [CrossRef]
- Obrador, E.; Benlloch, M.; Pellicer, J.A.; Asensi, M.; Estrela, J.M. Intertissue flow of glutathione (GSH) as a tumor growth-promoting mechanism: Interleukin 6 induces GSH release from hepatocytes in metastatic B16 melanoma-bearing mice. J. Biol. Chem. 2011, 286, 15716–15727. [Google Scholar] [CrossRef]
- Loscalzo, J. The oxidant stress of hyperhomocyst(e)inemia. J. Clin. Investig. 1996, 98, 5–7. [Google Scholar] [CrossRef] [PubMed]
- Domagała, T.B.; Undas, A.; Libura, M.; Szczeklik, A. Pathogenesis of vascular disease in hyperhomocysteinaemia. J. Cardiovasc. Risk 1998, 5, 239–247. [Google Scholar] [CrossRef]
- James, S.J.; Rose, S.; Melnyk, S.; Jernigan, S.; Blossom, S.; Pavliv, O.; Gaylor, D.W. Cellular and mitochondrial glutathione redox imbalance in lymphoblastoid cells derived from children with autism. FASEB J. 2009, 23, 2374–2383. [Google Scholar] [CrossRef] [PubMed]
- Tretter, V.; Hochreiter, B.; Zach, M.L.; Krenn, K.; Klein, K.U. Understanding Cellular Redox Homeostasis: A Challenge for Precision Medicine. Int. J. Mol. Sci. 2021, 23, 106. [Google Scholar] [CrossRef]
- Ahn, B.; Han, B.S.; Kim, D.J.; Ohshima, H. Immunohistochemical localization of inducible nitric oxide synthase and 3-nitrotyrosine in rat liver tumors induced by N-nitrosodiethylamine. Carcinogenesis 1999, 20, 1337–1344. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vermorken, A.J.; Zhu, J.; Andrès, E. Obesity and colorectal cancer risk: The role of oxidative stress. Gut 2014, 63, 529–530. [Google Scholar] [CrossRef] [PubMed]
- Hazen, S.L.; Heinecke, J.W. 3-Chlorotyrosine, a specific marker of myeloperoxidase-catalyzed oxidation, is markedly elevated in low density lipoprotein isolated from human atherosclerotic intima. J. Clin. Investig. 1997, 99, 2075–2081. [Google Scholar] [CrossRef] [PubMed]
- Buss, I.H.; Senthilmohan, R.; Darlow, B.A.; Mogridge, N.; Kettle, A.J.; Winterbourn, C.C. 3-Chlorotyrosine as a marker of protein damage by myeloperoxidase in tracheal aspirates from preterm infants: Association with adverse respiratory outcome. Pediatr. Res. 2003, 53, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Poli, G.; Schaur, R.J. 4-Hydroxynonenal in the pathomechanisms of oxidative stress. IUBMB Life 2000, 50, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Dalleau, S.; Baradat, M.; Guéraud, F.; Huc, L. Cell death and diseases related to oxidative stress: 4-hydroxynonenal (HNE) in the balance. Cell Death Differ. 2013, 20, 1615–1630. [Google Scholar] [CrossRef] [PubMed]
- de Carvalho, S.C.; Muniz, M.T.; Siqueira, M.D.; Siqueira, E.R.; Gomes, A.V.; Silva, K.A.; Bezerra, L.C.; D’Almeida, V.; de Oliveira, C.P.; Pereira, L.M. Plasmatic higher levels of homocysteine in non-alcoholic fatty liver disease (NAFLD). Nutr. J. 2013, 12, 37. [Google Scholar] [CrossRef] [PubMed]
- Pastore, A.; Alisi, A.; di Giovamberardino, G.; Crudele, A.; Ceccarelli, S.; Panera, N.; Dionisi-Vici, C.; Nobili, V. Plasma levels of homocysteine and cysteine increased in pediatric NAFLD and strongly correlated with severity of liver damage. Int. J. Mol. Sci. 2014, 15, 21202–21214. [Google Scholar] [CrossRef] [PubMed]
- Matté, C.; Stefanello, F.M.; Mackedanz, V.; Pederzolli, C.D.; Lamers, M.L.; Dutra-Filho, C.S.; Dos Santos, M.F.; Wyse, A.T. Homocysteine induces oxidative stress, inflammatory infiltration, fibrosis and reduces glycogen/glycoprotein content in liver of rats. Int. J. Dev. Neurosci. 2009, 27, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Bartoli, G.M.; Sies, H. Reduced and oxidized glutathione efflux from liver. FEBS Lett. 1978, 86, 89–91. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, J.; Li, Q.; Yang, W.; Song, Q.; Li, W.; Liu, J. KLF4 promotes hydrogen-peroxide-induced apoptosis of chronic myeloid leukemia cells involving the bcl-2/bax pathway. Cell Stress Chaperones 2010, 15, 905–912. [Google Scholar] [CrossRef]
- Zhang, G.; Zhu, H.; Wang, Y.; Yang, S.; Liu, M.; Zhang, W.; Quan, L.; Bai, J.; Liu, Z.; Xu, N. Krüppel-like factor 4 represses transcription of the survivin gene in esophageal cancer cell lines. Biol. Chem. 2009, 390, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Nagare, T.; Sakaue, H.; Matsumoto, M.; Cao, Y.; Inagaki, K.; Sakai, M.; Takashima, Y.; Nakamura, K.; Mori, T.; Okada, Y.; et al. Overexpression of KLF15 transcription factor in adipocytes of mice results in down-regulation of SCD1 protein expression in adipocytes and consequent enhancement of glucose-induced insulin secretion. J. Biol. Chem. 2011, 286, 37458–37469. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Zapico, M.E.; Mladek, A.; Ellenrieder, V.; Folch-Puy, E.; Miller, L.; Urrutia, R. An mSin3A interaction domain links the transcriptional activity of KLF11 with its role in growth regulation. EMBO J. 2003, 22, 4748–4758. [Google Scholar] [CrossRef]
- Knoedler, J.R.; Ávila-Mendoza, J.; Subramani, A.; Denver, R.J. The Paralogous Krüppel-like Factors 9 and 13 Regulate the Mammalian Cellular Circadian Clock Output Gene Dbp. J. Biol. Rhythms. 2020, 35, 257–274. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Wang, X.; Antonson, P.; Gustafsson, J.Å.; Li, Z. Genomics of sex hormone receptor signaling in hepatic sexual dimorphism. Mol. Cell. Endocrinol. 2018, 471, 33–41. [Google Scholar] [CrossRef] [PubMed]
- European Patent Office. Available online: https://patentimages.storage.googleapis.com/a3/e8/db/63ae2cfb9a1757/EP1757302B1.pdf (accessed on 14 March 2022).
- Turano, M.; Cammarota, F.; Duraturo, F.; Izzo, P.; De Rosa, M. A Potential Role of IL-6/IL-6R in the Development and Management of Colon Cancer. Membranes 2021, 11, 312. [Google Scholar] [CrossRef] [PubMed]


| Fold change in body weight (BW) after 12 wk on HFD; n = 8–9 mice/sex/genotype | |||
| WT males | Klf9−/− males | WT females | Klf9−/− females |
| 1.775 + 0.204 | 1.851 + 0.263 | 1.416 + 0.0984 | 1.490 + 0.0832 |
| Males (WT plus Klf9−/−) | Females (WT plus Klf9−/−) | ||
| 1.811 + 0.228 | 1.455 + 0.0957 1 | ||
| Retroperitoneal fat weight (normalized to BW); n = 8–9 mice/sex/genotype | |||
| WT males | Klf9−/− males | WT females | Klf9−/− females |
| 1 + 0.299 | 0.868 + 0.400 | 1 + 0.732 | 1.237 + 0.681 |
| Gonadal fat weight (normalized to BW); n = 8–9 mice/sex/genotype | |||
| WT males | Klf9−/− males | WT females | Klf9−/− females |
| 1 + 0.368 | 0.895 + 0.444 | 1 + 0.610 | 0.887 + 0.425 |
| Liver weight (normalized to BW); n = 8–9 mice/sex/genotype | |||
| WT males | Klf9−/− males | WT females | Klf9−/− females |
| 1 + 0.134 | 1.194 + 0.185 2 | 1 + 0.115 | 1.209 + 0.221 3 |
| Liver triglycerides (mg/dL) | Serum triglycerides (mg/dL) | ||
| WT females | Klf9−/− females | WT females | Klf9−/− females |
| 137.3 + 61.1 | 144.3 + 64.1 | 145.7 + 29.2 | 126.7 + 37.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brown, A.R.; Alhallak, I.; Simmen, R.C.M.; Melnyk, S.B.; Heard-Lipsmeyer, M.E.; Montales, M.T.E.; Habenicht, D.; Van, T.T.; Simmen, F.A. Krüppel-like Factor 9 (KLF9) Suppresses Hepatocellular Carcinoma (HCC)-Promoting Oxidative Stress and Inflammation in Mice Fed High-Fat Diet. Cancers 2022, 14, 1737. https://doi.org/10.3390/cancers14071737
Brown AR, Alhallak I, Simmen RCM, Melnyk SB, Heard-Lipsmeyer ME, Montales MTE, Habenicht D, Van TT, Simmen FA. Krüppel-like Factor 9 (KLF9) Suppresses Hepatocellular Carcinoma (HCC)-Promoting Oxidative Stress and Inflammation in Mice Fed High-Fat Diet. Cancers. 2022; 14(7):1737. https://doi.org/10.3390/cancers14071737
Chicago/Turabian StyleBrown, Adam R., Iad Alhallak, Rosalia C. M. Simmen, Stepan B. Melnyk, Melissa E. Heard-Lipsmeyer, Maria Theresa E. Montales, Daniel Habenicht, Trang T. Van, and Frank A. Simmen. 2022. "Krüppel-like Factor 9 (KLF9) Suppresses Hepatocellular Carcinoma (HCC)-Promoting Oxidative Stress and Inflammation in Mice Fed High-Fat Diet" Cancers 14, no. 7: 1737. https://doi.org/10.3390/cancers14071737
APA StyleBrown, A. R., Alhallak, I., Simmen, R. C. M., Melnyk, S. B., Heard-Lipsmeyer, M. E., Montales, M. T. E., Habenicht, D., Van, T. T., & Simmen, F. A. (2022). Krüppel-like Factor 9 (KLF9) Suppresses Hepatocellular Carcinoma (HCC)-Promoting Oxidative Stress and Inflammation in Mice Fed High-Fat Diet. Cancers, 14(7), 1737. https://doi.org/10.3390/cancers14071737









