Stacking Machine Learning Algorithms for Biomarker-Based Preoperative Diagnosis of a Pelvic Mass
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Clinical Patient Evaluation and Measurement of Biomarkers
2.2. Univariate Statistical Analysis and Logistic Regression
2.3. Data Splitting and Pre-Processing
2.4. Feature Selection
2.5. Supervised Machine Learning and Ensemble Stacking
2.6. Software
3. Results:
3.1. Patient Demographic, Univariate Statistical Analysis, and Logistic Regression
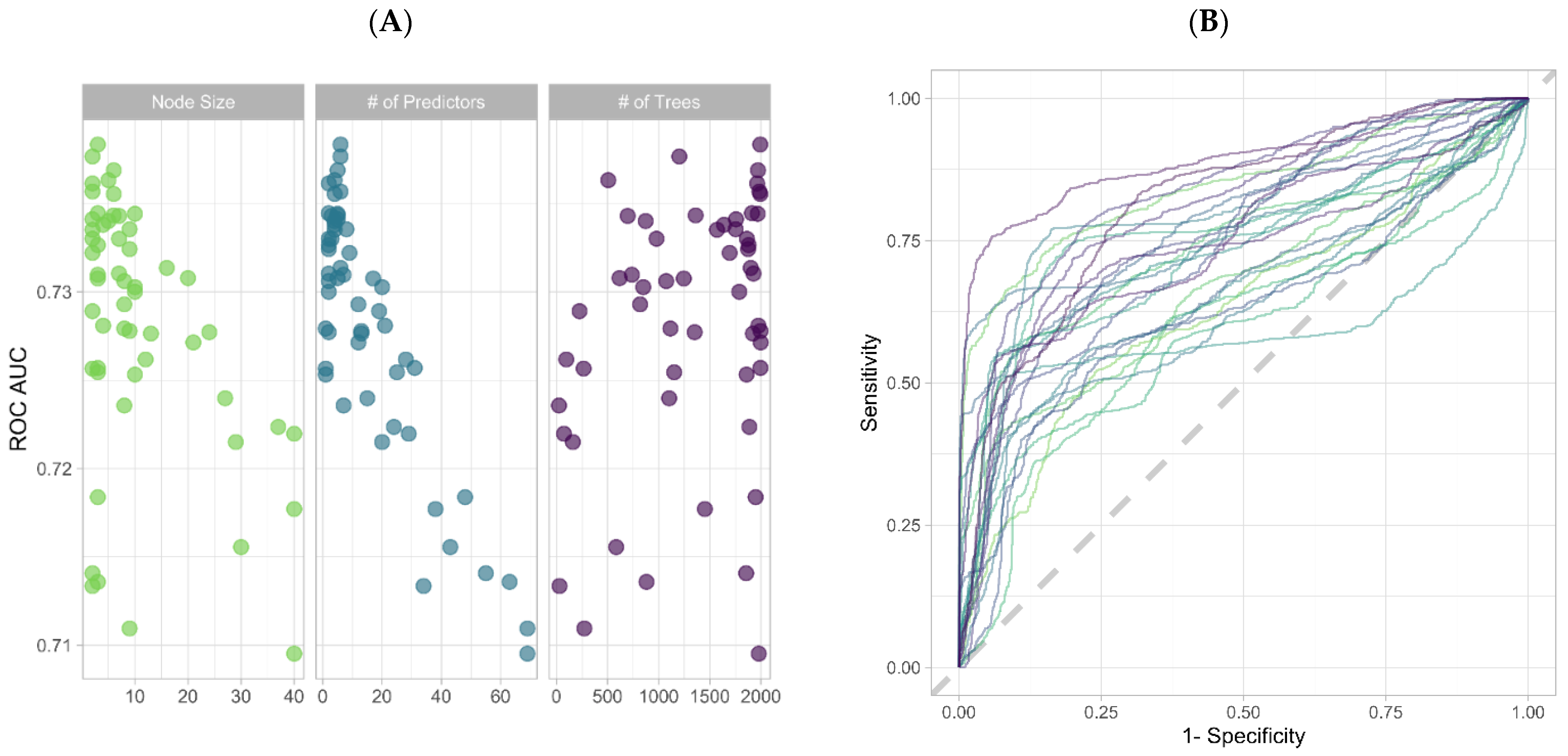
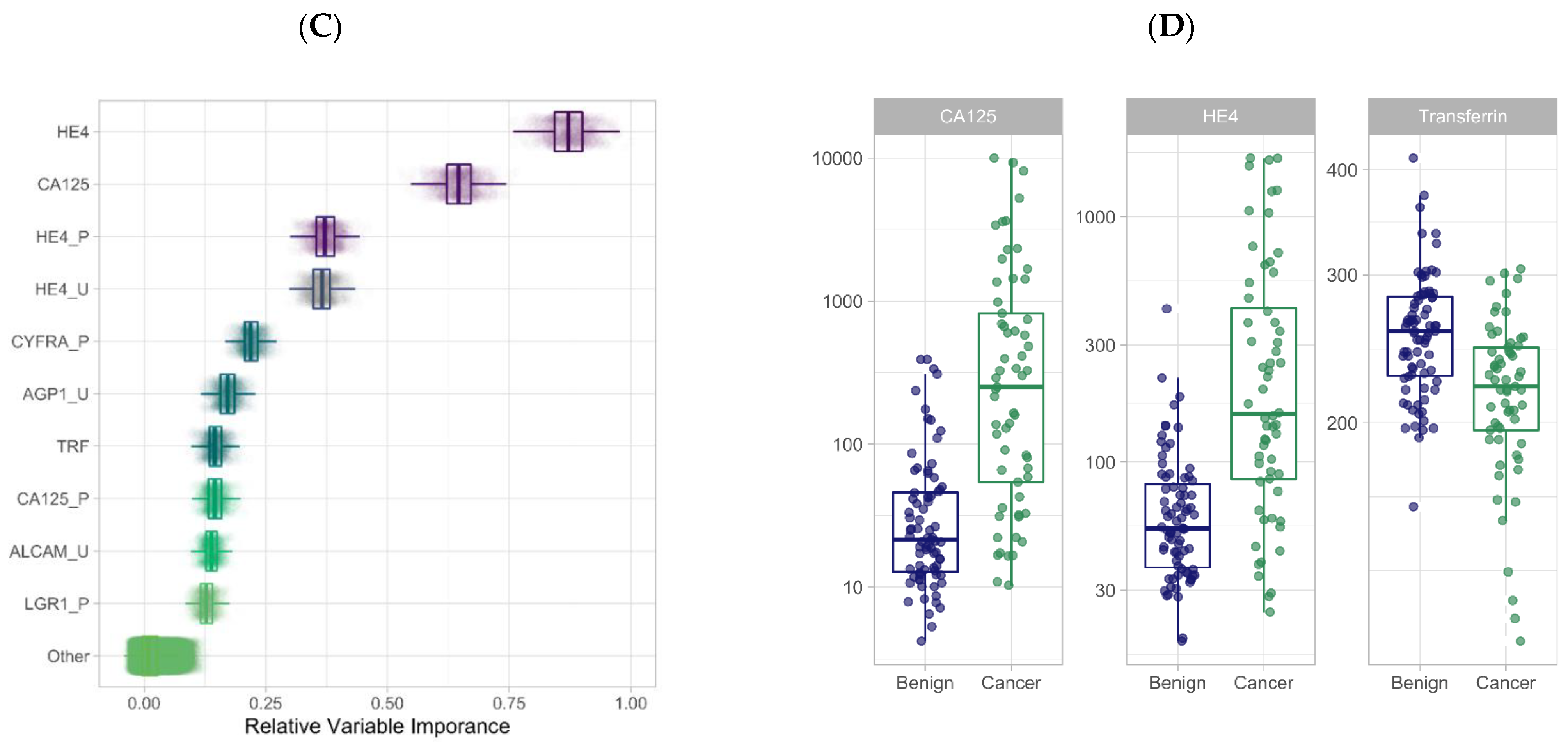
3.2. Feature Selection through Orthogonal Approaches
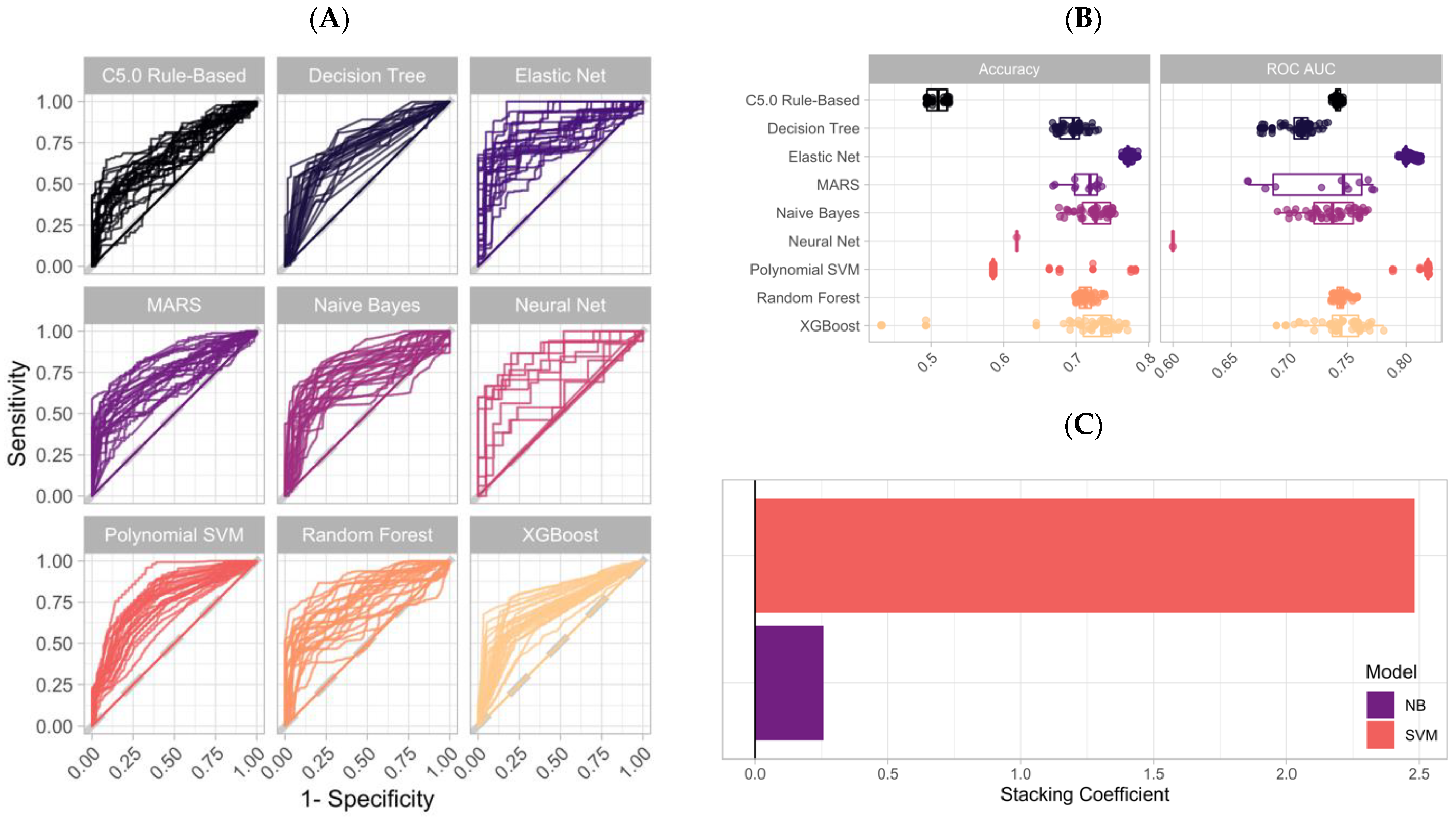
3.3. Training ML Classifiers and Generating an Ensemble Stack Using LASSO Regression
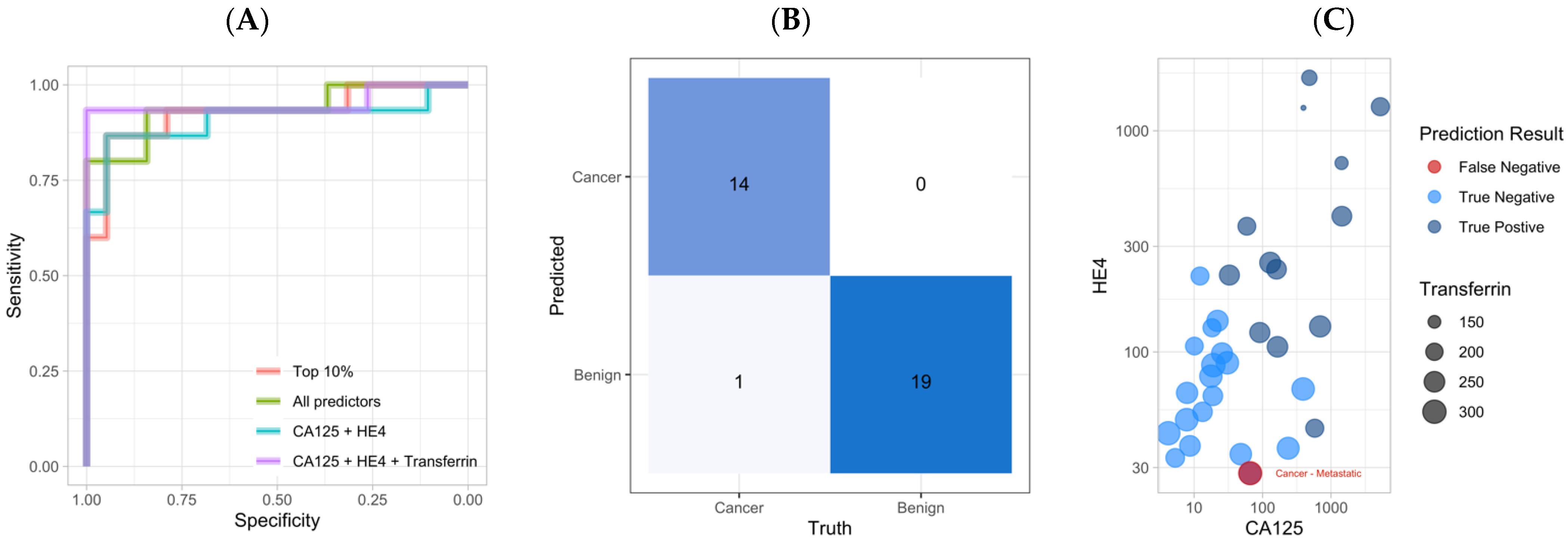
3.4. Application of an Ensemble Stack Generates Accurate Predictions on the Testing Dataset
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vernooij, F.; Heintz, A.P.M.; Coebergh, J.W.; Massuger, L.F.A.G.; Witteveen, P.O.; van der Graaf, Y. Specialized and high-volume care leads to better outcomes of ovarian cancer treatment in the Netherlands. Gynecol. Oncol. 2009, 112, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Goff, B.A.; Matthews, B.J.; Larson, E.H.; Andrilla, C.H.A.; Wynn, M.; Lishner, D.M.; Baldwin, L.M. Predictors of comprehensive surgical treatment in patients with ovarian cancer. Cancer 2007, 109, 2031–2042. [Google Scholar] [CrossRef] [PubMed]
- Aune, G.; Torp, S.H.; Syversen, U.; Hagen, B.; Tingulstad, S. Ten years’ experience with centralized surgery of ovarian cancer in one health region in Norway. Int. J. Gynecol. Cancer 2012, 22, 226–231. [Google Scholar] [CrossRef] [PubMed]
- Myers, E.R.; Bastian, L.A.; Havrilesky, L.J.; Kulasingam, S.L.; Terplan, M.S.; Cline, K.E.; Gray, R.N.; McCrory, D.C. Management of adnexal mass. Evid. Rep. Technol. Assess. 2006, 130, 1–145. [Google Scholar]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA. Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, I.; Oram, D.; Fairbanks, J.; Turner, J.; Frost, C.; Grudzinskas, J.G. A risk of malignancy index incorporating CA 125, ultrasound and menopausal status for the accurate preoperative diagnosis of ovarian cancer. BJOG Int. J. Obstet. Gynaecol. 1990, 97, 922–929. [Google Scholar] [CrossRef] [PubMed]
- van den Akker, P.A.J.; Aalders, A.L.; Snijders, M.P.L.M.; Kluivers, K.B.; Samlal, R.A.K.; Vollebergh, J.H.A.; Massuger, L.F.A.G. Evaluation of the risk of malignancy index in daily clinical management of adnexal masses. Gynecol. Oncol. 2010, 116, 384–388. [Google Scholar] [CrossRef]
- Moore, R.G.; Brown, A.K.; Miller, M.C.; Skates, S.; Allard, W.J.; Verch, T.; Steinhoff, M.; Messerlian, G.; DiSilvestro, P.; Granai, C.O.; et al. The use of multiple novel tumor biomarkers for the detection of ovarian carcinoma in patients with a pelvic mass. Gynecol. Oncol. 2008, 108, 402–408. [Google Scholar] [CrossRef]
- Moore, R.G.; McMeekin, D.S.; Brown, A.K.; DiSilvestro, P.; Miller, M.C.; Allard, W.J.; Gajewski, W.; Kurman, R.; Bast, R.C.; Skates, S.J. A novel multiple marker bioassay utilizing HE4 and CA125 for the prediction of ovarian cancer in patients with a pelvic mass. Gynecol. Oncol. 2009, 112, 40–46. [Google Scholar] [CrossRef]
- Moore, R.G.; Miller, M.C.; Disilvestro, P.; Landrum, L.M.; Gajewski, W.; Ball, J.J.; Skates, S.J. Evaluation of the diagnostic accuracy of the risk of ovarian malignancy algorithm in women with a pelvic mass. Obstet. Gynecol. 2011, 118, 280. [Google Scholar] [CrossRef]
- Bristow, R.E.; Smith, A.; Zhang, Z.; Chan, D.W.; Crutcher, G.; Fung, E.T.; Munroe, D.G. Ovarian malignancy risk stratification of the adnexal mass using a multivariate index assay. Gynecol. Oncol. 2013, 128, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Coleman, R.L.; Herzog, T.J.; Chan, D.W.; Munroe, D.G.; Pappas, T.C.; Smith, A.; Zhang, Z.; Wolf, J. Validation of a second-generation multivariate index assay for malignancy risk of adnexal masses. Am. J. Obstet. Gynecol. 2016, 215, 82-e1. [Google Scholar] [CrossRef] [PubMed]
- Bast, R.C.; Lu, Z.; Han, C.Y.; Lu, K.H.; Anderson, K.S.; Drescher, C.W.; Skates, S.J. Biomarkers and Strategies for Early Detection of Ovarian Cancer. Cancer Epidemiol. Biomark. Prev. 2020, 29, 2504–2512. [Google Scholar] [CrossRef]
- Nolen, B.M.; Lokshin, A.E. Biomarker testing for ovarian cancer: Clinical utility of multiplex assays. Mol. Diagn. Ther. 2013, 17, 139–146. [Google Scholar] [CrossRef][Green Version]
- Yurkovetsky, Z.; Skates, S.; Lomakin, A.; Nolen, B.; Pulsipher, T.; Modugno, F.; Marks, J.; Godwin, A.; Gorelik, E.; Jacobs, I.; et al. Development of a multimarker assay for early detection of ovarian cancer. J. Clin. Oncol. 2010, 28, 2159. [Google Scholar] [CrossRef] [PubMed]
- Snoek, J.; Larochelle, H.; Adams, R.P. Practical Bayesian optimization of machine learning algorithms. In Proceedings of the 2012 Advances in Neural Information Processing Systems, Lake Tahoe, NV, USA, 3–6 December 2012. [Google Scholar]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the Areas under Two or More Correlated Receiver Operating Characteristic Curves: A Nonparametric Approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, J.; Bithell, J. Bootstrap confidence intervals: When, which, what? A practical guide for medical statisticians. Stat. Med. 2000, 19, 1141–1164. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. 2019. Available online: https://cran.r-project.org/src/base/R-4/ (accessed on 1 April 2020).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Sun, X.; Xu, W. Fast implementation of DeLong’s algorithm for comparing the areas under correlated receiver operating characteristic curves. IEEE Signal Process. Lett. 2014, 21, 1389–1393. [Google Scholar] [CrossRef]
- Kawakami, E.; Tabata, J.; Yanaihara, N.; Ishikawa, T.; Koseki, K.; Iida, Y.; Saito, M.; Komazaki, H.; Shapiro, J.S.; Goto, C.; et al. Application of artificial intelligence for preoperative diagnostic and prognostic prediction in epithelial ovarian cancer based on blood biomarkers. Clin. Cancer Res. 2019, 25, 3006–3015. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J. Elements of Statistical Learning, 2nd ed.; Springer: New York, NY, USA, 2009. [Google Scholar]
- Liu, J.H.; Zanotti, K.M. Management of the adnexal mass. Obstet. Gynecol. 2011, 117, 1413–1428. [Google Scholar] [CrossRef] [PubMed]
- Nagy, B.; Krasznai, Z.T.; Balla, H.; Csobán, M.; Antal-Szalmás, P.; Hernádi, Z.; Kappelmayer, J. Elevated human epididymis protein 4 concentrations in chronic kidney disease. Ann. Clin. Biochem. 2012, 49, 377–380. [Google Scholar] [CrossRef] [PubMed]
- Sheng, J.Q.; Li, S.R.; Wu, Z.T.; Xia, C.H.; Wu, X.; Chen, J.; Rao, J. Transferrin dipstick as a potential novel test for colon cancer screening: A comparative study with immuno fecal occult blood test. Cancer Epidemiol. Biomark. Prev. 2009, 18, 2182–2185. [Google Scholar] [CrossRef] [PubMed]
- O’Neal, R.L.; Nam, K.T.; Lafleur, B.J.; Barlow, B.; Nozaki, K.; Lee, H.J.; Kim, W.H.; Yang, H.K.; Shi, C.; Maitra, A.; et al. Human epididymis protein 4 is up-regulated in gastric and pancreatic adenocarcinomas. Hum. Pathol. 2013, 44, 734–742. [Google Scholar] [CrossRef] [PubMed]
- Bell, R.; Koren, Y.; Volinsky, C. The BellKor 2008 Solution to the Netflix Prize. Netflix Prize Doc. 2009. Available online: https://www2.seas.gwu.edu/~simhaweb/champalg/cf/papers/KorenBellKor2009.pdf (accessed on 20 May 2021).
- Džeroski, S.; Ženko, B. Is combining classifiers with stacking better than selecting the best one? Mach. Learn. 2004, 54, 255–273. [Google Scholar] [CrossRef]
- Reid, S.; Grudic, G. Regularized linear models in stacked generalization. In Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Springer: Berlin/Heidelberg, Germany, 2009. [Google Scholar] [CrossRef]
- Guyon, I.; Weston, J.; Barnhill, S.; Vapnik, V. Gene selection for cancer classification using support vector machines. Mach. Learn. 2002, 46, 389–422. [Google Scholar] [CrossRef]
- Karabatak, M. A new classifier for breast cancer detection based on Naïve Bayesian. Meas. J. Int. Meas. Confed. 2015, 72, 32–36. [Google Scholar] [CrossRef]
- Parmar, C.; Grossmann, P.; Rietveld, D.; Rietbergen, M.M.; Lambin, P.; Aerts, H.J.W.L. Radiomic machine-learning classifiers for prognostic biomarkers of head and neck cancer. Front. Oncol. 2015, 5, 272. [Google Scholar] [CrossRef]
- Das, J.; Gayvert, K.M.; Bunea, F.; Wegkamp, M.H.; Yu, H. ENCAPP: Elastic-net-based prognosis prediction and biomarker discovery for human cancers. BMC Genom. 2015, 16, 263. [Google Scholar] [CrossRef]
- Díaz-Uriarte, R.; Alvarez de Andrés, S. Gene selection and classification of microarray data using random forest. BMC Bioinform. 2006, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Kwiatkowski, K.; Coe, K.; Bailar, J.C.; Swanson, G.M. Inclusion of minorities and women in cancer clinical trials, a decade later: Have we improved? Cancer 2013, 119, 2956–2963. [Google Scholar] [CrossRef] [PubMed]
| A. Patient demographics | |||||
| Training (n = 106) | Testing (n = 34) | All Patients (n = 140) | |||
| Age | |||||
| Years (range) | 54.7 (20–91) | 55.3 (25–82) | 54.9 (20–91) | ||
| Race | |||||
| Black | 2 (1.9%) | 1 (2.9%) | 3 (2.1%) | ||
| Hispanic | 1 (0.9%) | 1 (2.9%) | 2 (1.4%) | ||
| Other | 6 (5.7%) | 1 (2.9%) | 7 (5.0%) | ||
| White | 97 (91.5%) | 31 (91.2%) | 128 (91.4%) | ||
| Menopausal Status | |||||
| Post-menopausal | 68 (64.2%) | 22 (64.7%) | 90 (64.3%) | ||
| Pre-menopausal | 38 (35.8%) | 12 (35.3%) | 50 (35.7%) | ||
| Histological Diagnosis | |||||
| Benign | 60 (56.6%) | 19 (55.9%) | 79 (56.4%) | ||
| Borderline/LMP | 3 (2.8%) | 1 (2.9%) | 4 (2.9%) | ||
| Cancer—EOC I–II | 17 (16.0%) | 4 (11.8%) | 21 (15.0%) | ||
| Cancer—EOC III–IV | 15 (14.2%) | 9 (26.5%) | 24 (17.1%) | ||
| Cancer—Metastatic | 5 (4.7%) | 1 (2.9%) | 6 (4.3%) | ||
| Cancer—EOC Unstaged | 1 (0.9%) | 0 (0.0%) | 1 (0.7%) | ||
| Cancer—Non-EOC | 2 (1.9%) | 0 (0.0%) | 2 (1.4%) | ||
| Cancer—Other Gyn | 3 (2.8%) | 0 (0.0%) | 3 (2.1%) | ||
| Stage | |||||
| IA | 8 (7.5%) | 2 (5.9%) | 10 (7.1%) | ||
| IB | 1 (0.9%) | 0 (0.0%) | 1 (0.7%) | ||
| IC | 6 (5.7%) | 1 (2.9%) | 7 (5.0%) | ||
| II | 1 (0.9%) | 0 (0.0%) | 1 (0.7%) | ||
| IIA | 3 (2.8%) | 0 (0.0%) | 3 (2.1%) | ||
| IIB | 1 (0.9%) | 0 (0.0%) | 1 (0.7%) | ||
| IIC | 1 (0.9%) | 1 (2.9%) | 2 (1.4%) | ||
| IIIA | 5 (4.7%) | 2 (5.9%) | 7 (5.0%) | ||
| IIIB | 1 (0.9%) | 1 (2.9%) | 2 (1.4%) | ||
| IIIC | 12 (11.3%) | 7 (20.6%) | 19 (13.6%) | ||
| IV | 5 (4.7%) | 1 (2.9%) | 6 (4.3%) | ||
| Unstaged | 1 (0.9%) | 0 (0.0%) | 1 (0.7%) | ||
| B. Malignant tumor histology | |||||
| EOC Type | Stage I | Stage II | Stage III | Unstaged | All |
| Clear Cell | 2 (33.3%) | 1 (16.7%) | 3 (50.0%) | 0 (0.0%) | 6 (13.0%) |
| Endometrioid | 1 (50.0%) | 0 (0.0%) | 1 (50.0%) | 0 (0.0%) | 2 (4.3%) |
| Mucinous | 7 (87.5%) | 1 (12.5%) | 0 (0.0%) | 0 (0.0%) | 8 (17.4%) |
| Serous | 4 (13.8%) | 4 (13.8%) | 20 (69.0%) | 1 (3.4%) | 29 (63.0%) |
| Mixed | 0 (0.0%) | 1 (100%) | 0 (0.0%) | 0 (0.0%) | 1 (2.2%) |
| All EOC | 14 (30.4%) | 7 (15.2%) | 24 (52.2%) | 1 (2.2%) | 46 (100%) |
| C. Malignant tumor grade | |||||
| All patients | Pre-Menopausal | Post-Menopausal | |||
| EOC Grade | n (%) | n (%) | n (%) | ||
| Grade 1 | 9 (19.6%) | 3 (30.0%) | 6 (16.7%) | ||
| Grade 2 | 3 (6.5%) | 1 (10.0%) | 2 (5.6%) | ||
| Grade 3 | 34 (73.9%) | 6 (60.0%) | 28 (77.8%) | ||
| Total | 46 (100%) | 10 (21.7%) | 36 (78.3%) | ||
| Ensemble Model | AUC | Accuracy | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|---|
| CA125 + HE4 + TRF | 0.951 | 97.1% | 93.3% | 100.0% | 100.0% | 95.0% |
| CA125 + HE4 | 0.937 | 85.3% | 86.7% | 84.2% | 81.2% | 88.9% |
| Top 10% | 0.926 | 82.4% | 66.7% | 94.7% | 90.9% | 78.3% |
| All predictors | 0.909 | 88.2% | 80.0% | 94.7% | 92.3% | 85.7% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shaw, R.; Lokshin, A.E.; Miller, M.C.; Messerlian-Lambert, G.; Moore, R.G. Stacking Machine Learning Algorithms for Biomarker-Based Preoperative Diagnosis of a Pelvic Mass. Cancers 2022, 14, 1291. https://doi.org/10.3390/cancers14051291
Shaw R, Lokshin AE, Miller MC, Messerlian-Lambert G, Moore RG. Stacking Machine Learning Algorithms for Biomarker-Based Preoperative Diagnosis of a Pelvic Mass. Cancers. 2022; 14(5):1291. https://doi.org/10.3390/cancers14051291
Chicago/Turabian StyleShaw, Reid, Anna E. Lokshin, Michael C. Miller, Geralyn Messerlian-Lambert, and Richard G. Moore. 2022. "Stacking Machine Learning Algorithms for Biomarker-Based Preoperative Diagnosis of a Pelvic Mass" Cancers 14, no. 5: 1291. https://doi.org/10.3390/cancers14051291
APA StyleShaw, R., Lokshin, A. E., Miller, M. C., Messerlian-Lambert, G., & Moore, R. G. (2022). Stacking Machine Learning Algorithms for Biomarker-Based Preoperative Diagnosis of a Pelvic Mass. Cancers, 14(5), 1291. https://doi.org/10.3390/cancers14051291







