HER2–CDH1 Interaction via Wnt/B-Catenin Is Associated with Patients’ Survival in HER2-Positive Metastatic Gastric Adenocarcinoma
Abstract
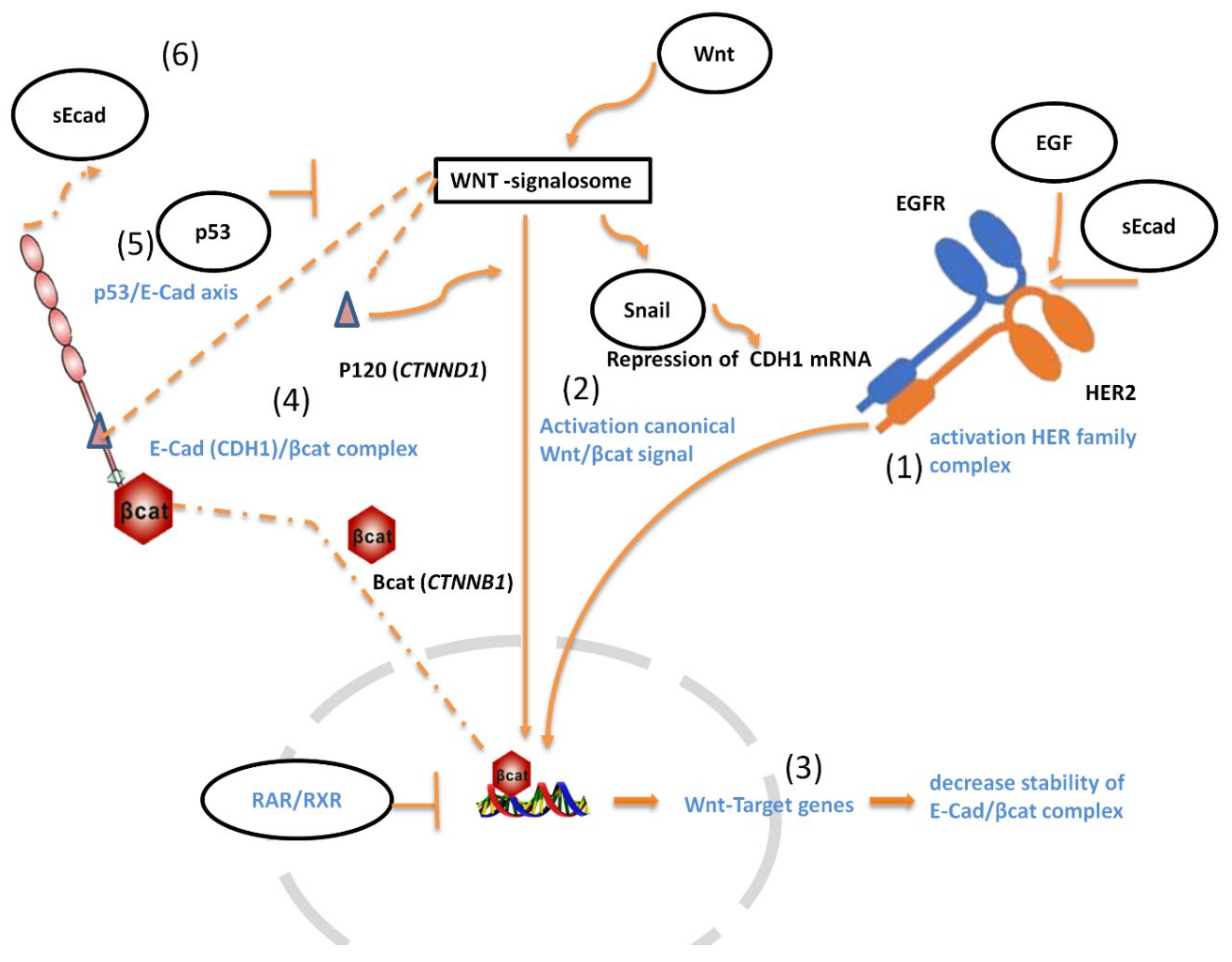
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Design of the Study
2.3. RNA Extraction and NanoString Quantification
2.4. Cell Line Assay
2.5. E-CAD and HER2 Histological Evaluation
2.6. Serum Sample Collection for Soluble HER2 (sHER2) and Soluble E-CAD (sCDH1) Evaluation
2.7. DNA Isolation and CDH1 Genotyping Assay
2.8. Statistical Analysis
3. Results
3.1. Patients’ and Series Characteristics
3.2. Analysis of the Impact of HER2 and E-CAD status on OS of Patients
3.3. Comparison between IHC versus mRNA for Detecting HER2 and E-CAD
3.4. Serum Soluble E-CAD Levels Are Reduced after Treatment Both in HER2-Negative and in HER2-Positive mGC
3.5. CDH1 rs16260 Genetic Variant Associated with E-CAD in HER2-Positive MGC
3.6. Genes Differentially Expressed according to the HER2 Status
3.7. Wnt/-β-Catenin Signature Joined CDH1 and HER2 mRNA Expression
3.8. External Validation of Genes Associated with CDH1 and HER2 Overexpression in the TCGA Dataset
3.9. Combination of Trastuzumab with EGF-Induced CDH1 mRNA Expression in NCI-N87 Gastric Cell Line
3.10. Identification of Genes Associated with Wnt/-β-Catenin Signature for Discriminating OS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| ATM | KLRG1 | RARA | TNFAIP8L3 | B2M |
| CDH1 | MDM2 | RARB | TP53 | GAPDH |
| CDKN1A | MDM4 | RARG | TWIST1 | HPRT1 |
| CTNNB1 | MLH1 | RXRA | VCL | RPL19 |
| CTNND1 | MMP2 | RXRB | WNT1 | |
| EBER1 | MMP9 | RXRG | WNT2 | |
| EGF | MSH2 | SNAI1 | WNT3 | |
| EGFR | MSH6 | SNAI2 | WNT5a | |
| GSK3B | PIK3CA | SRC | WNT7a | |
| HER2 | PMS2 | TNFAIP8L1 | ZEB1 |
References
- Van Cutsem, E.; Bang, Y.-J.; Feng-Yi, F.; Xu, J.M.; Lee, K.-W.; Jiao, S.-C.; Chong, J.L.; López-Sanchez, R.I.; Price, T.; Gladkov, O.; et al. HER2 screening data from ToGA: Targeting HER2 in gastric and gastroesophageal junction cancer. Gastric Cancer 2015, 18, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Petrova, Y.I.; Schecterson, L.; Gumbiner, B.M. Roles for E-cadherin cell surface regulation in cancer. Mol. Biol. Cell 2016, 27, 3233–3244. [Google Scholar] [CrossRef] [PubMed]
- Caggiari, L.; Miolo, G.; Buonadonna, A.; Basile, D.; Santeufemia, D.A.; Cossu, A.; Palmieri, G.; De Zorzi, M.; Fornasarig, M.; Alessandrini, L.; et al. Characterizing Metastatic HER2-Positive Gastric Cancer at the CDH1 Haplotype. Int. J. Mol. Sci. 2017, 19, 47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magnani, E.; Macchi, F.; Mancini, M.; Lomazzi, V.; Cogliati, S.; Pistore, C.; Mandruzzato, M.; Dock-Bregeon, A.-C.; Bonapace, I.M. UHRF1 regulates CDH1 via promoter associated non-coding RNAs in prostate cancer cells. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 258–270. [Google Scholar] [CrossRef]
- Pisignano, G.; Napoli, S.; Magistri, M.; Mapelli, S.N.; Pastori, C.; Di Marco, S.; Civenni, G.; Albino, D.; Enriquez, C.; Allegrini, S.; et al. A promoter-proximal transcript targeted by genetic polymorphism controls E-cadherin silencing in human cancers. Nat. Commun. 2017, 8, 15622. [Google Scholar] [CrossRef] [Green Version]
- Keirsebilck, A.; Van Hoorde, L.; Gao, Y.; De Bruyne, G.; Bruyneel, E.; Vermassen, P.; Mareel, M.; van Roy, F. Mechanisms of downregulation of transfected E-cadherin cDNA during formation of invasive tumors in syngeneic mice. Invasion Metastasis 1998, 18, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chu, K.-M. E-Cadherin and Gastric Cancer: Cause, Consequence, and Applications. BioMed Res. Int. 2014, 2014, 637308. [Google Scholar] [CrossRef] [Green Version]
- Nicolas, E.; Parisot, P.; Pinto-Monteiro, C.; de Walque, R.; De Vleeschouwer, C.; Lafontaine, D.L.J. Involvement of human ribosomal proteins in nucleolar structure and p53-dependent nucleolar stress. Nat. Commun. 2016, 7, 11390. [Google Scholar] [CrossRef] [PubMed]
- Hunsu, V.O.; Facey, C.O.B.; Fields, J.Z.; Boman, B.M. Retinoids as Chemo-Preventive and Molecular-Targeted Anti-Cancer Therapies. Int. J. Mol. Sci. 2021, 22, 7731. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network; Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.M.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
- Alessandrini, L.; Manchi, M.; De Re, V.; Dolcetti, R.; Canzonieri, V. Proposed Molecular and miRNA Classification of Gastric Cancer. Int. J. Mol. Sci. 2018, 19, 1683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shenoy, S. CDH1 (E-Cadherin) Mutation and Gastric Cancer: Genetics, Molecular Mechanisms and Guidelines for Management. Cancer Manag. Res. 2019, 11, 10477–10486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Repetto, O.; De Paoli, P.; De Re, V.; Canzonieri, V.; Cannizzaro, R. Levels of soluble E-cadherin in breast, gastric, and colorectal cancers. BioMed Res. Int. 2014, 2014, 408047. [Google Scholar] [CrossRef]
- Brouxhon, S.M.; Kyrkanides, S.; Teng, X.; O’Banion, M.K.; Clarke, R.; Byers, S.; Ma, L. Soluble-E-cadherin activates HER and IAP family members in HER2+ and TNBC human breast cancers. Mol. Carcinog. 2014, 53, 893–906. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Yang, Y.; Liu, S.; Tao, T.; Cai, J.; Wu, J.; Guan, H.; Zhu, X.; He, Z.; Li, J.; et al. EGF-induced nuclear localization of SHCBP1 activates β-catenin signaling and promotes cancer progression. Oncogene 2019, 38, 747–764. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Wang, J.; Nika, H.; Hawke, D.; Keezer, S.; Ge, Q.; Fang, B.; Fang, X.; Fang, D.; Litchfield, D.W.; et al. EGF-Induced ERK Activation Promotes CK2-Mediated Disassociation of alpha-Catenin from beta-Catenin and Transactivation of beta-Catenin. Mol. Cell 2009, 36, 547–559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Z.; Ghosh, S.; Wang, Z.; Hunter, T. Downregulation of caveolin-1 function by EGF leads to the loss of E-cadherin, increased transcriptional activity of β-catenin, and enhanced tumor cell invasion. Cancer Cell 2003, 4, 499–515. [Google Scholar] [CrossRef] [Green Version]
- Bachelder, R.E.; Yoon, S.-O.; Franci, C.; de Herreros, A.G.; Mercurio, A.M. Glycogen synthase kinase-3 is an endogenous inhibitor of Snail transcription. J. Cell Biol. 2005, 168, 29–33. [Google Scholar] [CrossRef] [Green Version]
- Sengupta, P.K.; Bouchie, M.P.; Kukuruzinska, M.A. N-glycosylation gene DPAGT1 is a target of the Wnt/beta-catenin signaling pathway. J. Biol. Chem. 2010, 285, 31164–31173. [Google Scholar] [CrossRef] [Green Version]
- Carvalho, S.; Catarino, T.A.; Dias, A.M.; Kato, M.; Almeida, A.; Hessling, B.; Figueiredo, J.; Gärtner, F.; Sanches, J.M.; Ruppert, T.; et al. Preventing E-cadherin aberrant N-glycosylation at Asn-554 improves its critical function in gastric cancer. Oncogene 2016, 35, 1619–1631. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.L. Involvement of Retinoic Acid Regulates Wnt Signaling Pathway in Cancer Metastasis. Available online: https://www.semanticscholar.org/paper/Involvement-of-Retinoic-Acid-Regulates-Wnt-Pathway-Zhu-Luo/bd5a639e12beefe6d619034e79c3816a41f62766 (accessed on 25 January 2022).
- Vinyoles, M.; Del Valle-Pérez, B.; Curto, J.; Viñas-Castells, R.; Alba-Castellón, L.; García de Herreros, A.; Duñach, M. Multivesicular GSK3 sequestration upon Wnt signaling is controlled by p120-catenin/cadherin interaction with LRP5/6. Mol. Cell 2014, 53, 444–457. [Google Scholar] [CrossRef] [Green Version]
- Schade, B.; Lesurf, R.; Sanguin-Gendreau, V.; Bui, T.; Deblois, G.; O’Toole, S.A.; Millar, E.K.A.; Zardawi, S.J.; Lopez-Knowles, E.; Sutherland, R.L.; et al. β-Catenin signaling is a critical event in ErbB2-mediated mammary tumor progression. Cancer Res. 2013, 73, 4474–4487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, N.H.; Kim, H.S.; Kim, N.-G.; Lee, I.; Choi, H.-S.; Li, X.-Y.; Kang, S.E.; Cha, S.Y.; Ryu, J.K.; Na, J.M.; et al. p53 and microRNA-34 are suppressors of canonical Wnt signaling. Sci. Signal. 2011, 4, ra71. [Google Scholar] [CrossRef] [Green Version]
- Oikawa, T.; Otsuka, Y.; Onodera, Y.; Horikawa, M.; Handa, H.; Hashimoto, S.; Suzuki, Y.; Sabe, H. Necessity of p53-binding to the CDH1 locus for its expression defines two epithelial cell types differing in their integrity. Sci. Rep. 2018, 8, 1595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaplan, M.; Narasimhan, S.; de Heus, C.; Mance, D.; van Doorn, S.; Houben, K.; Popov-Čeleketić, D.; Damman, R.; Katrukha, E.A.; Jain, P.; et al. EGFR Dynamics Change during Activation in Native Membranes as Revealed by NMR. Cell 2016, 167, 1241–1251.e11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Citri, A.; Yarden, Y. EGF-ERBB signalling: Towards the systems level. Nat. Rev. Mol. Cell Biol. 2006, 7, 505–516. [Google Scholar] [CrossRef]
- Ware, M.F.; Tice, D.A.; Parsons, S.J.; Lauffenburger, D.A. Overexpression of cellular Src in fibroblasts enhances endocytic internalization of epidermal growth factor receptor. J. Biol. Chem. 1997, 272, 30185–30190. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Bai, Z.-G.; Yin, J.; Wu, G.-C.; Zhang, Z.-T. Role of c-Src activity in the regulation of gastric cancer cell migration. Oncol. Rep. 2014, 32, 45–49. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Hirsch, D.S.; Sasiela, C.A.; Wu, W.J. Cdc42 regulates E-cadherin ubiquitination and degradation through an epidermal growth factor receptor to Src-mediated pathway. J. Biol. Chem. 2008, 283, 5127–5137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, W.J.; Nusse, R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science 2004, 303, 1483–1487. [Google Scholar] [CrossRef] [Green Version]
- Voloshanenko, O.; Gmach, P.; Winter, J.; Kranz, D.; Boutros, M. Mapping of Wnt-Frizzled interactions by multiplex CRISPR targeting of receptor gene families. FASEB J. 2017, 31, 4832–4844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, X.-H.; Gudas, L.J. Retinoids, retinoic acid receptors, and cancer. Annu. Rev. Pathol. 2011, 6, 345–364. [Google Scholar] [CrossRef]
- Osei-Sarfo, K.; Gudas, L.J. Retinoic acid suppresses the canonical Wnt signaling pathway in embryonic stem cells and activates the noncanonical Wnt signaling pathway. Stem Cells 2014, 32, 2061–2071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakamura, S.; Kuroki, K.; Ohki, I.; Sasaki, K.; Kajikawa, M.; Maruyama, T.; Ito, M.; Kameda, Y.; Ikura, M.; Yamamoto, K.; et al. Molecular basis for E-cadherin recognition by killer cell lectin-like receptor G1 (KLRG1). J. Biol. Chem. 2009, 284, 27327–27335. [Google Scholar] [CrossRef] [Green Version]
- Williams, A.B.; Schumacher, B. p53 in the DNA-Damage-Repair Process. Cold Spring Harb. Perspect. Med. 2016, 6, a026070. [Google Scholar] [CrossRef] [Green Version]
- Pestov, D.G.; Strezoska, Z.; Lau, L.F. Evidence of p53-dependent cross-talk between ribosome biogenesis and the cell cycle: Effects of nucleolar protein Bop1 on G(1)/S transition. Mol. Cell. Biol. 2001, 21, 4246–4255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deisenroth, C.; Zhang, Y. The ribosomal protein-mdm2-p53 pathway and energy metabolism: Bridging the gap between feast and famine. Genes Cancer 2011, 2, 392–403. [Google Scholar] [CrossRef] [Green Version]
- Smyth, E.C.; Nilsson, M.; Grabsch, H.I.; van Grieken, N.C.; Lordick, F. Gastric cancer. Lancet 2020, 396, 635–648. [Google Scholar] [CrossRef]
- Chiurillo, M.A. Role of the Wnt/β-catenin pathway in gastric cancer: An in-depth literature review. World J. Exp. Med. 2015, 5, 84–102. [Google Scholar] [CrossRef]
- Flanagan, D.J.; Austin, C.R.; Vincan, E.; Phesse, T.J. Wnt Signalling in Gastrointestinal Epithelial Stem Cells. Genes 2018, 9, 178. [Google Scholar] [CrossRef] [Green Version]
- Wen, Z.; Chen, M.; Guo, W.; Guo, K.; Du, P.; Fang, Y.; Gao, M.; Wang, Q. RORβ suppresses the stemness of gastric cancer cells by downregulating the activity of the Wnt signaling pathway. Oncol. Rep. 2021, 46, 180. [Google Scholar] [CrossRef] [PubMed]
- Jung, D.H.; Bae, Y.J.; Kim, J.-H.; Shin, Y.K.; Jeung, H.-C. HER2 Regulates Cancer Stem Cell Activities via the Wnt Signaling Pathway in Gastric Cancer Cells. Oncology 2019, 97, 311–318. [Google Scholar] [CrossRef]
- Wang, B.; Tian, T.; Kalland, K.-H.; Ke, X.; Qu, Y. Targeting Wnt/β-Catenin Signaling for Cancer Immunotherapy. Trends Pharmacol. Sci. 2018, 39, 648–658. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xiang, Y.; Li, F.; Yin, C.; Li, B.; Ke, X. WNT/β-Catenin Signaling Pathway Regulating T Cell-Inflammation in the Tumor Microenvironment. Front. Immunol. 2019, 10, 2293. [Google Scholar] [CrossRef] [Green Version]
- Colozza, G.; Koo, B.-K. Wnt/β-catenin signaling: Structure, assembly and endocytosis of the signalosome. Dev. Growth Differ. 2021, 63, 199–218. [Google Scholar] [CrossRef]
- Ramesh, S.; Nash, J.; McCulloch, P.G. Reduction in membranous expression of beta-catenin and increased cytoplasmic E-cadherin expression predict poor survival in gastric cancer. Br. J. Cancer 1999, 81, 1392–1397. [Google Scholar] [CrossRef] [Green Version]
- Gao, C.; Xiao, G.; Hu, J. Regulation of Wnt/β-catenin signaling by posttranslational modifications. Cell Biosci. 2014, 4, 13. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Yuan, J.; Liu, Z.; Zhang, J.; Chang, J. Label-Free Quantitative Proteomics Combined with Biological Validation Reveals Activation of Wnt/β-Catenin Pathway Contributing to Trastuzumab Resistance in Gastric Cancer. Int. J. Mol. Sci. 2018, 19, 1981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acebron, S.P.; Niehrs, C. β-Catenin-Independent Roles of Wnt/LRP6 Signaling. Trends Cell Biol. 2016, 26, 956–967. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.; Lee, S.-H.; Park, S.-Y.; Jeong, M.-H.; Lee, S.Y.; Kim, M.-J.; Yoo, J.-Y.; Jang, S.; Choi, K.-C.; Yoon, H.-G. GPR177 promotes gastric cancer proliferation by suppressing endoplasmic reticulum stress-induced cell death. J. Cell. Biochem. 2019, 120, 2532–2539. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Sentani, K.; Tanaka, H.; Yano, T.; Suzuki, K.; Oshima, M.; Yasui, W.; Tamura, A.; Tsukita, S. Deficiency of Stomach-Type Claudin-18 in Mice Induces Gastric Tumor Formation Independent of H. pylori Infection. Cell. Mol. Gastroenterol. Hepatol. 2019, 8, 119–142. [Google Scholar] [CrossRef]
- Hu, T.; Li, C. Convergence between Wnt-beta-catenin and EGFR signaling in cancer. Mol. Cancer 2010, 9, 236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, G.; Hossain, M.M.; Bhat, A.Q.; Ayaz, M.O.; Bano, N.; Eachkoti, R.; Dar, M.J. Identification of a cross-talk between EGFR and Wnt/beta-catenin signaling pathways in HepG2 liver cancer cells. Cell. Signal. 2021, 79, 109885. [Google Scholar] [CrossRef]
- Sampera, A.; Sánchez-Martín, F.J.; Arpí, O.; Visa, L.; Iglesias, M.; Menéndez, S.; Gaye, É.; Dalmases, A.; Clavé, S.; Gelabert-Baldrich, M.; et al. HER-Family Ligands Promote Acquired Resistance to Trastuzumab in Gastric Cancer. Mol. Cancer Ther. 2019, 18, 2135–2145. [Google Scholar] [CrossRef] [Green Version]
- Zheng, L.; Tan, W.; Zhang, J.; Yuan, D.; Yang, J.; Liu, H. Combining trastuzumab and cetuximab combats trastuzumab-resistant gastric cancer by effective inhibition of EGFR/ErbB2 heterodimerization and signaling. Cancer Immunol. Immunother. 2014, 63, 581–586. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.; Patel, H.; Alanazi, S.; Yuan, L.; Garrett, J.T. HER3 signaling and targeted therapy in cancer. Oncol. Rev. 2018, 12, 355. [Google Scholar] [CrossRef] [Green Version]
- Ma, F.; Ouyang, Q.; Li, W.; Jiang, Z.; Tong, Z.; Liu, Y.; Li, H.; Yu, S.; Feng, J.; Wang, S.; et al. Pyrotinib or Lapatinib Combined with Capecitabine in HER2-Positive Metastatic Breast Cancer with Prior Taxanes, Anthracyclines, and/or Trastuzumab: A Randomized, Phase II Study. J. Clin. Oncol. 2019, 37, 2610–2619. [Google Scholar] [CrossRef]
- Saura, C.; Oliveira, M.; Feng, Y.-H.; Dai, M.-S.; Chen, S.-W.; Hurvitz, S.A.; Kim, S.-B.; Moy, B.; Delaloge, S.; Gradishar, W.; et al. Neratinib Plus Capecitabine Versus Lapatinib Plus Capecitabine in HER2-Positive Metastatic Breast Cancer Previously Treated with ≥2 HER2-Directed Regimens: Phase III NALA Trial. J. Clin. Oncol. 2020, 38, 3138–3149. [Google Scholar] [CrossRef]
- Suurs, F.V.; Lub-de Hooge, M.N.; de Vries, E.G.E.; de Groot, D.J.A. A review of bispecific antibodies and antibody constructs in oncology and clinical challenges. Pharmacol. Ther. 2019, 201, 103–119. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, Y.; Shang, C.; Chen, L.; Chen, H.; Wang, D.; Zeng, X. Distinct Ring1b complexes defined by DEAD-box helicases and EMT transcription factors synergistically enhance E-cadherin silencing in breast cancer. Cell Death Dis. 2021, 12, 202. [Google Scholar] [CrossRef]
- Peinado, H.; Ballestar, E.; Esteller, M.; Cano, A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol. Cell. Biol. 2004, 24, 306–319. [Google Scholar] [CrossRef] [Green Version]
- Yook, J.I.; Li, X.-Y.; Ota, I.; Hu, C.; Kim, H.S.; Kim, N.H.; Cha, S.Y.; Ryu, J.K.; Choi, Y.J.; Kim, J.; et al. A Wnt-Axin2-GSK3 beta cascade regulates Snail1 activity in breast cancer cells. Nat. Cell Biol. 2006, 8, 1398–1406. [Google Scholar] [CrossRef] [PubMed]
- Najy, A.J.; Day, K.C.; Day, M.L. The Ectodomain Shedding of E-cadherin by ADAM15 Supports ErbB Receptor Activation. J. Biol. Chem. 2008, 283, 18393–18401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grabowska, M.M.; Sandhu, B.; Day, M.L. EGF promotes the shedding of soluble E-cadherin in an ADAM10-dependent manner in prostate epithelial cells. Cell. Signal. 2012, 24, 532–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oh, D.-Y.; Lee, K.-W.; Cho, J.Y.; Kang, W.K.; Im, S.-A.; Kim, J.W.; Bang, Y.-J. Phase II trial of dacomitinib in patients with HER2-positive gastric cancer. Gastric Cancer 2016, 19, 1095–1103. [Google Scholar] [CrossRef]
- Fang, C.; Kang, Y. E-Cadherin: Context-Dependent Functions of a Quintessential Epithelial Marker in Metastasis. Cancer Res. 2021, 81, 5800–5802. [Google Scholar] [CrossRef]






| Series 1 | Series 2 | Series 3 | |
|---|---|---|---|
| Characteristic | Number (%) | Number (%) | Number (%) |
| Patients | 38 | 36 | 354 |
| Country | Italy | Italy | USA |
| Etnia | Caucasic | Caucasic | White 224 (63.3%) Asian 73 (20.6%) Other 12 (3.4%) nv 45 (12.7%) |
| Median age (range) | 69 (46–83) | 61 (45–85) | 67 (35–90) |
| Gender | |||
| Male | 25 (65.8%) | 23 (63.9%) | 229 (64.7%) |
| Female | 13 (34.2%) | 13 (36.1%) | 125 (35.3%) |
| Lauren classification | |||
| Intestinal | 7 (18.4%) | 8 (22.2%) | --- |
| Diffuse | 18 (47.4%) | 23 (63.9%) | --- |
| Adenocarcinoma-mix | 13 (34.2%) | 5 (13.9%) | --- |
| GRADE | |||
| Low | 10 (26.3%) | 10 (27.8%) | --- |
| Moderate | 10 (26.3%) | 15 (41.7%) | --- |
| High | 10 (26.3%) | 9 (25.0%) | --- |
| Undifferentiate | 8 (21.1%) | 2 (5.6%) | --- |
| Initial TNM stage | |||
| Stage 1 | 1/34 (2.9%) | -- | 48 (14.2%) |
| Stage 2 | 3/34 (8.8%) | 8 (22.2%) | 110 (32.4%) |
| Stage 3 | 17/34(50.0%) | 18 (50.0%) | 146 (43.1%) |
| Stage 4 | 13/34 (38.2%) | 10 (27.8%) | 35 (10.3%) |
| missing | 4 | -- | |
| GC with metastasis | |||
| M0 | -- | 26 (72.2%) | --- |
| M1 | 38 (100.0%) | 10 (27.8%) | --- |
| Tumor location | |||
| Upper | 5 (13.2%) | 7 (19.4%) | --- |
| Middle | 17 (44.7%) | 17 (47.2%) | --- |
| Lower | 13 (34.2%) | 9 (25.0%) | --- |
| Whole-NAS | 3 (7.9%) | 3 (8.3%) | |
| HER2-positive # | |||
| Yes | 13 (34.2%) | -- | 71 (20.1%) °°° |
| No | 25 (65.8%) | 36 (100%) | 283 (79.9%) °°° |
| Median Overall Survival $ | |||
| at GC diagnosis | 17 (95%CI 15.2–21.7) | 36 (95%CI 16.0–83.0) | 29 (95%CI 25.5–56.2) |
| at metastasis diagnosis | 9 (95%CI 6.0–28.0) | 14 (95%CI 7.0–91.1) | --- |
| Series 1 | Series 2 | Series 3 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| HER2-Negative | HER2-Positive | mGC vs. GC | |||||||
| mRNA (SD) | mRNA (SD) | Difference | (95% CI) | p | Difference | p | Best Cut-Off (FPKM) | p *** | |
| HER2 | 662.63 (267.54) | 6494.43 (6611.22) | 5831.80 (3180.43 to 8483.16) | 0.0001 | 171.6005 | 0.3607 | 15.6 | 0.093 | |
| TP53 | 264.28 (100.17) | 162.95 (82.44) | −101.33 (−166.95 to −35.71) | 0.0034 | 248.4487 | 0.0280 | 26.26 | 0.008 | |
| RARA | 191.88 (91.46) | 363.31 (281.13) | 171.43 (47.53 to 295.33) | 0.0080 | 120.9811 | 0.3188 | 16.18 | 0.016 | |
| MLH1 | 279.30 (81.69) | 207.04 (67.18) | −72.26 (−125.77 to −18.75) | 0.0095 | −11.0855 | 0.7260 | 5.08 | 0.044 | |
| EGF | 4.25 (3.73) | 19.21 (28.11) | 14.95 (3.50 to 26.41) | 0.0119 | 1.8393 | 0.4567 | 0.016 | <0.0001 | |
| ZEB1 | 319.80 (232.65) | 142.66 (93.25) | −177.14 (−314.06 to −40.22) | 0.0127 | 71.8493 | 0.5166 | 6.15 | 0.0056 | |
| VCL | 1168.13 (816.28) | 611.60 (315.10) | −556.53 (−1035.64 to −77.42) | 0.0240 | −2.0464 | 0.9946 | 26.28 | 0.040 | |
| RXRA | 488.90 (317.90) | 271.43 (145.48) | −217.47 (−406.67 to −28.28) | 0.0255 | 120.9811 | 0.3188 | 7.62 | 0.012 | |
| ATM | 207.38 (120.23) | 125.66 (53.69) | −81.72 (−153.11 to −10.33) | 0.0260 | −39.7789 | 0.5616 | 2.7 | 0.310 | |
| RPL19 | 17,988 (5299.35) | 46,340.39 (62,340.74) | 28,351.44 (3211 to 53,491) | 0.0282 | 4555.8384 | 0.2742 | 338.18 | 0.131 | |
| WNT1 | 4.31 (2.52) | 14.51 (22.32) | 10.20 (1.15 to 19.25) | 0.0282 | 0.6250 | 0.1977 | 0.11 | 0.017 | |
| KLRG1 | 24.78 (17.99) | 12.29 (11.25) | −12.49 (−23.63 to −1.35) | 0.0290 | −11.8929 | 0.1798 | 0.86 | 0.021 | |
| MDM4 | 499.69 (266.15) | 314.51 (176.44) | −185.18 (−351.62 to −18.74) | 0.0302 | 45.8611 | 0.6586 | 3.45 | 0.120 | |
| SRC | 1137.41 (882.62) | 711.11 (335.50) | −426.30 (−816.04 to −36.55) | 0.0330 | 590.5061 | 0.0439 | 17.08 | 0.023 | |
| SNAIL1 | 117.15 (55.26) | 165.45 (84.07) | 48.30 (2.35 to 94.26) | 0.0399 | 112.8464 | 0.0009 | 3.8 | 0.004 | |
| SNAIL2 | 258.73 (182.49) | 141.57 (131.76) | −117.16 (−233.18 to −1.14) | 0.0479 | 56.8309 | 0.4574 | 4.85 | 0.002 | |
| MSH6 | 215.81 (44.97) | 185.66 (39.37) | −30.14 (−60.093 to −0.19) | 0.0486 | 181.88 | 0.035 | 3.2 | 0.120 | |
| WNT3 | 16.88 (9.55) | 33.67 (40.78) | 16.79 (−0.41 to 33.99) | 0.0554 | 9.4464 | 0.0725 | 0.74 | 0.180 | |
| CTNND1 | 1920 (775.98) | 1448.12 (537.48) | −471.79 (−961.05 to 17.46) | 0.0583 | 711.6163 | 0.1020 | 50.2 | 0.072 | |
| B2M | 49,959 (16,893) | 63,541.36 (27,058.56) | 13,581.78 (−870 to 28,034) | 0.0647 | 10,086.9989 | 0.3130 | 684.8 | 0.241 | |
| CTNNB1 | 2638.01 (817.46) | 2160.69 (664.70) | −477.32 (−1011 to 56) | 0.0782 | 946.5439 | 0.0893 | 59.93 | 0.018 | |
| MMP2 | 2480.42 (1475) | 1579.34 (1440.04) | −901.08 (−1916.38 to 114.22) | 0.0803 | 393.2500 | 0.6960 | 23.97 | 0.016 | |
| WNT7a | 8.75 (9.17) | 19.83 (28.57) | 11.080 (−1.48 to 23.64) | 0.0821 | −1.4464 | 0.6109 | 0.01 | <0.0001 | |
| CDH1 | 1224.33 (615.21) | 1618.05 (615.21) | 393.71 (−70.07 to 857.50) | 0.0937 | 683.5668 | 0.2511 | 91.73 | 0.120 | |
| Univariate Analysis | Multivariate Analysis | |||||||
|---|---|---|---|---|---|---|---|---|
| Variable | AUC | SE | 95% CI | Best Cut-Off | HR (95% CI) | p-Value | HR (95% CI) | p-Value |
| HER2 | 0.905 | 0.0999 | 0.616 to 0.996 | |||||
| EGF | 0.929 | 0.0696 | 0.647 to 0.999 | >2.51 | 0.053 (0.008 to 0.334) | 0.002 | -- | |
| CTNND1 | 0.881 | 0.105 | 0.587 to 0.991 | >1470.73 | 0.074(0.014 to 0.377) | 0.002 | 0.0217 (0.001 to 0.352) | 0.007 |
| SRC ** | 0.857 | 0.113 | 0.558 to 0.984 | -- | ||||
| TP53 ** | 0.857 | 0.123 | 0.558 to 0.984 | -- | ||||
| SNAIL1 ** | 0.833 | 0.128 | 0.531 to 0.976 | -- | ||||
| RPL19 | 0.833 | 0.124 | 0.531 to 0.976 | >17,216 | 0.111(0.028 to 0.447) | 0.002 | -- | |
| RARA | 0.81 | 0.137 | 0.505 to 0.967 | >260.21 | 0.173(0.044 to 0.68) | 0.012 | 0.0782 (0.009 to 0.68) | 0.0211 |
| RXRA | 0.738 | 0.152 | 0.430 to 0.934 | |||||
| EGFR | 0.69 | 0.162 | 0.384 to 0.908 | |||||
| SNAIL2 | 0.667 | 0.162 | 0.362 to 0.894 | |||||
| KLRG1 | 0.643 | 0.18 | 0.340 to 0.879 | |||||
| ZEB1 | 0.643 | 0.177 | 0.340 to 0.879 | |||||
| GSK3B | 0.643 | 0.184 | 0.340 to 0.879 | |||||
| CDH1 | 0.619 | 0.173 | 0.319 to 0.864 | |||||
| B2M | 0.595 | 0.172 | 0.298 to 0.848 | |||||
| WNT7A | 0.595 | 0.173 | 0.298 to 0.848 | |||||
| ATM | 0.595 | 0.181 | 0.298 to 0.848 | |||||
| CTNNB1 | 0.571 | 0.182 | 0.278 to 0.831 | |||||
| VCL | 0.571 | 0.183 | 0.278 to 0.831 | |||||
| WNT1 | 0.571 | 0.191 | 0.278 to 0.831 | |||||
| WNT3 | 0.56 | 0.206 | 0.268 to 0.823 | |||||
| MSH6 | 0.548 | 0.183 | 0.259 to 0.814 | |||||
| MLH1 | 0.548 | 0.176 | 0.259 to 0.814 | |||||
| MMP2 | 0.5 | 0.178 | 0.221 to 0.779 | |||||
| MDM4 | 0.5 | 0.224 | 0.221 to 0.779 | |||||
| Univariate Analysis | Multivariate Analysis | |||||||
|---|---|---|---|---|---|---|---|---|
| Variable | AUC | SE | 95% CI | Best Cut-Off | HR (95% CI) | p-Value | HR (95% CI) | p-Value |
| HER2 | 0.81 | 0.165 | 0.505 to 0.967 | |||||
| RARA | 0.801 | 0.083 | 0.640 to 0.912 | >285.16 | 0.426 (0.131 to 1.371) | ns | ||
| EGF | 0.747 | 0.101 | 0.580 to 0.874 | >3.83 | 0.177 (0.041 to 0.775) | 0.0207 | 0.220 (0.0053 to 0.9158) | 0.037 |
| RPL19 | 0.716 | 0.111 | 0.547 to 0.850 | >22,791 | 0.212 (0.0561 to 0.780) | 0.0220 | ||
| TP53 | 0.682 | 0.121 | 0.511 to 0.823 | |||||
| MLH1 | 0.643 | 0.168 | 0.340 to 0.879 | |||||
| SRC | 0.621 | 0.103 | 0.449 to 0.773 | |||||
| KLRG1 | 0.619 | 0.173 | 0.319 to 0.864 | |||||
| ZEB1 | 0.595 | 0.181 | 0.298 to 0.848 | |||||
| WNT3 | 0.595 | 0.181 | 0.298 to 0.848 | |||||
| B2M | 0.595 | 0.176 | 0.298 to 0.848 | |||||
| MSH6 | 0.595 | 0.176 | 0.298 to 0.848 | |||||
| CTNND1 | 0.59 | 0.096 | 0.419 to 0.747 | |||||
| WNT1 | 0.571 | 0.191 | 0.278 to 0.831 | |||||
| MMP2 | 0.571 | 0.175 | 0.278 to 0.831 | |||||
| CDH1 | 0.571 | 0.191 | 0.278 to 0.831 | |||||
| MDM4 | 0.569 | 0.127 | 0.399 to 0.728 | |||||
| MDM2 | 0.548 | 0.121 | 0.378 to 0.710 | |||||
| VCL | 0.548 | 0.179 | 0.259 to 0.814 | |||||
| RXRA | 0.548 | 0.187 | 0.259 to 0.814 | |||||
| SNAIL2 | 0.548 | 0.209 | 0.259 to 0.814 | |||||
| CTNNB1 | 0.548 | 0.176 | 0.259 to 0.814 | |||||
| EGFR | 0.536 | 0.112 | 0.368 to 0.699 | |||||
| ATM | 0.524 | 0.179 | 0.240 to 0.797 | |||||
| GSK3B | 0.524 | 0.187 | 0.240 to 0.797 | |||||
| SNAI1 | 0.519 | 0.122 | 0.352 to 0.684 | |||||
| WNT7A | 0.5 | 0.190 | 0.221 to 0.779 | |||||
| Univariate Analysis | Multivariate Analysis | |||||
|---|---|---|---|---|---|---|
| Covariate | b | SE | p | b | SE | p |
| RPL19 | 0.0003035 | 0.0002348 | 0.1962 | |||
| RARA | −0.0004683 | 0.004318 | 0.9136 | |||
| KLRG1 | 0.05449 | 0.6961 | 0.9376 | |||
| WNT1 | 0.05449 | 0.6961 | 0.9376 | |||
| ZEB1 | 0.0508 | 0.0529 | 0.3369 | |||
| SNAI2 | 0.06641 | 0.08788 | 0.4498 | |||
| EGF | 0.3819 | 0.1358 | 0.0049 | 0.4320 | 0.1454 | 0.0030 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Re, V.; Alessandrini, L.; Brisotto, G.; Caggiari, L.; De Zorzi, M.; Casarotto, M.; Miolo, G.; Puglisi, F.; Garattini, S.K.; Lonardi, S.; et al. HER2–CDH1 Interaction via Wnt/B-Catenin Is Associated with Patients’ Survival in HER2-Positive Metastatic Gastric Adenocarcinoma. Cancers 2022, 14, 1266. https://doi.org/10.3390/cancers14051266
De Re V, Alessandrini L, Brisotto G, Caggiari L, De Zorzi M, Casarotto M, Miolo G, Puglisi F, Garattini SK, Lonardi S, et al. HER2–CDH1 Interaction via Wnt/B-Catenin Is Associated with Patients’ Survival in HER2-Positive Metastatic Gastric Adenocarcinoma. Cancers. 2022; 14(5):1266. https://doi.org/10.3390/cancers14051266
Chicago/Turabian StyleDe Re, Valli, Lara Alessandrini, Giulia Brisotto, Laura Caggiari, Mariangela De Zorzi, Mariateresa Casarotto, Gianmaria Miolo, Fabio Puglisi, Silvio Ken Garattini, Sara Lonardi, and et al. 2022. "HER2–CDH1 Interaction via Wnt/B-Catenin Is Associated with Patients’ Survival in HER2-Positive Metastatic Gastric Adenocarcinoma" Cancers 14, no. 5: 1266. https://doi.org/10.3390/cancers14051266
APA StyleDe Re, V., Alessandrini, L., Brisotto, G., Caggiari, L., De Zorzi, M., Casarotto, M., Miolo, G., Puglisi, F., Garattini, S. K., Lonardi, S., Cannizzaro, R., Canzonieri, V., Fassan, M., & Steffan, A. (2022). HER2–CDH1 Interaction via Wnt/B-Catenin Is Associated with Patients’ Survival in HER2-Positive Metastatic Gastric Adenocarcinoma. Cancers, 14(5), 1266. https://doi.org/10.3390/cancers14051266










