Sphingosine Kinase-1 Is Overexpressed and Correlates with Hypoxia in Osteosarcoma: Relationship with Clinicopathological Parameters
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient and Tumor Characteristics
2.2. Immunohistochemistry
2.3. Chemicals and Reagents
2.4. Cell Lines
2.5. Sphingosine Kinase-1 Enzymatic Activity
2.6. Western-Blot Analysis and Antibodies
2.7. Quantitative Real Time PCR
2.8. Cell Viability Assay
2.9. RNA Interference Experiments
2.10. Statistical Analysis
3. Results
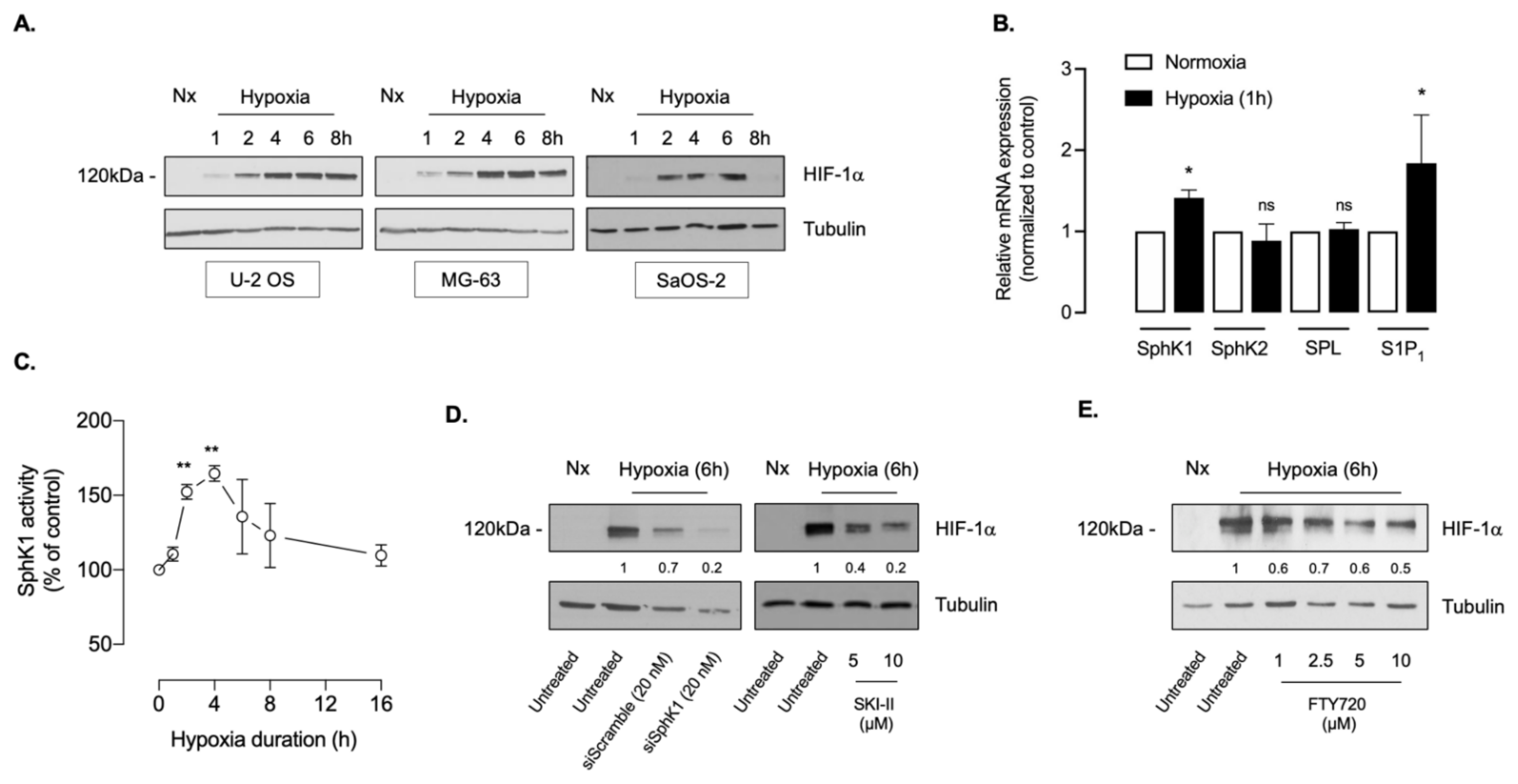
3.1. The SphK1/S1P Signaling Regulates HIF-1α Accumulation in Osteosarcoma Cell Lines
3.2. Sphingosine Kinase-1 Activity Is Overexpressed in Osteosarcoma Biopsy Samples
3.3. Description of the Patient Population
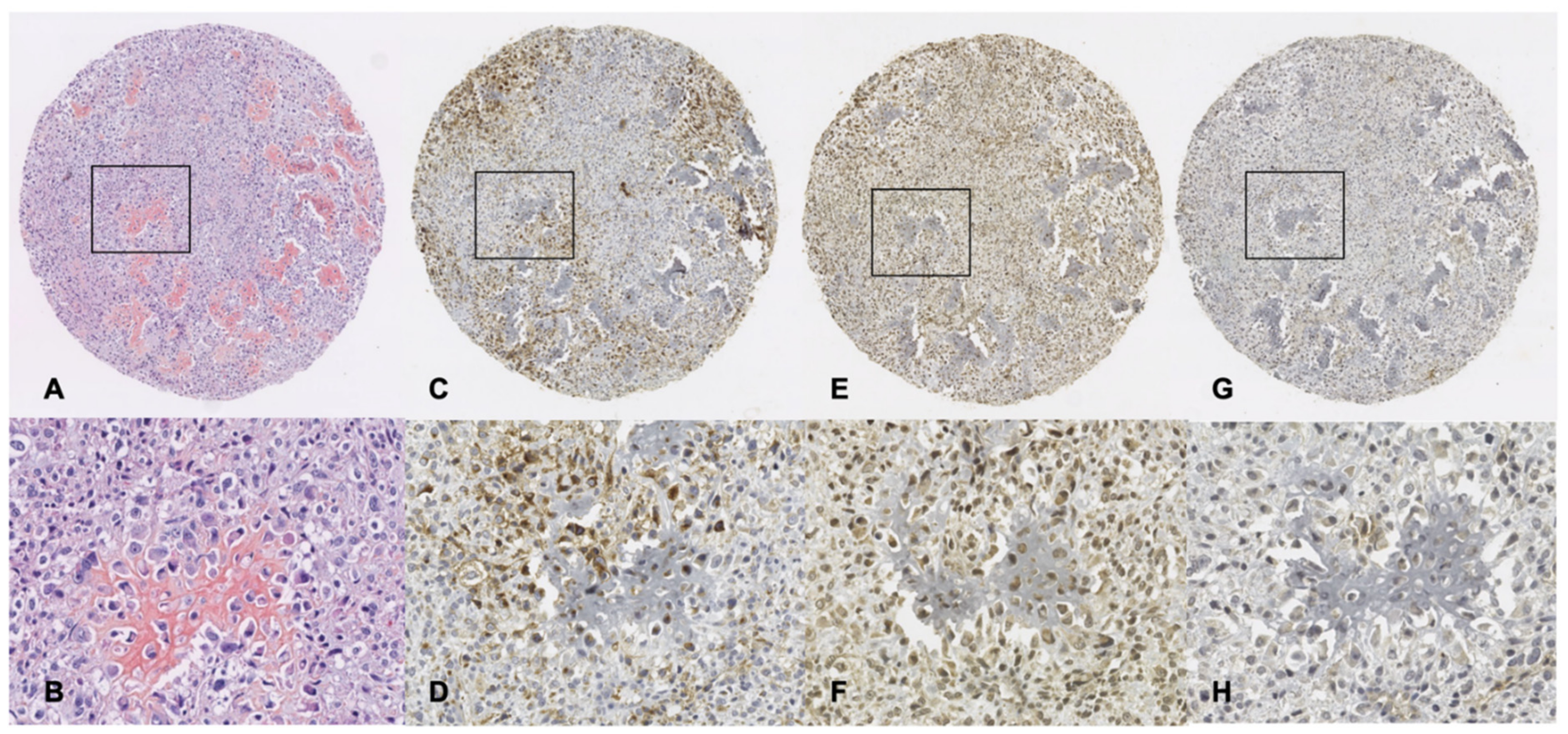
3.4. Immunohistochemistry Analysis of GLUT-1, SphK1 and S1P1 Expression
3.5. Clinical Parameters and Biomarkers Association with Treatment Response
3.6. Clinical Parameters and Biomarkers Associated with Survival in the Different Subgroups (Long Bones Versus Flat Bones and Younger Versus Older Patients)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kansara, M.; Teng, M.; Smyth, M.; Thomas, D. Translational biology of osteosarcoma. Nat. Rev. Cancer 2014, 14, 722–735. [Google Scholar] [CrossRef]
- Fletcher, C.D.M.; Bridge, J.A.; Hogendoorn, P.C.W.; Mertens, F. WHO Classification of Tumours of Soft Tissue and Bone; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Bertin, H.; Gomez-Brouchet, A.; Rédini, F. Osteosarcoma of the jaws: An overview of the pathophysiological mechanisms. Crit. Rev. Oncol. Hematol. 2020, 156, 103126. [Google Scholar] [CrossRef] [PubMed]
- Perry, J.A.; Kiezun, A.; Tonzi, P.; Van Allen, E.M.; Carter, S.L.; Baca, S.C.; Cowley, G.S.; Bhatt, A.S.; Rheinbay, E.; Pedamallu, C.S.; et al. Complementary genomic approaches highlight the PI3K/mTOR pathway as a common vulnerability in osteosarcoma. Proc. Natl. Acad. Sci. USA 2014, 111, E5564–E5573. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Bahrami, A.; Pappo, A.; Easton, J.; Dalton, J.; Hedlund, E.; Ellison, D.; Shurtleff, S.; Wu, G.; Wei, L.; et al. Recurrent Somatic Structural Variations Contribute to Tumorigenesis in Pediatric Osteosarcoma. Cell Rep. 2014, 7, 104–112. [Google Scholar] [CrossRef] [Green Version]
- Bousquet, M.; Noirot, C.; Accadbled, F.; de Gauzy, J.S.; Castex, M.; Brousset, P.; Gomez-Brouchet, A. Whole-exome sequencing in osteosarcoma reveals important heterogeneity of genetic alterations. Ann. Oncol. 2016, 27, 738–744. [Google Scholar] [CrossRef]
- Trama, A.; Botta, L.; Foschi, R.; Ferrari, A.; Stiller, C.; Desandes, E.; Maule, M.M.; Merletti, F.; Gatta, G.; Group, E.-W. Survival of European adolescents and young adults diagnosed with cancer in 2000–07: Population-based data from EUROCARE-5. Lancet Oncol. 2016, 17, 896–906. [Google Scholar] [CrossRef] [Green Version]
- Bielack, S.S.; Smeland, S.; Whelan, J.S.; Marina, N.; Jovic, G.; Hook, J.M.; Krailo, M.D.; Gebhardt, M.; Papai, Z.; Meyer, J.; et al. Methotrexate, Doxorubicin, and Cisplatin (MAP) Plus Maintenance Pegylated Interferon Alfa-2b Versus MAP Alone in Patients with Resectable High-Grade Osteosarcoma and Good Histologic Response to Preoperative MAP: First Results of the EURAMOS-1 Good Response Randomized Controlled Trial. J. Clin. Oncol. 2015, 33, 2279–2287. [Google Scholar] [CrossRef]
- Marina, N.M.; Smeland, S.; Bielack, S.S.; Bernstein, M.; Jovic, G.; Krailo, M.D.; Hook, J.M.; Arndt, C.; van den Berg, H.; Brennan, B.; et al. Comparison of MAPIE versus MAP in patients with a poor response to preoperative chemotherapy for newly diagnosed high-grade osteosarcoma (EURAMOS-1): An open-label, international, randomised controlled trial. Lancet Oncol. 2016, 17, 1396–1408. [Google Scholar] [CrossRef] [Green Version]
- Meyers, P.A.; Schwartz, C.L.; Krailo, M.D.; Healey, J.H.; Bernstein, M.L.; Betcher, D.; Ferguson, W.S.; Gebhardt, M.C.; Goorin, A.M.; Harris, M.; et al. Osteosarcoma: The addition of muramyl tripeptide to chemotherapy improves overall survival--a report from the Children’s Oncology Group. J. Clin. Oncol. 2008, 26, 633–638. [Google Scholar] [CrossRef]
- Piperno-Neumann, S.; Le Deley, M.-C.; Rédini, F.; Pacquement, H.; Marec-Bérard, P.; Petit, P.; Brisse, H.J.; Lervat, C.; Gentet, J.-C.; Entz-Werlé, N.; et al. Zoledronate in combination with chemotherapy and surgery to treat osteosarcoma (OS2006): A randomised, multicentre, open-label, phase 3 trial. Lancet Oncol. 2016, 17, 1070–1080. [Google Scholar] [CrossRef]
- Prudowsky, Z.D.; Yustein, J.T. Recent Insights into Therapy Resistance in Osteosarcoma. Cancers 2020, 13, 83. [Google Scholar] [CrossRef] [PubMed]
- Harris, A.L. Hypoxia—A key regulatory factor in tumour growth. Nat. Rev. Cancer 2002, 2, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Kang, Y. Hypoxia and Hypoxia-Inducible Factors: Master Regulators of Metastasis. Clin. Cancer Res. 2010, 16, 5928–5935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trédan, O.; Galmarini, C.M.; Patel, K.; Tannock, I.F. Drug Resistance and the Solid Tumor Microenvironment. J. Natl. Cancer Inst. 2007, 99, 1441–1454. [Google Scholar] [CrossRef] [Green Version]
- Semenza, G.L. Hypoxia-Inducible Factors in Physiology and Medicine. Cell 2012, 148, 399–408. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zhang, W.; Li, S.; Tu, C. Prognosis value of Hypoxia-inducible factor-1α expression in patients with bone and soft tissue sarcoma: A meta-analysis. SpringerPlus 2016, 5, 1370. [Google Scholar] [CrossRef] [Green Version]
- Ren, H.Y.; Zhang, Y.H.; Li, H.Y.; Xie, T.; Sun, L.L.; Zhu, T.; Wang, S.D.; Ye, Z.M. Prognostic role of hypoxia-inducible factor-1 alpha expression in osteosarcoma: A meta-analysis. OncoTargets Ther. 2016, 9, 1477–1487. [Google Scholar] [CrossRef] [Green Version]
- Ouyang, Y.; Li, H.; Bu, J.; Li, X.; Chen, Z.; Xiao, T. Hypoxia-Inducible Factor-1 Expression Predicts Osteosarcoma Patients’ Survival: A Meta-Analysis. Int. J. Biol. Markers 2016, 31, 229–234. [Google Scholar] [CrossRef]
- Kunkel, G.T.; Maceyka, M.; Milstien, S.; Spiegel, S. Targeting the sphingosine-1-phosphate axis in cancer, inflammation and beyond. Nat. Rev. Drug Discov. 2013, 12, 688–702. [Google Scholar] [CrossRef] [Green Version]
- Brizuela, L.; Ader, I.; Mazerolles, C.; Bocquet, M.; Malavaud, B.; Cuvillier, O. First Evidence of Sphingosine 1-Phosphate Lyase Protein Expression and Activity Downregulation in Human Neoplasm: Implication for Resistance to Therapeutics in Prostate Cancer. Mol. Cancer Ther. 2012, 11, 1841–1851. [Google Scholar] [CrossRef] [Green Version]
- Malavaud, B.; Pchejetski, D.; Mazerolles, C.; de Paiva, G.R.; Calvet, C.; Doumerc, N.; Pitson, S.; Rischmann, P.; Cuvillier, O. Sphingosine kinase-1 activity and expression in human prostate cancer resection specimens. Eur. J. Cancer 2010, 46, 3417–3424. [Google Scholar] [CrossRef]
- Gachechiladze, M.; Tichy, T.; Kolek, V.; Grygarkova, I.; Klein, J.; Mgebrishvili, G.; Kharaishvili, G.; Janikova, M.; Smickova, P.; Cierna, L.; et al. Sphingosine kinase-1 predicts overall survival outcomes in non-small cell lung cancer patients treated with carboplatin and navelbine. Oncol. Lett. 2019, 18, 1259–1266. [Google Scholar] [CrossRef] [Green Version]
- Cuvillier, O.; Ader, I.; Bouquerel, P.; Brizuela, L.; Malavaud, B.; Mazerolles, C.; Rischmann, P. Activation of Sphingosine Kinase-1 in Cancer: Implications for Therapeutic Targeting. Curr. Mol. Pharmacol. 2010, 3, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Ader, I.; Brizuela, L.; Bouquerel, P.; Malavaud, B.; Cuvillier, O. Sphingosine Kinase 1: A New Modulator of Hypoxia Inducible Factor 1α during Hypoxia in Human Cancer Cells. Cancer Res. 2008, 68, 8635–8642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ader, I.; Gstalder, C.; Bouquerel, P.; Golzio, M.; Andrieu, G.; Zalvidea, S.; Richard, S.; Sabbadini, R.A.; Malavaud, B.; Cuvillier, O. Neutralizing S1P inhibits intratumoral hypoxia, induces vascular remodelling and sensitizes to chemotherapy in prostate cancer. Oncotarget 2015, 6, 13803–13821. [Google Scholar] [CrossRef] [Green Version]
- Bouquerel, P.; Gstalder, C.; Muller, D.; Laurent, J.; Brizuela, L.; Sabbadini, R.; Malavaud, B.; Pyronnet, S.; Martineau, Y.; Ader, I.; et al. Essential role for SphK1/S1P signaling to regulate hypoxia-inducible factor 2alpha expression and activity in cancer. Oncogenesis 2016, 5, e209. [Google Scholar] [CrossRef] [Green Version]
- Cohen, J.A.; Barkhof, F.; Comi, G.; Hartung, H.-P.; Khatri, B.O.; Montalban, X.; Pelletier, J.; Capra, R.; Gallo, P.; Izquierdo, G.; et al. Oral Fingolimod or Intramuscular Interferon for Relapsing Multiple Sclerosis. N. Engl. J. Med. 2010, 362, 402–415. [Google Scholar] [CrossRef]
- Kappos, L.; Radue, E.-W.; O’Connor, P.; Polman, C.; Hohlfeld, R.; Calabresi, P.; Selmaj, K.; Agoropoulou, C.; Leyk, M.; Zhang-Auberson, L.; et al. A Placebo-Controlled Trial of Oral Fingolimod in Relapsing Multiple Sclerosis. N. Engl. J. Med. 2010, 362, 387–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graler, M.H.; Goetzl, E.J. The immunosuppressant FTY720 down-regulates sphingosine 1-phosphate G-protein-coupled receptors. FASEB J. 2004, 18, 551–553. [Google Scholar] [CrossRef] [PubMed]
- Vessey, D.A.; Kelley, M.; Zhang, J.; Li, L.; Tao, R.; Karliner, J.S. Dimethylsphingosine and FTY720 inhibit the SK1 form but activate the SK2 form of sphingosine kinase from rat heart. J. Biochem. Mol. Toxicol. 2007, 21, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Pchejetski, D.; Bohler, T.; Brizuela, L.; Sauer, L.; Doumerc, N.; Golzio, M.; Salunkhe, V.; Teissié, J.; Malavaud, B.; Waxman, J.; et al. FTY720 (Fingolimod) Sensitizes Prostate Cancer Cells to Radiotherapy by Inhibition of Sphingosine Kinase-1. Cancer Res. 2010, 70, 8651–8661. [Google Scholar] [CrossRef] [Green Version]
- Lim, K.G.; Tonelli, F.; Li, Z.; Lu, X.; Bittman, R.; Pyne, S.; Pyne, N.J. FTY720 analogues as sphingosine kinase 1 inhibitors: Enzyme inhibition kinetics, allosterism, proteasomal degradation, and actin rearrangement in MCF-7 breast cancer cells. J. Biol. Chem. 2011, 286, 18633–18640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azuma, H.; Takahara, S.; Ichimaru, N.; Wang, J.D.; Itoh, Y.; Otsuki, Y.; Morimoto, J.; Fukui, R.; Hoshiga, M.; Ishihara, T.; et al. Marked prevention of tumor growth and metastasis by a novel immunosuppressive agent, FTY720, in mouse breast cancer models. Cancer Res. 2002, 62, 1410–1419. [Google Scholar] [PubMed]
- Lamontagne, K.; Littlewood-Evans, A.; Schnell, C.; O’Reilly, T.; Wyder, L.; Sanchez, T.; Probst, B.; Butler, J.; Wood, A.; Liau, G.; et al. Antagonism of Sphingosine-1-Phosphate Receptors by FTY720 Inhibits Angiogenesis and Tumor Vascularization. Cancer Res. 2006, 66, 221–231. [Google Scholar] [CrossRef] [Green Version]
- Li, M.-H.; Hla, T.; Ferrer, F. FTY720 inhibits tumor growth and enhances the tumor-suppressive effect of topotecan in neuroblastoma by interfering with the sphingolipid signaling pathway. Pediatr. Blood Cancer 2013, 60, 1418–1423. [Google Scholar] [CrossRef] [Green Version]
- Rosa, R.; Marciano, R.; Malapelle, U.; Formisano, L.; Nappi, L.; D’Amato, C.; D’Amato, V.; Damiano, V.; Marfè, G.; Del Vecchio, S.; et al. Sphingosine Kinase 1 Overexpression Contributes to Cetuximab Resistance in Human Colorectal Cancer Models. Clin. Cancer Res. 2013, 19, 138–147. [Google Scholar] [CrossRef] [Green Version]
- Gstalder, C.; Ader, I.; Cuvillier, O. FTY720 (Fingolimod) Inhibits HIF1 and HIF2 Signaling, Promotes Vascular Remodeling, and Chemosensitizes in Renal Cell Carcinoma Animal Model. Mol. Cancer Ther. 2016, 15, 2465–2474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ader, I.; Malavaud, B.; Cuvillier, O. When the sphingosine kinase-1/sphingosine 1-phosphate pathway meets hypoxia signaling: New targets for cancer therapy. Cancer Res. 2009, 69, 3723–3726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuvillier, O.; Ader, I.; Bouquerel, P.; Brizuela, L.; Gstalder, C.; Malavaud, B. Hypoxia, Therapeutic Resistance, and Sphingosine 1-Phosphate. Adv. Cancer Res. 2013, 117, 117–141. [Google Scholar] [CrossRef]
- Pitson, S.M.; Moretti, P.A.; Zebol, J.R.; Lynn, H.E.; Xia, P.; Vadas, M.A.; Wattenberg, B.W. Activation of sphingosine kinase 1 by ERK1/2-mediated phosphorylation. EMBO. J. 2003, 22, 5491–5500. [Google Scholar] [CrossRef] [Green Version]
- Talmont, F.; Moulédous, L.; Baranger, M.; Gomez-Brouchet, A.; Zajac, J.-M.; Deffaud, C.; Cuvillier, O.; Hatzoglou, A. Development and characterization of sphingosine 1-phosphate receptor 1 monoclonal antibody suitable for cell imaging and biochemical studies of endogenous receptors. PLoS ONE 2019, 14, e0213203. [Google Scholar] [CrossRef]
- Fedchenko, N.; Reifenrath, J. Different approaches for interpretation and reporting of immunohistochemistry analysis results in the bone tissue—A review. Diagn. Pathol. 2014, 9, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Specht, E.; Kaemmerer, D.; Sanger, J.; Wirtz, R.M.; Schulz, S.; Lupp, A. Comparison of immunoreactive score, HER2/neu score and H score for the immunohistochemical evaluation of somatostatin receptors in bronchopulmonary neuroendocrine neoplasms. Histopathology 2015, 67, 368–377. [Google Scholar] [CrossRef]
- Brizuela, L.; Cuvillier, O. Biochemical Methods for Quantifying Sphingolipids: Ceramide, Sphingosine, Sphingosine Kinase-1 Activity, and Sphingosine-1-Phosphate. Methods Mol. Biol. 2012, 874, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Cuvillier, O.; Nava, V.E.; Murthy, S.K.; Edsall, L.C.; Levade, T.; Milstien, S.; Spiegel, S. Sphingosine generation, cytochrome c release, and activation of caspase-7 in doxorubicin-induced apoptosis of MCF7 breast adenocarcinoma cells. Cell Death Differ. 2001, 8, 162–171. [Google Scholar] [CrossRef] [Green Version]
- Knowles, H.J.; Schaefer, K.L.; Dirksen, U.; Athanasou, N.A. Hypoxia and hypoglycaemia in Ewing’s sarcoma and osteosarcoma: Regulation and phenotypic effects of Hypoxia-Inducible Factor. BMC Cancer 2010, 10, 372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Naggar, A.; Clarkson, P.; Zhang, F.; Mathers, J.; Tognon, C.; Sorensen, P.H. Expression and stability of hypoxia inducible factor 1α in osteosarcoma. Pediatr. Blood Cancer 2012, 59, 1215–1222. [Google Scholar] [CrossRef] [PubMed]
- Adamski, J.; Price, A.; Dive, C.; Makin, G. Hypoxia–Induced Cytotoxic Drug Resistance in Osteosarcoma Is Independent of HIF-1Alpha. PLoS ONE 2013, 8, e65304. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Wu, Y.; Chen, Y.; Liu, H. Clinical significance of hypoxia-inducible factor 1 and VEGF-A in osteosarcoma. Int. J. Clin. Oncol. 2015, 20, 1233–1243. [Google Scholar] [CrossRef] [PubMed]
- Guan, G.; Zhang, Y.; Lu, Y.; Liu, L.; Shi, D.; Wen, Y.; Yang, L.; Ma, Q.; Liu, T.; Zhu, X.; et al. The HIF-1alpha/CXCR4 pathway supports hypoxia-induced metastasis of human osteosarcoma cells. Cancer Lett. 2015, 357, 254–264. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, Q.; Yu, T.; Sun, S.; Wang, W.; Liu, G. Hypoxia promotes drug resistance in osteosarcoma cells via activating AMP-activated protein kinase (AMPK) signaling. J. Bone Oncol. 2016, 5, 22–29. [Google Scholar] [CrossRef] [Green Version]
- French, K.J.; Schrecengost, R.S.; Lee, B.D.; Zhuang, Y.; Smith, S.N.; Eberly, J.L.; Yun, J.; Smith, C.D. Discovery and evaluation of inhibitors of human sphingosine kinase. Cancer Res. 2003, 63, 5962–5969. [Google Scholar] [PubMed]
- Salzer-Kuntschik, M.; Brand, G.; Delling, G. Determination of the degree of morphological regression following chemotherapy in malignant bone tumors. Der Pathol. 1983, 4, 135–141. [Google Scholar]
- Vaupel, P.; Mayer, A. Hypoxia in cancer: Significance and impact on clinical outcome. Cancer Metastasis Rev. 2007, 26, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Kaya, M.; Wada, T.; Akatsuka, T.; Kawaguchi, S.; Nagoya, S.; Shindoh, M.; Higashino, F.; Mezawa, F.; Okada, F.; Ishii, S. Vascular endothelial growth factor expression in untreated osteosarcoma is predictive of pulmonary metastasis and poor prognosis. Clin. Cancer Res. 2000, 6, 572–577. [Google Scholar]
- Mizobuchi, H.; García-Castellano, J.M.; Philip, S.; Healey, J.H.; Gorlick, R. Hypoxia Markers in Human Osteosarcoma: An Exploratory Study. Clin. Orthop. Relat. Res. 2008, 466, 2052–2059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Yang, Y.; Yuan, Z.; Wang, C.; Shi, Y. Predicting chemosensitivity in osteosarcoma prior to chemotherapy: An investigational study of biomarkers with immunohistochemistry. Oncol. Lett. 2012, 3, 1011–1016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Q.-C.; Zeng, B.-F.; Dong, Y.; Shi, Z.-M.; Jiang, Z.-M.; Huang, J. Overexpression of Hypoxia-Inducible Factor-1α in Human Osteosarcoma: Correlation with Clinicopathological Parameters and Survival Outcome. Jpn. J. Clin. Oncol. 2007, 37, 127–134. [Google Scholar] [CrossRef]
- Chen, W.-L.; Feng, H.-J.; Li, H.-G. Expression and significance of hypoxemia-inducible factor-1α in osteosarcoma of the jaws. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontol. 2008, 106, 254–257. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.; Cai, C.; Zhao, G.; Qiu, X.; Zhao, H.; Ma, Q.; Tian, L.; Li, X.; Hu, Y.; Liao, B.; et al. Hypoxia Promotes Migration and Induces CXCR4 Expression via HIF-1α Activation in Human Osteosarcoma. PLoS ONE 2014, 9, e90518. [Google Scholar] [CrossRef]
- Hu, T.; He, N.; Yang, Y.; Yin, C.; Sang, N.; Yang, Q. DEC2 expression is positively correlated with HIF-1 activation and the invasiveness of human osteosarcomas. J. Exp. Clin. Cancer Res. 2015, 34, 22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Ren, T.; Huang, Y.; Bao, X.; Sun, K.; Shen, D.; Guo, W. BMPR2 and HIF1-α overexpression in resected osteosarcoma correlates with distant metastasis and patient survival. Chin. J. Cancer Res. 2017, 29, 447–454. [Google Scholar] [CrossRef] [Green Version]
- Petty, J.C.; Lana, S.E.; Thamm, D.H.; Charles, J.B.; Bachand, A.M.; Bush, J.M.; Ehrhart, E.J. Glucose transporter 1 expression in canine osteosarcoma. Vet. Comp. Oncol. 2008, 6, 133–140. [Google Scholar] [CrossRef]
- Cifuentes, M.; García, M.A.; Arrabal, P.M.; Martínez, F.; Yañez, M.J.; Jara, N.; Weil, B.; Domínguez, D.; Medina, R.A.; Nualart, F. Insulin regulates GLUT1-mediated glucose transport in MG-63 human osteosarcoma cells. J. Cell. Physiol. 2011, 226, 1425–1432. [Google Scholar] [CrossRef]
- Kubo, T.; Shimose, S.; Fujimori, J.; Furuta, T.; Arihiro, K.; Ochi, M. Does Expression of Glucose Transporter Protein-1 Relate to Prognosis and Angiogenesis in Osteosarcoma? Clin. Orthop. Relat. Res. 2015, 473, 305–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geng, X.; Wang, H.; Xu, L.; Han, Y.; Liu, Y. MicroRNA-140-5p is Downregulated in Osteosarcoma and Overexpression of MicroRNA-140-5p Inhibits Cancer Cell Proliferation by Downregulating GLUT-1. OncoTargets Ther. 2021, 14, 995–1002. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, Y.; Wan, Z.; Liu, S.; Cao, Y.; Zeng, Z. Sphingosine Kinase 1 and Cancer: A Systematic Review and Meta-Analysis. PLoS ONE 2014, 9, e90362. [Google Scholar] [CrossRef]
- Cortini, M.; Armirotti, A.; Columbaro, M.; Longo, D.; Pompo, G.; Cannas, E.; Maresca, A.; Errani, C.; Longhi, A.; Righi, A.; et al. Exploring Metabolic Adaptations to the Acidic Microenvironment of Osteosarcoma Cells Unveils Sphingosine 1-Phosphate as a Valuable Therapeutic Target. Cancers 2021, 13, 311. [Google Scholar] [CrossRef] [PubMed]

| Variable | All | Long Bones | Flat Bones | p Value | |||
|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | ||
| 130 | 100 | 71 | 54.6 | 59 | 45.4 | ||
| Age (Years) | |||||||
| Median (Range) | 22.4 (5.7–83.9) | 15.8 (6.6–82.8) | 39 (5.7–83.9) | <0.0001 | |||
| <25 (Years) | 69 | 53.1 | 53 | 40.8 | 16 | 12.3 | <0.0001 |
| ≥25 (Years) | 61 | 46.9 | 18 | 13.8 | 43 | 33.1 | |
| Gender | 0.0068 | ||||||
| Male | 80 | 61.5 | 51 | 39.2 | 29 | 22.3 | |
| Female | 49 | 37.7 | 20 | 15.4 | 29 | 22.3 | |
| Unknown | 1 | 0.8 | 0 | 0.0 | 1 | 0.8 | |
| Site | |||||||
| Flat bones | 59 | 45.4 | |||||
| Long bones | 71 | 54.6 | |||||
| Histological type | 0.8099 | ||||||
| Chondroblastic | 32 | 24.6 | 11 | 8.5 | 21 | 16.2 | |
| Osteoblastic | 60 | 46.2 | 47 | 36.2 | 13 | 10.0 | |
| Fibroblastic | 25 | 19.2 | 7 | 5.4 | 18 | 13.8 | |
| Other | 10 | 7.7 | 4 | 3.1 | 6 | 4.6 | |
| Unknown | 3 | 2.3 | 2 | 1.5 | 1 | 0.8 | |
| Metastatic vs Non Metastatic | 0.012 | ||||||
| Localized | 89 | 68.5 | 42 | 32.3 | 47 | 36.2 | |
| Metastatic | 41 | 31.5 | 29 | 22.3 | 12 | 9.2 | |
| Response to chemotherapy | 0.0064 | ||||||
| Good | 53 | 40.8 | 39 | 30.0 | 14 | 10.8 | |
| Poor | 43 | 33.1 | 20 | 15.4 | 23 | 17.7 | |
| Unknown | 34 | 26.2 | 12 | 9.2 | 22 | 16.9 | |
| Variable | Doxorubicin | FOLD | FTY720 | FOLD | ||
|---|---|---|---|---|---|---|
| Normoxia | Hypoxia | Normoxia | Hypoxia | |||
| U-2 OS | 0.066 | 0.195 | 3 | 4.21 | 4.21 | 1 |
| SaOS-2 | 0.073 | 1.00 | 14 | 3.20 | 3.23 | 1 |
| MG-63 | 0.025 | 0.507 | 20 | 1.52 | 1.50 | 1 |
| 143B | 0.017 | 0.134 | 8 | 1.11 | 1.22 | 1 |
| Variable | All | Long Bones | Flat Bones | p Value | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AVG | SD | Median | Min | Max | AVG | SD | Median | Min | Max | AVG | SD | Median | Min | Max | ||
| GLUT1 | ||||||||||||||||
| n = 98 | n = 54 | n = 44 | ||||||||||||||
| % of stained cells | 49.29 | 31.73 | 60 | 0 | 100 | 55.37 | 32.95 | 65 | 0 | 100 | 41.82 | 28.8 | 50 | 0 | 90 | 0.0116 |
| IRS score | 6.31 | 4.03 | 6 | 0 | 12 | 7.06 | 4.03 | 9 | 0 | 12 | 5.39 | 3.88 | 6 | 0 | 12 | 0.0353 |
| SphK1 | ||||||||||||||||
| n = 96 | n = 52 | n = 44 | ||||||||||||||
| % of stained cells | 67.28 | 28.29 | 80 | 0 | 100 | 70.77 | 26.7 | 80 | 0 | 100 | 63.16 | 29.83 | 80 | 0 | 90 | 0.1835 |
| IRS score | 4.57 | 2.7 | 4 | 0 | 12 | 4.85 | 2.55 | 6 | 0 | 12 | 4.25 | 2.86 | 3 | 0 | 12 | 0.1828 |
| S1P1 | ||||||||||||||||
| n = 111 | n = 63 | n = 48 | ||||||||||||||
| % of stained cells | 64.23 | 23.14 | 70 | 0 | 100 | 64.29 | 23.6 | 70 | 0 | 100 | 64.17 | 22.77 | 70 | 0 | 90 | 0.9279 |
| IRS score | 4.3 | 2.52 | 3 | 0 | 12 | 4.25 | 2.67 | 3 | 0 | 12 | 4.35 | 2.33 | 4 | 0 | 12 | 0.6881 |
| All OS | |||||||
| correlation coefficient (r) | GLUT1 % cells | SphK1 % cells | S1P1 % cells | correlation coefficient (r) | GLUT1 IRS score | SphK1 IRS score | S1P1 IRS score |
| GLUT1 % cells | NA | 0.3019 | 0.009351 | GLUT1 IRS score | NA | 0.3498 | −0.01708 |
| SphK1 % cells | NA | 0.1387 | SphK1 IRS score | NA | −0.01275 | ||
| S1P1 % cells | NA | S1P1 IRS score | NA | ||||
| p value | GLUT1 % cells | SphK1 % cells | S1P1 % cells | p value | GLUT1 IRS score | SphK1 IRS score | S1P1 IRS score |
| GLUT1 % cells | NA | 0.005 | 0.9287 | GLUT1 IRS score | NA | 0.001 | 0.8702 |
| SphK1 % cells | NA | 0.1922 | SphK1 IRS score | NA | 0.9051 | ||
| S1P1 % cells | NA | S1P1 IRS score | NA | ||||
| Long Bones | |||||||
| correlation coefficient (r) | GLUT1 % cells | SphK1 % cells | S1P1 % cells | correlation coefficient (r) | GLUT1 IRS score | SphK1 IRS score | S1P1 IRS score |
| GLUT1 % cells | NA | 0.3608 | −0.1556 | GLUT1 IRS score | NA | 0.2902 | −0.1024 |
| SphK1 % cells | NA | 0.1765 | SphK1 IRS score | NA | −0.1345 | ||
| S1P1 % cells | NA | S1P1 IRS score | NA | ||||
| p value | GLUT1 % cells | SphK1 % cells | S1P1 % cells | p value | GLUT1 IRS score | SphK1 IRS score | S1P1 IRS score |
| GLUT1 % cells | NA | 0.0127 | 0.2706 | GLUT1 IRS score | NA | 0.0478 | 0.4699 |
| SphK1 % cells | NA | 0.2153 | SphK1 IRS score | NA | 0.3466 | ||
| S1P1 % cells | NA | S1P1 IRS score | NA | ||||
| Flat Bones | |||||||
| correlation coefficient (r) | GLUT1 % cells | SphK1 % cells | S1P1 %cells | correlation coefficient (r) | GLUT1 IRS score | SphK1 IRS score | S1P1 IRS score |
| GLUT1 % cells | NA | 0.1979 | 0.174 | GLUT1 IRS score | NA | 0.3948 | 0.09562 |
| SphK1 % cells | NA | 0.1661 | SphK1 IRS score | NA | 0.2053 | ||
| S1P1 % cells | NA | S1P1 IRS score | NA | ||||
| p value | GLUT1 % cells | SphK1 % cells | S1P1 %cells | p value | GLUT1 IRS score | SphK1 IRS score | S1P1 IRS score |
| GLUT1 % cells | NA | 0.2336 | 0.2703 | GLUT1 IRS score | NA | 0.0142 | 0.5469 |
| SphK1 % cells | NA | 0.3058 | SphK1 IRS score | NA | 0.2039 | ||
| S1P1 % cells | NA | S1P1 IRS score | NA |
| All OS | Good | Poor | p Value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| n = 40 | n = 30 | ||||||||||
| GLUT1 | AVG | SD | Median | Min | Max | AVG | SD | Median | Min | Max | |
| % of stained cells | 57.75 | 31.73 | 70 | 0 | 100 | 45 | 29.8 | 50 | 0 | 90 | 0.1945 |
| IRS score | 7.4 | 3.85 | 9 | 0 | 12 | 5.67 | 3.94 | 6 | 0 | 12 | 0.0587 |
| SphK1 | n = 38 | n = 31 | |||||||||
| % of stained cells | 69.74 | 25.09 | 80 | 0 | 100 | 68.39 | 25.96 | 80 | 0 | 90 | 0.9506 |
| IRS score | 4.76 | 2.8 | 5 | 0 | 12 | 4.45 | 2.39 | 4 | 0 | 8 | 0.7955 |
| S1P1 | n = 45 | n = 36 | |||||||||
| % of stained cells | 57.78 | 23.54 | 60 | 0 | 90 | 66.11 | 24.06 | 70 | 0 | 90 | 0.2325 |
| IRS score | 3.69 | 2.2 | 3 | 0 | 12 | 4.58 | 2.71 | 3 | 0 | 12 | 0.0787 |
| Long Bones | n = 39 | n = 20 | p value | ||||||||
| GLUT1 | AVG | SD | Median | Min | Max | AVG | SD | Median | Min | Max | |
| % of stained cells | 58.97 | 31.89 | 70 | 0 | 100 | 56.67 | 30.86 | 60 | 0 | 90 | 0.8059 |
| IRS score | 7.59 | 3.78 | 9 | 0 | 12 | 7 | 4.05 | 9 | 0 | 12 | 0.6482 |
| SphK1 | n = 28 | n = 13 | |||||||||
| % of stained cells | 75 | 17.53 | 80 | 0 | 100 | 69.23 | 29.85 | 80 | 0 | 90 | 0.744 |
| IRS score | 5 | 2.33 | 6 | 0 | 12 | 4.54 | 2.37 | 4 | 0 | 8 | 0.6947 |
| S1P1 | n = 34 | n = 18 | |||||||||
| % of stained cells | 56.18 | 23.1 | 60 | 0 | 90 | 71.67 | 23.33 | 80 | 0 | 90 | 0.0038 |
| IRS score | 3.47 | 2.22 | 3 | 0 | 12 | 5.22 | 2.94 | 6 | 0 | 12 | 0.0098 |
| Flat Bones | n = 11 | n = 15 | p value | ||||||||
| GLUT1 | AVG | SD | Median | Min | Max | AVG | SD | Median | Min | Max | |
| % of stained cells | 54.55 | 32.67 | 60 | 0 | 90 | 33.33 | 24.4 | 40 | 0 | 70 | 0.0569 |
| IRS score | 6.91 | 4.18 | 9 | 0 | 12 | 4.33 | 3.44 | 6 | 0 | 9 | 0.0993 |
| SphK1 | n = 10 | n = 18 | |||||||||
| % of stained cells | 55 | 36.59 | 70 | 0 | 90 | 67.78 | 23.65 | 75 | 0 | 90 | 0.5847 |
| IRS score | 4.1 | 3.93 | 3 | 0 | 12 | 4.39 | 2.48 | 3 | 0 | 8 | 0.4539 |
| S1P1 | n = 11 | n = 18 | |||||||||
| % of stained cells | 62.73 | 25.33 | 80 | 20 | 90 | 60.56 | 24.13 | 65 | 0 | 90 | 0.7043 |
| IRS score | 4.36 | 2.11 | 4 | 2 | 8 | 3.94 | 2.36 | 3 | 0 | 9 | 0.6521 |
| Variable | Death (Overall Survival) | Death or Metastatic Progression (MPFS) | Good Response to Chemotherapy | ||||||
|---|---|---|---|---|---|---|---|---|---|
| All OS | Long Bones | Flat Bones | All OS | Long Bones | Flat Bones | All OS | Long Bones | Flat Bones | |
| GLUT1 (% of cells) * | |||||||||
| n | 98 | 54 | 44 | 98 | 54 | 44 | 70 | 44 | 26 |
| HRa | 1.03 | 0.92 | 1.2 | 1.05 | 0.97 | 1.16 | 1.09 | 0.92 | 1.32 |
| [IC 95%] ** | [0.93–1.15] | [0.80–1.06] | [1.00–1.44] | [0.96–1.15] | [0.87–1.08] | [0.98–1.38] | [0.92–1.29] | [0.72–1.19] | [0.95–1.84] |
| p | 0.57 | 0.24 | 0.05 | 0.31 | 0.58 | 0.08 | 0.34 | 0.53 | 0.1 |
| SphK1 (% of cells) * | |||||||||
| n | 96 | 52 | 44 | 96 | 52 | 44 | 69 | 41 | 28 |
| HRa [IC 95%] ** | 1.04 | 1.1 | 0.9 | 1.03 | 1.06 | 0.95 | 0.98 | 1.04 | 0.82 |
| [0.88–1.15] | [0.90–1.36] | [0.74–1.11] | [0.91–1.16] | [0.90–1.25] | [0.79–1.16] | [0.79–1.21] | [0.73–1.47] | [0.62–1.10] | |
| p | 0.96 | 0.36 | 0.33 | 0.68 | 0.46 | 0.63 | 0.82 | 0.83 | 0.19 |
| S1P1 (% of cells) * | |||||||||
| n | 111 | 63 | 48 | 111 | 63 | 48 | 81 | 52 | 29 |
| HRa [IC 95%] ** | 1.09 | 1.29 | 0.92 | 1.01 | 1.23 | 0.91 | 0.88 | 0.72 | 1.07 |
| [0.94–1.27] | [1.00–1.66] | [0.75–1.13] | [0.96–1.26] | [1.02–1.48] | [0.74–1.11] | [0.71–1.09] | [0.52–1.01] | [0.6–1.51] | |
| p | 0.27 | 0.05 | 0.42 | 0.17 | 0.03 | 0.35 | 0.24 | 0.06 | 0.7 |
| GLUT1 (IRS Score) * | |||||||||
| n | 98 | 54 | 44 | 98 | 54 | 44 | 70 | 44 | 26 |
| HRa [IC 95%] ** | 1.05 | 0.97 | 1.15 | 1.05 | 0.99 | 1.14 | 1.09 | 0.98 | 1.2 |
| [0.96–1.14] | [0.86–1.09] | [1.00–1.32] | [0.98–1.13] | [0.90–1.08] | [1.00–1.29] | [0.95–1.25] | [0.80–1.19] | [0.94–1.54] | |
| p | 0.27 | 0.58 | 0.04 | 0.17 | 0.87 | 0.06 | 0.23 | 0.82 | 0.13 |
| SphK1 (IRS Score) * | |||||||||
| n | 96 | 52 | 44 | 96 | 52 | 44 | 69 | 41 | 28 |
| HRa [IC 95%] ** | 1.07 | 1.25 | 0.9 | 1.03 | 1.13 | 0.94 | 0.95 | 0.94 | 0.93 |
| [0.93–1.23] | [1.03–1.51] | [0.71–1.14] | [0.92–1.16] | [0.96–1.35] | [0.77–1.16] | [0.77–1.17] | [0.66–1.34] | [0.71–1.23] | |
| p | 0.35 | 0.02 | 0.39 | 0.6 | 0.15 | 0.58 | 0.63 | 0.73 | 0.62 |
| S1P1 (IRS Score) * | |||||||||
| n | 111 | 63 | 48 | 111 | 63 | 48 | 81 | 52 | 29 |
| HRa [IC 95%] ** | 1.05 | 1.07 | 1.02 | 1.07 | 1.09 | 1.01 | 0.89 | 0.8 | 1.1 |
| [0.94–1.18] | [0.92–1.23] | [0.84–1.24] | [0.97–1.18] | [0.97–1.21] | [0.83–1.24] | [0.73–1.08] | [0.62–1.03] | [0.78–1.55] | |
| p | 0.4 | 0.39 | 0.86 | 0.18 | 0.14 | 0.89 | 0.22 | 0.08 | 0.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gomez-Brouchet, A.; Illac, C.; Ledoux, A.; Fortin, P.-Y.; de Barros, S.; Vabre, C.; Despas, F.; Peries, S.; Casaroli, C.; Bouvier, C.; et al. Sphingosine Kinase-1 Is Overexpressed and Correlates with Hypoxia in Osteosarcoma: Relationship with Clinicopathological Parameters. Cancers 2022, 14, 499. https://doi.org/10.3390/cancers14030499
Gomez-Brouchet A, Illac C, Ledoux A, Fortin P-Y, de Barros S, Vabre C, Despas F, Peries S, Casaroli C, Bouvier C, et al. Sphingosine Kinase-1 Is Overexpressed and Correlates with Hypoxia in Osteosarcoma: Relationship with Clinicopathological Parameters. Cancers. 2022; 14(3):499. https://doi.org/10.3390/cancers14030499
Chicago/Turabian StyleGomez-Brouchet, Anne, Claire Illac, Adeline Ledoux, Pierre-Yves Fortin, Sandra de Barros, Clémentine Vabre, Fabien Despas, Sophie Peries, Christelle Casaroli, Corinne Bouvier, and et al. 2022. "Sphingosine Kinase-1 Is Overexpressed and Correlates with Hypoxia in Osteosarcoma: Relationship with Clinicopathological Parameters" Cancers 14, no. 3: 499. https://doi.org/10.3390/cancers14030499
APA StyleGomez-Brouchet, A., Illac, C., Ledoux, A., Fortin, P.-Y., de Barros, S., Vabre, C., Despas, F., Peries, S., Casaroli, C., Bouvier, C., Aubert, S., de Pinieux, G., Larousserie, F., Galmiche, L., Talmont, F., Pitson, S., Maddelein, M.-L., & Cuvillier, O. (2022). Sphingosine Kinase-1 Is Overexpressed and Correlates with Hypoxia in Osteosarcoma: Relationship with Clinicopathological Parameters. Cancers, 14(3), 499. https://doi.org/10.3390/cancers14030499








