The Role of Radiomics and AI Technologies in the Segmentation, Detection, and Management of Hepatocellular Carcinoma
Abstract
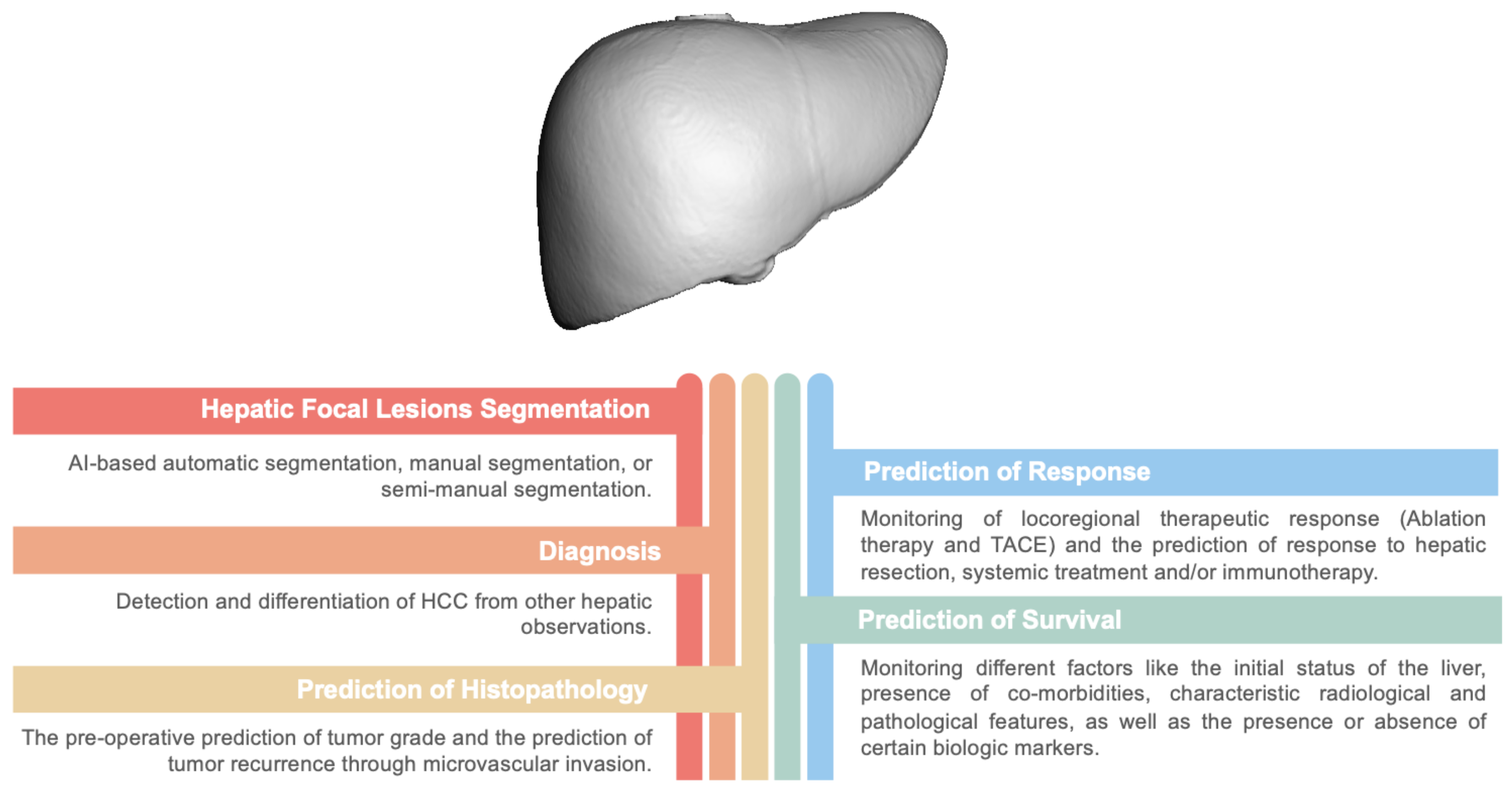
Simple Summary
Abstract
1. Introduction
2. Background on AI and Radiomics Techniques
2.1. Artificial Intelligence (AI)
2.2. Radiomics
2.2.1. Feature-Based Radiomics
- Shape characteristics, such as sphericity, compactness, surface area, and maximum dimensions, reflect the geometric characteristics of the segmented VOI [26].
- First-order statistical features or histogram-based features describe how the intensity signals of pixels/voxels are distributed over the segmented ROI/VOI. These features neglect the spatial orientation and spatial relationship between pixels/voxels [23].
- Second-order statistical features or textural features are statistical relationships between the signal intensity of adjacent pixels/voxels or groups of pixels or voxels. These features serve to quantify intratumoral heterogeneity. Textural features are created by numerically characterizing matrices that encode the exact spatial connections between the pixels/voxels in the source image. The gray-level co-occurrence matrix (GLCM) [31] is the most widely used texture analysis matrix. The GLCM shows how many times two intensity levels appear in adjacent pixels or voxels within a given distance and in a defined direction. Multiple textural characteristics, including energy, contrast, correlation, variance, homogeneity, cluster prominence, dissimilarity, cluster inclination, and maximum likelihood, can be measured using the GLCM. The difference in intensity levels between one pixel/voxel and its 26-pixel 3D neighborhood is represented by the neighborhood gray-level different matrix (NGLDM). For each image intensity, the gray-level run length matrix (GLRLM) encodes the size of homogeneous runs [32]. Long-run emphasis (LRE), short-run emphasis (SRE), low gray-level run emphasis (LGRE), run percentage (RP), and high gray-level run emphasis (HGRE) can all be derived from the GLRLM. There are other matrices that capture pixel-wise spatial relationships and can be used to compute additional texture-based features [31].
- Higher-order statistical features are quantified using statistical methods after applying complex mathematical transformations (filters), such as for pattern recognition, noise reduction, local binary patterns (LBP), histogram-oriented gradients, or edge enhancement. Minkowski functionals, fractal analysis, wavelet or Fourier transforms, and Laplacian transforms of Gaussian-filtered images (Laplacian-of-Gaussian) are examples of these mathematical transformations or filters [28].
- Filter methods (univariate methods) examine how features and labels are related without taking into account their redundancy, or correlation. Minimum redundancy maximum significance, Student’s t-test, Chi-squared score, Fisher score, and the Wilcoxon rank sum test are among the most commonly used filter methods. While these feature selection methods are widely used, they do not take the associations and interactions between features into account [33,34].
- Wrapper methods (multivariate methods), known as greedy algorithms, avoid the filter method constraint by looking at the entire space of features and considering the relationships between each feature and other features in the dataset. A predictive model is used to evaluate the output of a group of features. The consistency of a given technique’s output is used to test each new subset of features. Wrapper approaches are computationally intensive, as they strive to find the best-performing functional group of features. Forward feature selection, backward feature exclusion, exhaustive feature selection, and bidirectional search are all examples of wrapper methods [33,34].
- Embedded approaches carry out the feature selection process as part of the ML model’s development; in other words, the best group of features is chosen in the model’s training phase. In this way, embedded approaches incorporate the benefits of both the filter and wrapper methods. Embedded approaches provide more reliability than filter methods, have a lower execution time than wrapper methods, and are not very susceptibility to data overfitting, as they take into account the interactions between features. The least absolute shrinkage and selection operator (LASSO), tree-based algorithms such as the random forest classifier, and ridge regression are examples of commonly used embedded methods [33,34].
2.2.2. DL-Based Radiomics
2.3. Evaluation Metrics in AI and Radiomics Techniques
- Total Score: evaluation of different segmentation measures, namely, the overlap error, the relative absolute volume difference, and the surface distance (in terms of mean, RMS, and maximum).
- Dice similarity coefficient (Dice score) =
- Accuracy =
- Sensitivity =
- Specificity =
- Area under the curve (AUC): The area under the receiver operating characteristics (ROC) curve, which connects the true positive rate (sensitivity) and the false positive rate (1-specificity); the AUC value ranges from 0 to 1, with 1 being the best performance.
2.4. Clinical Application of AI and Radiomic Techniques in Liver Cancer
3. Segmentation of Hepatic Focal Lesions
4. Detection and Differentiation of HCC from Other Hepatic Masses
5. Managment of HCC
5.1. Prediction of HCC Histopathology
5.1.1. Microvascular Invasion
5.1.2. HCC Grade and Molecular Signature
5.2. Monitoring of Locoregional Therapeutic Response
5.2.1. Prediction of Response to Ablation Therapy (MWA & RFA)
5.2.2. Prediction of Response to TACE
5.3. Prediction of Response to Hepatic Resection
5.4. Prediction of Response to Systemic Treatment and/or Immunotherapy
5.5. Prediction of the Survival of HCC Patients
6. Limitations and Future Perspectives
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ADC | apparent diffusion coefficient |
| AFP | alpha fetoprotein |
| AI | artificial intelligence |
| ANN | artificial neural network |
| AUC | area under the curve |
| CAD | computer-aided diagnosis |
| CC | cholangiocarcinoma |
| CDN | convolutional dense network |
| CNN | convolutional neural network |
| CRF | conditional random fields |
| CT | computed tomography |
| DEB-TACE | drug-eluting bead trans-arterial chemoembolization |
| DFS | disease-free survival |
| DIR | deformable image registration |
| DL | deep learning |
| DPHCC | dual-phenotype HCC |
| DWI | diffusion-weighted images |
| FNH | focal nodular hyperplasia |
| FNN | Focal neural network |
| GBDT | gradient boosting decision tree |
| Gd-EOB-DTPA | gadolinium-ethoxybenzyl-diethylenetriamine pentaacetic acid |
| GLCM | gray-level co-occurrence matrix |
| HA | hepatocellular adenoma |
| HCC | hepatocellular carcinoma |
| HGRE | high gray-level run emphasis |
| HH | hepatic hemangioma |
| ICI | immune-checkpoint inhibitors |
| KNN | K-nearest neighbor |
| LASSO | logistic regression with the least absolute shrinkage and selection operator |
| LBP | local binary pattern |
| LCT | locoregional therapy |
| LGRE | low gray-level run emphasis |
| LiTS | liver tumor segmentation |
| LR | logistic regression |
| LRE | long-run emphasis |
| LRT | locoregional therapy |
| LTP | local tumor progression |
| ML | machine learning |
| MLP | multi-layer perception |
| MRI | magnetic resonance imaging |
| mRMR | minimum redundancy maximum relevance |
| MVI | microvascular invasion |
| MWA | microwave ablation |
| NGLDM | neighborhood gray-level different matrix |
| OS | overall survival |
| PACS | picture archives and communication systems |
| PCA | principal component analysis |
| PEI | percutaneous ethanol injection |
| PFS | progression free survival |
| RFA | radiofrequency ablation |
| ROC | receiver operating characteristics |
| ROI | region of interest |
| RP | run percentage |
| RRC | CT radiomics, radiological data, clinical data |
| RSF | random survival forest |
| SBRT | stereotactic body radiotherapy |
| SRE | Short-run emphasis |
| SVM | support vector machine |
| SWI | susceptibility weighted images |
| TACE | trans-arteria chemoembolization |
| TARE | trans-arterial radio-embolization |
| VOI | volume of interest |
| XG Boost | extreme gradient boost |
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Global Hepatitis Report 2017; World Health Organization: Geneva, Switzerland, 2017. [Google Scholar]
- Lewis, S.; Hectors, S.; Taouli, B. Radiomics of hepatocellular carcinoma. Abdom. Radiol. 2021, 46, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Gillies, R.J.; Kinahan, P.E.; Hricak, H. Radiomics: Images are more than pictures, they are data. Radiology 2016, 278, 563–577. [Google Scholar] [CrossRef] [PubMed]
- Russell, S.; Norvig, P. Artificial Intelligence: A Modern Approach; Prentice Hall Press: Upper Saddle River, NJ, USA, 2003. [Google Scholar]
- Obermeyer, Z.; Emanuel, E.J. Predicting the future—Big data, machine learning, and clinical medicine. N. Engl. J. Med. 2016, 375, 1216. [Google Scholar] [CrossRef] [PubMed]
- Castellino, R.A. Computer aided detection (CAD): An overview. Cancer Imaging 2005, 5, 17. [Google Scholar] [CrossRef]
- Rosenblatt, F. The perceptron: A probabilistic model for information storage and organization in the brain. Psychol. Rev. 1958, 65, 386. [Google Scholar] [CrossRef]
- SM, C.M. Artificial intelligence in radiology—Are we treating the image or the patient? Indian J. Radiol. Imaging 2018, 28, 137–139. [Google Scholar] [CrossRef]
- Bishop, C.M.; Nasrabadi, N.M. Pattern Recognition and Machine Learning; Springer: New York, NY, USA, 2006; Volume 4. [Google Scholar]
- Alpaydin, E. Introduction to Machine Learning; MIT Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Shen, D.; Wu, G.; Suk, H.I. Deep learning in medical image analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221. [Google Scholar] [CrossRef]
- Saba, L.; Dey, N.; Ashour, A.S.; Samanta, S.; Nath, S.S.; Chakraborty, S.; Sanches, J.; Kumar, D.; Marinho, R.; Suri, J.S. Automated stratification of liver disease in ultrasound: An online accurate feature classification paradigm. Comput. Methods Programs Biomed. 2016, 130, 118–134. [Google Scholar] [CrossRef]
- Saba, L.; Sanfilippo, R.; Tallapally, N.; Molinari, F.; Montisci, R.; Mallarini, G.; Suri, J.S. Evaluation of carotid wall thickness by using computed tomography and semiautomated ultrasonographic software. J. Vasc. Ultrasound 2011, 35, 136–142. [Google Scholar] [CrossRef]
- Dey, D.; Gaur, S.; Ovrehus, K.A.; Slomka, P.J.; Betancur, J.; Goeller, M.; Hell, M.M.; Gransar, H.; Berman, D.S.; Achenbach, S.; et al. Integrated prediction of lesion-specific ischaemia from quantitative coronary CT angiography using machine learning: A multicentre study. Eur. Radiol. 2018, 28, 2655–2664. [Google Scholar] [CrossRef] [PubMed]
- Kooi, T.; Litjens, G.; Van Ginneken, B.; Gubern-Mérida, A.; Sánchez, C.I.; Mann, R.; den Heeten, A.; Karssemeijer, N. Large scale deep learning for computer aided detection of mammographic lesions. Med. Image Anal. 2017, 35, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Handelman, G.; Kok, H.; Chandra, R.; Razavi, A.; Lee, M.; Asadi, H. eD octor: Machine learning and the future of medicine. J. Intern. Med. 2018, 284, 603–619. [Google Scholar] [CrossRef] [PubMed]
- Aerts, H.J. The potential of radiomic-based phenotyping in precision medicine: A review. JAMA Oncol. 2016, 2, 1636–1642. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Gu, Y.; Basu, S.; Berglund, A.; Eschrich, S.A.; Schabath, M.B.; Forster, K.; Aerts, H.J.; Dekker, A.; Fenstermacher, D.; et al. Radiomics: The process and the challenges. Magn. Reson. Imaging 2012, 30, 1234–1248. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; Van Stiphout, R.G.; Granton, P.; Zegers, C.M.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef]
- Abdel Razek, A.A.K.; Alksas, A.; Shehata, M.; AbdelKhalek, A.; Abdel Baky, K.; El-Baz, A.; Helmy, E. Clinical applications of artificial intelligence and radiomics in neuro-oncology imaging. Insights Imaging 2021, 12, 1–17. [Google Scholar] [CrossRef]
- Mazurowski, M.A. Radiogenomics: What it is and why it is important. J. Am. Coll. Radiol. 2015, 12, 862–866. [Google Scholar] [CrossRef]
- Lambin, P.; Leijenaar, R.T.; Deist, T.M.; Peerlings, J.; De Jong, E.E.; Van Timmeren, J.; Sanduleanu, S.; Larue, R.T.; Even, A.J.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Aerts, H.J.; Velazquez, E.R.; Leijenaar, R.T.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 1–9. [Google Scholar] [CrossRef]
- Rizzo, S.; Botta, F.; Raimondi, S.; Origgi, D.; Fanciullo, C.; Morganti, A.G.; Bellomi, M. Radiomics: The facts and the challenges of image analysis. Eur. Radiol. Exp. 2018, 2, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Alksas, A.; Shehata, M.; Saleh, G.A.; Shaffie, A.; Soliman, A.; Ghazal, M.; Khelifi, A.; Khalifeh, H.A.; Razek, A.A.; Giridharan, G.A.; et al. A novel computer-aided diagnostic system for accurate detection and grading of liver tumors. Sci. Rep. 2021, 11, 13148. [Google Scholar] [CrossRef] [PubMed]
- Yip, S.S.; Aerts, H.J. Applications and limitations of radiomics. Phys. Med. Biol. 2016, 61, R150. [Google Scholar] [CrossRef]
- Wu, J.; Liu, A.; Cui, J.; Chen, A.; Song, Q.; Xie, L. Radiomics-based classification of hepatocellular carcinoma and hepatic haemangioma on precontrast magnetic resonance images. BMC Med. Imaging 2019, 19, 23. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.W.; Lai, Y.H. Effective segmentation and classification for HCC biopsy images. Pattern Recognit. 2010, 43, 1550–1563. [Google Scholar] [CrossRef]
- Kim, D.W.; Lee, G.; Kim, S.Y.; Ahn, G.; Lee, J.G.; Lee, S.S.; Kim, K.W.; Park, S.H.; Lee, Y.J.; Kim, N. Deep learning–based algorithm to detect primary hepatic malignancy in multiphase CT of patients at high risk for HCC. Eur. Radiol. 2021, 31, 7047–7057. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I.H. Textural features for image classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Xu, D.H.; Kurani, A.S.; Furst, J.D.; Raicu, D.S. Run-length encoding for volumetric texture. Heart 2004, 27, 452–458. [Google Scholar]
- Parekh, V.; Jacobs, M.A. Radiomics: A new application from established techniques. Expert Rev. Precis. Med. Drug Dev. 2016, 1, 207–226. [Google Scholar] [CrossRef]
- Kuhn, M.; Johnson, K. Applied Predictive Modeling; Springer: New York, NY, USA, 2013; Volume 26. [Google Scholar]
- Yasaka, K.; Akai, H.; Abe, O.; Kiryu, S. Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast-enhanced CT: A preliminary study. Radiology 2018, 286, 887–896. [Google Scholar] [CrossRef]
- Tan, C.; Sun, F.; Kong, T.; Zhang, W.; Yang, C.; Liu, C. A survey on deep transfer learning. In Proceedings of the International Conference on Artificial Neural Networks, Rhodes, Greece, 4–7 October 2018; Springer: New York, NY, USA, 2018; pp. 270–279. [Google Scholar]
- Oliver 3rd, J.; Baron, R.L. Helical biphasic contrast-enhanced CT of the liver: Technique, indications, interpretation, and pitfalls. Radiology 1996, 201, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Häme, Y. Liver tumor segmentation using implicit surface evolution. The Midas J. 2008, 1–10. [Google Scholar] [CrossRef]
- Smeets, D.; Stijnen, B.; Loeckx, D.; De Dobbelaer, B.; Suetens, P. Segmentation of liver metastases using a level set method with spiral-scanning technique and supervised fuzzy pixel classification. In Proceedings of the MICCAI Workshop, New York, NY, USA, 6–10 September 2008; Volume 42, p. 43. [Google Scholar]
- Choudhary, A.; Moretto, N.; Ferrarese, F.P.; Zamboni, G.A. An entropy based multi-thresholding method for semi-automatic segmentation of liver tumors. In Proceedings of the MICCAI Workshop, New York, NY, USA, 6–10 September 2008; Volume 41, pp. 43–49. [Google Scholar]
- Moltz, J.H.; Bornemann, L.; Dicken, V.; Peitgen, H. Segmentation of liver metastases in CT scans by adaptive thresholding and morphological processing. In Proceedings of the MICCAI Workshop, New York, NY, USA, 6–10 September 2008; Volume 41, p. 195. [Google Scholar]
- Kadoury, S.; Vorontsov, E.; Tang, A. Metastatic liver tumour segmentation from discriminant Grassmannian manifolds. Phys. Med. Biol. 2015, 60, 6459. [Google Scholar] [CrossRef] [PubMed]
- Linguraru, M.G.; Richbourg, W.J.; Liu, J.; Watt, J.M.; Pamulapati, V.; Wang, S.; Summers, R.M. Tumor burden analysis on computed tomography by automated liver and tumor segmentation. IEEE Trans. Med Imaging 2012, 31, 1965–1976. [Google Scholar] [CrossRef] [PubMed]
- Christ, P.F.; Ettlinger, F.; Grün, F.; Elshaera, M.E.A.; Lipkova, J.; Schlecht, S.; Ahmaddy, F.; Tatavarty, S.; Bickel, M.; Bilic, P.; et al. Automatic liver and tumor segmentation of CT and MRI volumes using cascaded fully convolutional neural networks. arXiv 2017, arXiv:1702.05970. [Google Scholar]
- Han, X. Automatic liver lesion segmentation using a deep convolutional neural network method. arXiv 2017, arXiv:1704.07239. [Google Scholar]
- Vorontsov, E.; Tang, A.; Pal, C.; Kadoury, S. Liver lesion segmentation informed by joint liver segmentation. In Proceedings of the 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018); Washington, DC, USA, 4–7 April 2018, IEEE: Hoboken, NJ, USA, 2018; pp. 1332–1335. [Google Scholar]
- Li, X.; Chen, H.; Qi, X.; Dou, Q.; Fu, C.W.; Heng, P.A. H-DenseUNet: Hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans. Med Imaging 2018, 37, 2663–2674. [Google Scholar] [CrossRef]
- Chlebus, G.; Schenk, A.; Moltz, J.H.; van Ginneken, B.; Hahn, H.K.; Meine, H. Automatic liver tumor segmentation in CT with fully convolutional neural networks and object-based postprocessing. Sci. Rep. 2018, 8, 15497. [Google Scholar] [CrossRef]
- Meng, L.; Tian, Y.; Bu, S. Liver tumor segmentation based on 3D convolutional neural network with dual scale. J. Appl. Clin. Med. Phys. 2020, 21, 144–157. [Google Scholar] [CrossRef]
- Lubner, M.G.; Smith, A.D.; Sandrasegaran, K.; Sahani, D.V.; Pickhardt, P.J. CT texture analysis: Definitions, applications, biologic correlates, and challenges. Radiographics 2017, 37, 1483–1503. [Google Scholar] [CrossRef]
- Wakabayashi, T.; Ouhmich, F.; Gonzalez-Cabrera, C.; Felli, E.; Saviano, A.; Agnus, V.; Savadjiev, P.; Baumert, T.F.; Pessaux, P.; Marescaux, J.; et al. Radiomics in hepatocellular carcinoma: A quantitative review. Hepatol. Int. 2019, 13, 546–559. [Google Scholar] [CrossRef] [PubMed]
- Miranda Magalhaes Santos, J.M.; Clemente Oliveira, B.; Araujo-Filho, J.d.A.B.; Assuncao, A.N., Jr.; de M. Machado, F.A.; Carlos Tavares Rocha, C.; Horvat, J.V.; Menezes, M.R.; Horvat, N. State-of-the-art in radiomics of hepatocellular carcinoma: A review of basic principles, applications, and limitations. Abdom. Radiol. 2020, 45, 342–353. [Google Scholar] [CrossRef] [PubMed]
- Alksas, A.; Shehata, M.; Saleh, G.A.; Shaffie, A.; Soliman, A.; Ghazal, M.; Khalifeh, H.A.; Razek, A.A.; El-Baz, A. A novel computer-aided diagnostic system for early assessment of hepatocellular carcinoma. In Proceedings of the 2020 25th International Conference on Pattern Recognition (ICPR), Milan, Italy, 10–15 January 2021; IEEE: Hoboken, NJ, USA, 2021; pp. 10375–10382. [Google Scholar]
- Nie, P.; Yang, G.; Guo, J.; Chen, J.; Li, X.; Ji, Q.; Wu, J.; Cui, J.; Xu, W. A CT-based radiomics nomogram for differentiation of focal nodular hyperplasia from hepatocellular carcinoma in the non-cirrhotic liver. Cancer Imaging 2020, 20, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Nie, P.; Wang, N.; Pang, J.; Yang, G.; Duan, S.; Chen, J.; Xu, W. CT-based radiomics nomogram: A potential tool for differentiating hepatocellular adenoma from hepatocellular carcinoma in the noncirrhotic liver. Acad. Radiol. 2021, 28, 799–807. [Google Scholar] [CrossRef]
- Mokrane, F.Z.; Lu, L.; Vavasseur, A.; Otal, P.; Peron, J.M.; Luk, L.; Yang, H.; Ammari, S.; Saenger, Y.; Rousseau, H.; et al. Radiomics machine-learning signature for diagnosis of hepatocellular carcinoma in cirrhotic patients with indeterminate liver nodules. Eur. Radiol. 2020, 30, 558–570. [Google Scholar] [CrossRef]
- Ponnoprat, D.; Inkeaw, P.; Chaijaruwanich, J.; Traisathit, P.; Sripan, P.; Inmutto, N.; Na Chiangmai, W.; Pongnikorn, D.; Chitapanarux, I. Classification of hepatocellular carcinoma and intrahepatic cholangiocarcinoma based on multi-phase CT scans. Med. Biol. Eng. Comput. 2020, 58, 2497–2515. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Khalvati, F.; Namdar, K.; Fischer, S.; Lewis, S.; Taouli, B.; Haider, M.A.; Jhaveri, K.S. Can machine learning radiomics provide pre-operative differentiation of combined hepatocellular cholangiocarcinoma from hepatocellular carcinoma and cholangiocarcinoma to inform optimal treatment planning? Eur. Radiol. 2021, 31, 244–255. [Google Scholar] [CrossRef]
- Shi, W.; Kuang, S.; Cao, S.; Hu, B.; Xie, S.; Chen, S.; Chen, Y.; Gao, D.; Chen, Y.; Zhu, Y.; et al. Deep learning assisted differentiation of hepatocellular carcinoma from focal liver lesions: Choice of four-phase and three-phase CT imaging protocol. Abdom. Radiol. 2020, 45, 2688–2697. [Google Scholar] [CrossRef]
- Cao, S.E.; Zhang, L.Q.; Kuang, S.C.; Shi, W.Q.; Hu, B.; Xie, S.D.; Chen, Y.N.; Liu, H.; Chen, S.M.; Jiang, T.; et al. Multiphase convolutional dense network for the classification of focal liver lesions on dynamic contrast-enhanced computed tomography. World J. Gastroenterol. 2020, 26, 3660. [Google Scholar] [CrossRef]
- Hamm, C.A.; Wang, C.J.; Savic, L.J.; Ferrante, M.; Schobert, I.; Schlachter, T.; Lin, M.; Duncan, J.S.; Weinreb, J.C.; Chapiro, J.; et al. Deep learning for liver tumor diagnosis part I: Development of a convolutional neural network classifier for multi-phasic MRI. Eur. Radiol. 2019, 29, 3338–3347. [Google Scholar] [CrossRef]
- Wang, C.J.; Hamm, C.A.; Savic, L.J.; Ferrante, M.; Schobert, I.; Schlachter, T.; Lin, M.; Weinreb, J.C.; Duncan, J.S.; Chapiro, J.; et al. Deep learning for liver tumor diagnosis part II: Convolutional neural network interpretation using radiologic imaging features. Eur. Radiol. 2019, 29, 3348–3357. [Google Scholar] [CrossRef] [PubMed]
- Zhen, S.H.; Cheng, M.; Tao, Y.B.; Wang, Y.F.; Juengpanich, S.; Jiang, Z.Y.; Jiang, Y.K.; Yan, Y.Y.; Lu, W.; Lue, J.M.; et al. Deep learning for accurate diagnosis of liver tumor based on magnetic resonance imaging and clinical data. Front. Oncol. 2020, 10, 680. [Google Scholar] [CrossRef]
- Jian, W.; Ju, H.; Cen, X.; Cui, M.; Zhang, H.; Zhang, L.; Wang, G.; Gu, L.; Zhou, W. Improving the malignancy characterization of hepatocellular carcinoma using deeply supervised cross modal transfer learning for non-enhanced MR. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; IEEE: Hoboken, NJ, USA, 2019; pp. 853–856. [Google Scholar]
- Sun, K.; Shi, L.; Qiu, J.; Pan, Y.; Wang, X.; Wang, H. Multi-phase contrast-enhanced magnetic resonance image-based radiomics-combined machine learning reveals microscopic ultra-early hepatocellular carcinoma lesions. Eur. J. Nucl. Med. Mol. Imaging 2022, 49, 2917–2928. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Peralvarez, M.; Luong, T.V.; Andreana, L.; Meyer, T.; Dhillon, A.P.; Burroughs, A.K. A systematic review of microvascular invasion in hepatocellular carcinoma: Diagnostic and prognostic variability. Ann. Surg. Oncol. 2013, 20, 325–339. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, H.L.; Liu, Q.P.; Sun, S.W.; Zhang, J.; Zhu, F.P.; Yang, G.; Yan, X.; Zhang, Y.D.; Liu, X.S. Radiomic analysis of contrast-enhanced CT predicts microvascular invasion and outcome in hepatocellular carcinoma. J. Hepatol. 2019, 70, 1133–1144. [Google Scholar] [CrossRef]
- Bakr, S.H.; Echegaray, S.; Shah, R.P.; Kamaya, A.; Louie, J.; Napel, S.; Kothary, N.; Gevaert, O. Noninvasive radiomics signature based on quantitative analysis of computed tomography images as a surrogate for microvascular invasion in hepatocellular carcinoma: A pilot study. J. Med. Imaging 2017, 4, 041303. [Google Scholar] [CrossRef]
- Peng, J.; Zhang, J.; Zhang, Q.; Xu, Y.; Zhou, J.; Liu, L. A radiomics nomogram for preoperative prediction of microvascular invasion risk in hepatitis B virus-related hepatocellular carcinoma. Diagn. Interv. Radiol. 2018, 24, 121. [Google Scholar] [CrossRef]
- Zheng, J.; Chakraborty, J.; Chapman, W.C.; Gerst, S.; Gonen, M.; Pak, L.M.; Jarnagin, W.R.; DeMatteo, R.P.; Do, R.K.; Simpson, A.L.; et al. Preoperative prediction of microvascular invasion in hepatocellular carcinoma using quantitative image analysis. J. Am. Coll. Surg. 2017, 225, 778–788. [Google Scholar] [CrossRef]
- Cucchetti, A.; Piscaglia, F.; Grigioni, A.D.; Ravaioli, M.; Cescon, M.; Zanello, M.; Grazi, G.L.; Golfieri, R.; Grigioni, W.F.; Pinna, A.D. Preoperative prediction of hepatocellular carcinoma tumour grade and micro-vascular invasion by means of artificial neural network: A pilot study. J. Hepatol. 2010, 52, 880–888. [Google Scholar] [CrossRef]
- Jiang, Y.Q.; Cao, S.E.; Cao, S.; Chen, J.N.; Wang, G.Y.; Shi, W.Q.; Deng, Y.N.; Cheng, N.; Ma, K.; Zeng, K.N.; et al. Preoperative identification of microvascular invasion in hepatocellular carcinoma by XGBoost and deep learning. J. Cancer Res. Clin. Oncol. 2021, 147, 821–833. [Google Scholar] [CrossRef]
- Ni, M.; Zhou, X.; Lv, Q.; Li, Z.; Gao, Y.; Tan, Y.; Liu, J.; Liu, F.; Yu, H.; Jiao, L.; et al. Radiomics models for diagnosing microvascular invasion in hepatocellular carcinoma: Which model is the best model? Cancer Imaging 2019, 19, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Huang, S.; Xu, Y.; Wu, J. Diagnostic Accuracy of Artificial Intelligence Based on Imaging Data for Preoperative Prediction of Microvascular Invasion in Hepatocellular Carcinoma: A Systematic Review and Meta-Analysis. Front. Oncol. 2022, 12, 763842. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Jian, W.; Cen, X.; Zhang, L.; Guo, H.; Liu, Z.; Liang, C.; Wang, G. Prediction of microvascular invasion of hepatocellular carcinoma based on contrast-enhanced MR and 3D convolutional neural networks. Front. Oncol. 2021, 11, 588010. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Sun, Y.; Zhou, X.; Shen, Z.; Zhang, H.; Xing, J.; Zhou, Y. Histogram peritumoral enhanced features on MRI arterial phase with extracellular contrast agent can improve prediction of microvascular invasion of hepatocellular carcinoma. Quant. Imaging Med. Surg. 2022, 12, 1372–1384. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Lu, M.; Huang, B.; Tang, M.; Pang, T.; Liao, B.; Cai, H.; Huang, M.; Zhou, Y.; Chen, X.; et al. Considerable effects of imaging sequences, feature extraction, feature selection, and classifiers on radiomics-based prediction of microvascular invasion in hepatocellular carcinoma using magnetic resonance imaging. Quant. Imaging Med. Surg. 2021, 11, 1836. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Jian, W.; Cen, X.; Zhang, L.; Guo, H.; Liu, Z.; Liang, C.; Zhou, W. Prediction of microvascular invasion of hepatocellular carcinoma based on preoperative diffusion-weighted MR using deep learning. Acad. Radiol. 2021, 28, S118–S127. [Google Scholar] [CrossRef]
- Meng, X.P.; Wang, Y.C.; Zhou, J.Y.; Yu, Q.; Lu, C.Q.; Xia, C.; Tang, T.Y.; Xu, J.; Sun, K.; Xiao, W.; et al. Comparison of MRI and CT for the Prediction of Microvascular Invasion in Solitary Hepatocellular Carcinoma Based on a Non-Radiomics and Radiomics Method: Which Imaging Modality Is Better? J. Magn. Reson. Imaging 2021, 54, 526–536. [Google Scholar] [CrossRef]
- Okusaka, T.; Okada, S.; Ueno, H.; Ikeda, M.; Shimada, K.; Yamamoto, J.; Kosuge, T.; Yamasaki, S.; Fukushima, N.; Sakamoto, M. Satellite lesions in patients with small hepatocellular carcinoma with reference to clinicopathologic features. Cancer Interdiscip. Int. J. Am. Cancer Soc. 2002, 95, 1931–1937. [Google Scholar] [CrossRef]
- Bruix, J.; Sherman, M. Management of hepatocellular carcinoma: An update. Hepatology 2011, 53, 1020. [Google Scholar] [CrossRef]
- Gong, X.Q.; Tao, Y.Y.; Wu, Y.K.; Liu, N.; Yu, X.; Wang, R.; Zheng, J.; Liu, N.; Huang, X.H.; Li, J.D.; et al. Progress of MRI radiomics in hepatocellular carcinoma. Front. Oncol. 2021, 11, 698373. [Google Scholar] [CrossRef]
- Yao, S.; Ye, Z.; Wei, Y.; Jiang, H.Y.; Song, B. Radiomics in hepatocellular carcinoma: A state-of-the-art review. World J. Gastrointest. Oncol. 2021, 13, 1599. [Google Scholar] [CrossRef] [PubMed]
- Mao, B.; Zhang, L.; Ning, P.; Ding, F.; Wu, F.; Lu, G.; Geng, Y.; Ma, J. Preoperative prediction for pathological grade of hepatocellular carcinoma via machine learning–based radiomics. Eur. Radiol. 2020, 30, 6924–6932. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.; Tan, H.; Gao, F.; Hai, J.; Ning, P.; Chen, J.; Zhu, S.; Wang, M.; Dou, S.; Shi, D. Predicting the grade of hepatocellular carcinoma based on non-contrast-enhanced MRI radiomics signature. Eur. Radiol. 2019, 29, 2802–2811. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Wang, J.; Zhu, Y.; Chen, J.; Mao, L.; Kong, W.; Qiu, Y.; Wu, X.; Guan, Y.; He, J. Gd-EOB-DTPA-enhanced MRI radiomic features for predicting histological grade of hepatocellular carcinoma. Hepatobiliary Surg. Nutr. 2022, 11, 13. [Google Scholar] [CrossRef]
- Zhou, W.; Wang, G.; Xie, G.; Zhang, L. Grading of hepatocellular carcinoma based on diffusion weighted images with multiple b-values using convolutional neural networks. Med. Phys. 2019, 46, 3951–3960. [Google Scholar] [CrossRef]
- Lee, J.I.; Lee, J.W.; Kim, J.M.; Kim, J.K.; Chung, H.J.; Kim, Y.S. Prognosis of hepatocellular carcinoma expressing cytokeratin 19: Comparison with other liver cancers. World J. Gastroenterol. WJG 2012, 18, 4751. [Google Scholar] [CrossRef]
- Lee, C.W.; Kuo, W.L.; Yu, M.C.; Chen, T.C.; Tsai, C.N.; Lee, W.C.; Chen, M.F. The expression of cytokeratin 19 in lymph nodes was a poor prognostic factor for hepatocellular carcinoma after hepatic resection. World J. Surg. Oncol. 2013, 11, 1–11. [Google Scholar] [CrossRef]
- Huang, X.; Long, L.; Wei, J.; Li, Y.; Xia, Y.; Zuo, P.; Chai, X. Radiomics for diagnosis of dual-phenotype hepatocellular carcinoma using Gd-EOB-DTPA-enhanced MRI and patient prognosis. J. Cancer Res. Clin. Oncol. 2019, 145, 2995–3003. [Google Scholar] [CrossRef]
- Geng, Z.; Zhang, Y.; Wang, S.; Li, H.; Zhang, C.; Yin, S.; Xie, C.; Dai, Y. Radiomics analysis of susceptibility weighted imaging for hepatocellular carcinoma: Exploring the correlation between histopathology and radiomics features. Magn. Reson. Med. Sci. 2021, 20, 253. [Google Scholar] [CrossRef]
- Yang, F.; Xu, L.; Wan, Y.; Wu, Y.; Wang, J.; Shen, X.; Shao, C.; Zheng, S.; Niu, T.; Xu, X. MRI-Radiomics Prediction for Cytokeratin 19 Positive Hepatocellular Carcinoma, a Multi-Center Based Study. Front. Oncol. 2021, 11, 672126. [Google Scholar] [CrossRef]
- Wang, W.; Gu, D.; Wei, J.; Ding, Y.; Yang, L.; Zhu, K.; Luo, R.; Rao, S.X.; Tian, J.; Zeng, M. A radiomics-based biomarker for cytokeratin 19 status of hepatocellular carcinoma with gadoxetic acid–enhanced MRI. Eur. Radiol. 2020, 30, 3004–3014. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Yu, Y.; Wang, X.; Hu, M.; Hu, C. Radiomic analysis of Gd-EOB-DTPA-enhanced MRI predicts Ki-67 expression in hepatocellular carcinoma. BMC Med. Imaging 2021, 21, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Kampalath, R.; Tran-Harding, K.; Do, R.K.; Mendiratta-Lala, M.; Yaghmai, V. Evaluation of Hepatocellular Carcinoma Treatment Response After Locoregional Therapy. Magn. Reson. Imaging Clin. 2021, 29, 389–403. [Google Scholar] [CrossRef] [PubMed]
- Spieler, B.; Sabottke, C.; Moawad, A.W.; Gabr, A.M.; Bashir, M.R.; Do, R.K.G.; Yaghmai, V.; Rozenberg, R.; Gerena, M.; Yacoub, J.; et al. Artificial intelligence in assessment of hepatocellular carcinoma treatment response. Abdom. Radiol. 2021, 46, 3660–3671. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, S.M.; Nikolaidis, P.; Miller, F.H.; Lewandowski, R.J.; Ryu, R.K.; Sato, K.T.; Senthilnathan, S.; Riaz, A.; Kulik, L.; Mulcahy, M.F.; et al. Radiologic findings following Y90 radioembolization for primary liver malignancies. Abdom. Imaging 2009, 34, 566–581. [Google Scholar] [CrossRef]
- Joo, I.; Kim, H.C.; Kim, G.M.; Paeng, J.C. Imaging evaluation following 90Y radioembolization of liver tumors: What radiologists should know. Korean J. Radiol. 2018, 19, 209–222. [Google Scholar] [CrossRef]
- Riaz, A.; Kulik, L.; Lewandowski, R.J.; Ryu, R.K.; Giakoumis Spear, G.; Mulcahy, M.F.; Abecassis, M.; Baker, T.; Gates, V.; Nayar, R.; et al. Radiologic–pathologic correlation of hepatocellular carcinoma treated with internal radiation using yttrium-90 microspheres. Hepatology 2009, 49, 1185–1193. [Google Scholar] [CrossRef]
- Yip, C.; Hennedige, T.P.; Cook, G.J.; Goh, V. Imaging assessment after SBRT for hepatocellular carcinoma. Hepatoma Res. 2020, 6, 44. [Google Scholar] [CrossRef]
- An, C.; Jiang, Y.; Huang, Z.; Gu, Y.; Zhang, T.; Ma, L.; Huang, J. Assessment of Ablative Margin after Microwave Ablation for Hepatocellular Carcinoma Using Deep Learning-Based Deformable Image Registration. Front. Oncol. 2020, 10, 573316. [Google Scholar] [CrossRef]
- Hu, C.; Song, Y.; Zhang, J.; Dai, L.; Tang, C.; Li, M.; Liao, W.; Zhou, Y.; Xu, Y.; Zhang, Y.Y.; et al. Preoperative Gadoxetic Acid-Enhanced MRI Based Nomogram Improves Prediction of Early HCC Recurrence After Ablation Therapy. Front. Oncol. 2021, 11, 649682. [Google Scholar] [CrossRef]
- Liang, J.D.; Ping, X.O.; Tseng, Y.J.; Huang, G.T.; Lai, F.; Yang, P.M. Recurrence predictive models for patients with hepatocellular carcinoma after radiofrequency ablation using support vector machines with feature selection methods. Comput. Methods Programs Biomed. 2014, 117, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Liu, D.; Wang, K.; Xie, X.; Su, L.; Kuang, M.; Huang, G.; Peng, B.; Wang, Y.; Lin, M.; et al. Deep learning radiomics based on contrast-enhanced ultrasound might optimize curative treatments for very-early or early-stage hepatocellular carcinoma patients. Liver Cancer 2020, 9, 397–413. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Choi, S.J.; Lee, S.H.; Lee, H.Y.; Park, H. Predicting survival using pretreatment CT for patients with hepatocellular carcinoma treated with transarterial chemoembolization: Comparison of models using radiomics. Am. J. Roentgenol. 2018, 211, 1026–1034. [Google Scholar] [CrossRef] [PubMed]
- Morshid, A.; Elsayes, K.M.; Khalaf, A.M.; Elmohr, M.M.; Yu, J.; Kaseb, A.O.; Hassan, M.; Mahvash, A.; Wang, Z.; Hazle, J.D.; et al. A machine learning model to predict hepatocellular carcinoma response to transcatheter arterial chemoembolization. Radiol. Artif. Intell. 2019, 1, e180021. [Google Scholar] [CrossRef]
- Meng, X.P.; Wang, Y.C.; Ju, S.; Lu, C.Q.; Zhong, B.Y.; Ni, C.F.; Zhang, Q.; Yu, Q.; Xu, J.; Ji, J.; et al. Radiomics analysis on multiphase contrast-enhanced CT: A survival prediction tool in patients with hepatocellular carcinoma undergoing transarterial chemoembolization. Front. Oncol. 2020, 10, 1196. [Google Scholar] [CrossRef]
- Peng, J.; Kang, S.; Ning, Z.; Deng, H.; Shen, J.; Xu, Y.; Zhang, J.; Zhao, W.; Li, X.; Gong, W.; et al. Residual convolutional neural network for predicting response of transarterial chemoembolization in hepatocellular carcinoma from CT imaging. Eur. Radiol. 2020, 30, 413–424. [Google Scholar] [CrossRef]
- Liu, Q.P.; Xu, X.; Zhu, F.P.; Zhang, Y.D.; Liu, X.S. Prediction of prognostic risk factors in hepatocellular carcinoma with transarterial chemoembolization using multi-modal multi-task deep learning. EClinicalMedicine 2020, 23, 100379. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, N.; Wu, J.; Zhang, Q.; Lin, T.; Yao, Y.; Chen, Z.; Wang, M.; Sheng, L.; Liu, J.; et al. Radiomics analysis based on contrast-enhanced MRI for prediction of therapeutic response to transarterial chemoembolization in Hepatocellular Carcinoma. Front. Oncol. 2021, 11, 582788. [Google Scholar] [CrossRef]
- Sun, Y.; Bai, H.; Xia, W.; Wang, D.; Zhou, B.; Zhao, X.; Yang, G.; Xu, L.; Zhang, W.; Liu, P.; et al. Predicting the outcome of transcatheter arterial embolization therapy for unresectable hepatocellular carcinoma based on radiomics of preoperative multiparameter MRI. J. Magn. Reson. Imaging 2020, 52, 1083–1090. [Google Scholar] [CrossRef]
- Song, W.; Yu, X.; Guo, D.; Liu, H.; Tang, Z.; Liu, X.; Zhou, J.; Zhang, H.; Liu, Y.; Liu, X. MRI-based Radiomics: Associations with the recurrence-free survival of patients with hepatocellular carcinoma treated with conventional transcatheter arterial chemoembolization. J. Magn. Reson. Imaging 2020, 52, 461–473. [Google Scholar] [CrossRef]
- Kong, C.; Zhao, Z.; Chen, W.; Lv, X.; Shu, G.; Ye, M.; Song, J.; Ying, X.; Weng, Q.; Weng, W.; et al. Prediction of tumor response via a pretreatment MRI radiomics-based nomogram in HCC treated with TACE. Eur. Radiol. 2021, 31, 7500–7511. [Google Scholar] [CrossRef] [PubMed]
- Kuang, Y.; Li, R.; Jia, P.; Ye, W.; Zhou, R.; Zhu, R.; Wang, J.; Lin, S.; Pang, P.; Ji, W. MRI-Based Radiomics: Nomograms predicting the short-term response after transcatheter arterial chemoembolization (TACE) in hepatocellular carcinoma patients with diameter less than 5 cm. Abdom. Radiol. 2021, 46, 3772–3789. [Google Scholar] [CrossRef] [PubMed]
- Abajian, A.; Murali, N.; Savic, L.J.; Laage-Gaupp, F.M.; Nezami, N.; Duncan, J.S.; Schlachter, T.; Lin, M.; Geschwind, J.F.; Chapiro, J. Predicting treatment response to intra-arterial therapies for hepatocellular carcinoma with the use of supervised machine learning—An artificial intelligence concept. J. Vasc. Interv. Radiol. 2018, 29, 850–857. [Google Scholar] [CrossRef] [PubMed]
- Park, H.J.; Kim, J.H.; Choi, S.y.; Lee, E.S.; Park, S.J.; Byun, J.Y.; Choi, B.I. Prediction of therapeutic response of hepatocellular carcinoma to transcatheter arterial chemoembolization based on pretherapeutic dynamic CT and textural findings. Am. J. Roentgenol. 2017, 209, W211–W220. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Pei, Y.; Zhang, Y.; Wu, Y.; Liu, F.; Gu, S. Predicting the prognosis of hepatocellular carcinoma with the treatment of transcatheter arterial chemoembolization combined with microwave ablation using pretreatment MR imaging texture features. Abdom. Radiol. 2021, 46, 3748–3757. [Google Scholar] [CrossRef]
- Yu, J.; Zhang, H.; Tang, Z.; Zhou, J.; He, X.; Liu, Y.; Liu, X.; Guo, D. Value of texture analysis based on enhanced MRI for predicting an early therapeutic response to transcatheter arterial chemoembolisation combined with high-intensity focused ultrasound treatment in hepatocellular carcinoma. Clin. Radiol. 2018, 73, 758-e9. [Google Scholar] [CrossRef]
- Bruix, J.; Reig, M.; Sherman, M. Evidence-based diagnosis, staging, and treatment of patients with hepatocellular carcinoma. Gastroenterology 2016, 150, 835–853. [Google Scholar] [CrossRef]
- Kulik, L.; El-Serag, H.B. Epidemiology and management of hepatocellular carcinoma. Gastroenterology 2019, 156, 477–491. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Z.; Mo, Y.; Li, B.; Zhou, Q.; Peng, S.; Li, S.; Kuang, M. Prediction of post-hepatectomy liver failure in patients with hepatocellular carcinoma based on radiomics using Gd-EOB-DTPA-enhanced MRI: The liver failure model. Front. Oncol. 2021, 11, 605296. [Google Scholar] [CrossRef]
- Zhu, W.S.; Shi, S.Y.; Yang, Z.H.; Song, C.; Shen, J. Radiomics model based on preoperative gadoxetic acid-enhanced MRI for predicting liver failure. World J. Gastroenterol. 2020, 26, 1208. [Google Scholar] [CrossRef]
- Zheng, B.H.; Liu, L.Z.; Zhang, Z.Z.; Shi, J.Y.; Dong, L.Q.; Tian, L.Y.; Ding, Z.B.; Ji, Y.; Rao, S.X.; Zhou, J.; et al. Radiomics score: A potential prognostic imaging feature for postoperative survival of solitary HCC patients. BMC Cancer 2018, 18, 1148. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; He, L.; Huang, Y.; Chen, S.; Wu, P.; Ye, W.; Liu, Z.; Liang, C. CT-based radiomics signature: A potential biomarker for preoperative prediction of early recurrence in hepatocellular carcinoma. Abdom. Radiol. 2017, 42, 1695–1704. [Google Scholar] [CrossRef] [PubMed]
- Akai, H.; Yasaka, K.; Kunimatsu, A.; Nojima, M.; Kokudo, T.; Kokudo, N.; Hasegawa, K.; Abe, O.; Ohtomo, K.; Kiryu, S. Predicting prognosis of resected hepatocellular carcinoma by radiomics analysis with random survival forest. Diagn. Interv. Imaging 2018, 99, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Kiryu, S.; Akai, H.; Nojima, M.; Hasegawa, K.; Shinkawa, H.; Kokudo, N.; Yasaka, K.; Ohtomo, K. Impact of hepatocellular carcinoma heterogeneity on computed tomography as a prognostic indicator. Sci. Rep. 2017, 7, 12689. [Google Scholar] [CrossRef] [PubMed]
- Brenet Defour, L.; Mulé, S.; Tenenhaus, A.; Piardi, T.; Sommacale, D.; Hoeffel, C.; Thiéfin, G. Hepatocellular carcinoma: CT texture analysis as a predictor of survival after surgical resection. Eur. Radiol. 2019, 29, 1231–1239. [Google Scholar] [CrossRef]
- Oh, J.; Lee, J.M.; Park, J.; Joo, I.; Yoon, J.H.; Lee, D.H.; Ganeshan, B.; Han, J.K. Hepatocellular carcinoma: Texture analysis of preoperative computed tomography images can provide markers of tumor grade and disease-free survival. Korean J. Radiol. 2019, 20, 569–579. [Google Scholar] [CrossRef]
- Shan, Q.Y.; Hu, H.T.; Feng, S.T.; Peng, Z.P.; Chen, S.L.; Zhou, Q.; Li, X.; Xie, X.Y.; Lu, M.D.; Wang, W.; et al. CT-based peritumoral radiomics signatures to predict early recurrence in hepatocellular carcinoma after curative tumor resection or ablation. Cancer Imaging 2019, 19, 1–11. [Google Scholar] [CrossRef]
- Kim, S.; Shin, J.; Kim, D.Y.; Choi, G.H.; Kim, M.J.; Choi, J.Y. Radiomics on gadoxetic acid–enhanced magnetic resonance imaging for prediction of postoperative early and late recurrence of single hepatocellular carcinoma. Clin. Cancer Res. 2019, 25, 3847–3855. [Google Scholar] [CrossRef]
- Ji, G.W.; Zhu, F.P.; Xu, Q.; Wang, K.; Wu, M.Y.; Tang, W.W.; Li, X.C.; Wang, X.H. Radiomic features at contrast-enhanced CT predict recurrence in early stage hepatocellular carcinoma: A multi-institutional study. Radiology 2020, 294, 568–579. [Google Scholar] [CrossRef]
- Ji, G.W.; Zhu, F.P.; Xu, Q.; Wang, K.; Wu, M.Y.; Tang, W.W.; Li, X.C.; Wang, X.H. Machine-learning analysis of contrast-enhanced CT radiomics predicts recurrence of hepatocellular carcinoma after resection: A multi-institutional study. EBioMedicine 2019, 50, 156–165. [Google Scholar] [CrossRef]
- Wen, L.; Weng, S.; Yan, C.; Ye, R.; Zhu, Y.; Zhou, L.; Gao, L.; Li, Y. A radiomics nomogram for preoperative prediction of early recurrence of small hepatocellular carcinoma after surgical resection or radiofrequency ablation. Front. Oncol. 2021, 11, 657039. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Jiang, H.; Chen, J.; Wei, Y.; Cao, L.; Ye, Z.; Li, X.; Ma, L.; Song, B. Hepatocellular carcinoma: Radiomics nomogram on gadoxetic acid-enhanced MR imaging for early postoperative recurrence prediction. Cancer Imaging 2019, 19, 1–10. [Google Scholar] [CrossRef]
- Ahn, S.J.; Kim, J.H.; Park, S.J.; Kim, S.T.; Han, J.K. Hepatocellular carcinoma: Preoperative gadoxetic acid–enhanced MR imaging can predict early recurrence after curative resection using image features and texture analysis. Abdom. Radiol. 2019, 44, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, J.; Jiang, H.; Wei, Y.; Zhang, X.; Cao, L.; Duan, T.; Ye, Z.; Yao, S.; Pan, X.; et al. Gadoxetic acid-enhanced MRI radiomics signature: Prediction of clinical outcome in hepatocellular carcinoma after surgical resection. Ann. Transl. Med. 2020, 8. [Google Scholar] [CrossRef] [PubMed]
- Hui, T.; Chuah, T.; Low, H.; Tan, C. Predicting early recurrence of hepatocellular carcinoma with texture analysis of preoperative MRI: A radiomics study. Clin. Radiol. 2018, 73, 1056–e11. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, X.; Zhang, H.; He, X.; Liu, Y.; Zhou, J.; Guo, D. Texture analysis based on preoperative magnetic resonance imaging (MRI) and conventional MRI features for predicting the early recurrence of single hepatocellular carcinoma after hepatectomy. Acad. Radiol. 2019, 26, 1164–1173. [Google Scholar] [CrossRef]
- Zhao, Y.; Wu, J.; Zhang, Q.; Hua, Z.; Qi, W.; Wang, N.; Lin, T.; Sheng, L.; Cui, D.; Liu, J.; et al. Radiomics analysis based on multiparametric MRI for predicting early recurrence in hepatocellular carcinoma after partial hepatectomy. J. Magn. Reson. Imaging 2021, 53, 1066–1079. [Google Scholar] [CrossRef]
- Llovet, J.M.; Ricci, S.; Mazzaferro, V.; Hilgard, P.; Gane, E.; Blanc, J.F.; De Oliveira, A.C.; Santoro, A.; Raoul, J.L.; Forner, A.; et al. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008, 359, 378–390. [Google Scholar] [CrossRef]
- Kudo, M.; Finn, R.S.; Qin, S.; Han, K.H.; Ikeda, K.; Piscaglia, F.; Baron, A.; Park, J.W.; Han, G.; Jassem, J.; et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma: A randomised phase 3 non-inferiority trial. Lancet 2018, 391, 1163–1173. [Google Scholar] [CrossRef]
- Bouattour, M.; Mehta, N.; He, A.R.; Cohen, E.I.; Nault, J.C. Systemic treatment for advanced hepatocellular carcinoma. Liver Cancer 2019, 8, 341–358. [Google Scholar] [CrossRef]
- Brown, Z.J.; Greten, T.F.; Heinrich, B. Adjuvant treatment of hepatocellular carcinoma: Prospect of immunotherapy. Hepatology 2019, 70, 1437–1442. [Google Scholar] [CrossRef] [PubMed]
- Rimassa, L.; Pressiani, T.; Merle, P. Systemic treatment options in hepatocellular carcinoma. Liver Cancer 2019, 8, 427–446. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.S.; Qin, S.; Ikeda, M.; Galle, P.R.; Ducreux, M.; Kim, T.Y.; Kudo, M.; Breder, V.; Merle, P.; Kaseb, A.O.; et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N. Engl. J. Med. 2020, 382, 1894–1905. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.S.; Zhu, A.X. Evolution of systemic therapy for hepatocellular carcinoma. Hepatology 2021, 73, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Kelley, R.K. Atezolizumab plus bevacizumab—A landmark in liver cancer. N. Engl. J. Med. 2020, 382, 1953–1955. [Google Scholar] [CrossRef]
- Lee, M.S.; Ryoo, B.Y.; Hsu, C.H.; Numata, K.; Stein, S.; Verret, W.; Hack, S.P.; Spahn, J.; Liu, B.; Abdullah, H.; et al. Atezolizumab with or without bevacizumab in unresectable hepatocellular carcinoma (GO30140): An open-label, multicentre, phase 1b study. Lancet Oncol. 2020, 21, 808–820. [Google Scholar] [CrossRef]
- Liu, Z.; Lin, Y.; Zhang, J.; Zhang, Y.; Li, Y.; Liu, Z.; Li, Q.; Luo, M.; Liang, R.; Ye, J. Molecular targeted and immune checkpoint therapy for advanced hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2019, 38, 1–13. [Google Scholar] [CrossRef]
- Chen, D.S.; Mellman, I. Elements of cancer immunity and the cancer–immune set point. Nature 2017, 541, 321–330. [Google Scholar] [CrossRef]
- Liao, H.; Zhang, Z.; Chen, J.; Liao, M.; Xu, L.; Wu, Z.; Yuan, K.; Song, B.; Zeng, Y. Preoperative radiomic approach to evaluate tumor-infiltrating CD8+ T cells in hepatocellular carcinoma patients using contrast-enhanced computed tomography. Ann. Surg. Oncol. 2019, 26, 4537–4547. [Google Scholar] [CrossRef]
- Yuan, G.; Song, Y.; Li, Q.; Hu, X.; Zang, M.; Dai, W.; Cheng, X.; Huang, W.; Yu, W.; Chen, M.; et al. Development and validation of a contrast-enhanced CT-based radiomics nomogram for prediction of therapeutic efficacy of anti-PD-1 antibodies in advanced HCC patients. Front. Immunol. 2021, 11, 613946. [Google Scholar] [CrossRef]
- Hectors, S.J.; Lewis, S.; Besa, C.; King, M.J.; Said, D.; Putra, J.; Ward, S.; Higashi, T.; Thung, S.; Yao, S.; et al. MRI radiomics features predict immuno-oncological characteristics of hepatocellular carcinoma. Eur. Radiol. 2020, 30, 3759–3769. [Google Scholar] [CrossRef] [PubMed]
- Kudo, M. Gd-EOB-DTPA-MRI could predict WNT/β-catenin mutation and resistance to immune checkpoint inhibitor therapy in hepatocellular carcinoma. Liver Cancer 2020, 9, 479–490. [Google Scholar] [CrossRef]
- Aoki, T.; Nishida, N.; Kudo, M. Clinical significance of the duality of Wnt/β-catenin signaling in human hepatocellular carcinoma. Cancers 2022, 14, 444. [Google Scholar] [CrossRef] [PubMed]
- Aoki, T.; Nishida, N.; Ueshima, K.; Morita, M.; Chishina, H.; Takita, M.; Hagiwara, S.; Ida, H.; Minami, Y.; Yamada, A.; et al. Higher enhancement intrahepatic nodules on the hepatobiliary phase of Gd-EOB-DTPA-enhanced MRI as a poor responsive marker of Anti-PD-1/PD-L1 monotherapy for unresectable hepatocellular carcinoma. Liver Cancer 2021, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Lai, Q.; Spoletini, G.; Mennini, G.; Laureiro, Z.L.; Tsilimigras, D.I.; Pawlik, T.M.; Rossi, M. Prognostic role of artificial intelligence among patients with hepatocellular cancer: A systematic review. World J. Gastroenterol. 2020, 26, 6679. [Google Scholar] [CrossRef] [PubMed]
- Vitale, A.; Lai, Q.; Farinati, F.; Bucci, L.; Giannini, E.G.; Napoli, L.; Ciccarese, F.; Rapaccini, G.L.; Di Marco, M.; Caturelli, E.; et al. Utility of tumor burden score to stratify prognosis of patients with hepatocellular cancer: Results of 4759 cases from ITA. LI. CA study group. J. Gastrointest. Surg. 2018, 22, 859–871. [Google Scholar] [CrossRef]
- Lai, Q.; Vitale, A.; Halazun, K.; Iesari, S.; Viveiros, A.; Bhangui, P.; Mennini, G.; Wong, T.; Uemoto, S.; Lin, C.C.; et al. Identification of an upper limit of tumor burden for downstaging in candidates with hepatocellular cancer waiting for liver transplantation: A west–east collaborative effort. Cancers 2020, 12, 452. [Google Scholar] [CrossRef]
- Lai, Q.; Nicolini, D.; Inostroza Nunez, M.; Iesari, S.; Goffette, P.; Agostini, A.; Giovagnoni, A.; Vivarelli, M.; Lerut, J. A Novel Prognostic Index in Patients With Hepatocellular Cancer Waiting for Liver Transplantation. Ann. Surg. 2016, 264, 787–796. [Google Scholar] [CrossRef]
- Cleophas, T.J.; Cleophas, T.F. Artificial intelligence for diagnostic purposes: Principles, procedures and limitations. Clin. Chem. Lab. Med. 2010, 48, 159–165. [Google Scholar] [CrossRef]
- Hamamoto, I.; Okada, S.; Hashimoto, T.; Wakabayashi, H.; Maeba, T.; Maeta, H. Prediction of the early prognosis of the hepatectomized patient with hepatocellular carcinoma with a neural network. Comput. Biol. Med. 1995, 25, 49–59. [Google Scholar] [CrossRef]
- Ho, W.H.; Lee, K.T.; Chen, H.Y.; Ho, T.W.; Chiu, H.C. Disease-free survival after hepatic resection in hepatocellular carcinoma patients: A prediction approach using artificial neural network. PLoS ONE 2012, 7, e29179. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.Y.; Lee, K.T.; Lee, H.H.; Ho, W.H.; Sun, D.P.; Wang, J.J.; Chiu, C.C. Comparison of artificial neural network and logistic regression models for predicting in-hospital mortality after primary liver cancer surgery. PLoS ONE 2012, 7, e35781. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.Y.; Lee, K.T.; Wang, J.J.; Sun, D.P.; Lee, H.H.; Chiu, C.C. Artificial neural network model for predicting 5-year mortality after surgery for hepatocellular carcinoma: A nationwide study. J. Gastrointest. Surg. 2012, 16, 2126–2131. [Google Scholar] [CrossRef] [PubMed]
- Chiu, H.C.; Ho, T.W.; Lee, K.T.; Chen, H.Y.; Ho, W.H. Mortality predicted accuracy for hepatocellular carcinoma patients with hepatic resection using artificial neural network. Sci. World J. 2013, 2013, 201976. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xia, W.; Yan, Z.P.; Sun, J.H.; Zhong, B.Y.; Hou, Z.H.; Yang, M.J.; Zhou, G.H.; Wang, W.S.; Zhao, X.Y.; et al. Deep learning predicts overall survival of patients with unresectable hepatocellular carcinoma treated by transarterial chemoembolization plus sorafenib. Front. Oncol. 2020, 10, 593292. [Google Scholar] [CrossRef]
- Yuan, C.; Wang, Z.; Gu, D.; Tian, J.; Zhao, P.; Wei, J.; Yang, X.; Hao, X.; Dong, D.; He, N.; et al. Prediction early recurrence of hepatocellular carcinoma eligible for curative ablation using a Radiomics nomogram. Cancer Imaging 2019, 19, 1–12. [Google Scholar] [CrossRef]
- Zhu, H.B.; Zheng, Z.Y.; Zhao, H.; Zhang, J.; Zhu, H.; Li, Y.H.; Dong, Z.Y.; Xiao, L.S.; Kuang, J.J.; Zhang, X.L.; et al. Radiomics-based nomogram using CT imaging for noninvasive preoperative prediction of early recurrence in patients with hepatocellular carcinoma. Diagn. Interv. Radiol. 2020, 26, 411. [Google Scholar] [CrossRef]
- Weibin, W.; Qingqing, C.; Iwamoto, Y.; Xianhua, H.; Zhang, Q.; Hongjie, H.; Lanfen, L.; Yen-Wei, C. Deep learning-based radiomics models for early recurrence prediction of hepatocellular carcinoma with multi-phase CT images and clinical data. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; IEEE: Hoboken, NJ, USA, 2019; pp. 4881–4884. [Google Scholar]
- Hu, H.T.; Shan, Q.Y.; Chen, S.L.; Li, B.; Feng, S.T.; Xu, E.J.; Li, X.; Long, J.Y.; Xie, X.Y.; Lu, M.D.; et al. CT-based radiomics for preoperative prediction of early recurrent hepatocellular carcinoma: Technical reproducibility of acquisition and scanners. Radiol. Med. 2020, 125, 697–705. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The image biomarker standardization initiative: Standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 2020, 295, 328. [Google Scholar] [CrossRef]

| Study | Method | CT Data | Performance |
|---|---|---|---|
| Hame [38] |
| 10 subjects | Qualitative evaluation |
| Smeets et al. [39] | Supervised statistical pixel classification | 10 subjects | Total score = 69% |
| Choudhary et al. [40] | Adaptive multi-thresholding | 10 subjects | Total score = 70% |
| Moltz et al. [41] |
| 10 subjects | Total score = 72% |
| Kadoury et al. [42] | Appearance analysis using discriminant grassmannian manifolds | 43 subjects | Dice score = 91 |
| Linguraru et al. [43] | Morphology-based parameterization | 101 subjects | AUC = 0.85 |
| Christ et al. [44] | CNN | 100 subjects | Dice score = 94% |
| Han [45] | Deep CNN | 200 subjects | Dice score = 67% |
| Vorontsov et al. [46] | Fully connected CNN | 200 subjects | Dice score = 77.3% |
| Li et al. [47] | Hybrid CNN | 221 subjects | Dice score = 82.4% |
| Chlebus et al. [48] | Fully connected CNN and object-based refinement | 131 subjects | Accuracy = 77% |
| Meng et al. [49] | CNN and post-refinement | 131 subjects | Dice score = 68.9% |
| Study | Aim | Method | Data | Performance |
|---|---|---|---|---|
| Wu et al. [28] | Discrimination of HCC and HH | Radiomics-based classification | Multi-modal MRI (446 lesions) | Sensitivity = 82.2% and Specificity = 71.4% |
| Yasaka et al. [35] | Differentiation of typical HCC from indeterminate hepatic lesions, HH, and cysts | CNN | CE-CT (560 Subject) | Accuracy = 84% |
| Nie et al. [54] | Discrimination of HCC and FHN | Logistic regression analysis for radiomics-based features and clinical markers | CE-CT (119 subjects) | AUC = 0.92% |
| Nie et al. [55] | Discrimination of HCC and HA | Logistic regression analysis for radiomics-based features and clinical markers | CE-CT (119 subjects) | AUC = 0.94 |
| Mokrane et al. [56] | Detection of HCC | Imaging features-based classification | CE-CT (178 subjects) | AUC = 0.66 |
| Ponnoprat et al. [57] | Discrimination of HCC and CC | Histogram-based classification | CE-CT (257 subjects) | Accuracy = 88% |
| Liu et al. [58] | Detection of combined HCC-CC | Radiomics-based classification | CT and MRI (86 lesions) | AUC = 0.77 |
| Shi et al. [59] | Detection of HCC | 3 CDNs | CE-CT (342 subjects) | Accuracy = 85.6% |
| Cap et al. [60] | Differentiation of different types of hepatic lesions | CDN | CE-CT (517 subjects) | Accuracy = 81.3% |
| Hamm et al. [61] | Differentiation of different types of hepatic lesions | CNN | CE-MRI (494 subjects) | Accuracy = 91.9% |
| Wang et al. [62] | Differentiation of different types of hepatic lesions | CNN-based interpretation and feature maps | CE-MRI (494 subjects) | Accuracy = 88% |
| Zhen et al. [63] | Differentiation of different types of hepatic lesions | CNN | MRI (1411 subjects) | AUC = 0.98 |
| Jian et al. [64] | Detection of HCC | Transfer-learning | CE-MRI (150 subjects) | Accuracy = 77% |
| Sun et al. [65] | Detection of small HCC lesions | Radiomics-based classification | CE-MRI (124 subjects) | Accuracy = 90.40% |
| Study | Method | Data | Performance |
|---|---|---|---|
| Xu et al. [67] | Radiomics-based features | CE-CT (619 subjects) | AUC = 0.889 |
| Bakr et al. [68] | Morphological and textural features | CE-CT (28 subjects) | AUC = 0.76 |
| Peng et al. [69] | Radiomics-based features and clinical biomarkers | CE-CT (304 subjects) | Qualitative evaluation |
| Zheng et al. [70] | Morphological features and clinical biomarkers | CT (120 subjects) | AUC = 0.88 |
| Cucchetti et al. [71] | Clinical biomarkers | Radiology and histopathology (250 subjects) | Accuracy = 88% |
| Jiang et al. [72] | Radiomics-based features and CNN | CE-CT (405 subjects) | Accuracy = 85.2% and AUC = 0.906 |
| Ni et al. [73] | Radiomics-based features | CE-CT (206 subjects) | Accuracy = 84.4% |
| Zhang et al. [74] | Statistical analysis | CT or MRI (4759 subjects) | AUC = 0.86 |
| Zhou et al. [75] | CNN | CE-MRI (114 subjects) | AUC = 0.926 |
| Wang et al. [76] | Histogram-based features and clinical biomarkers | CE-MRI (113 subjects) | AUC = 0.798 |
| Dai et al. [77] | Radiomics-based features | CE-MRI (69 subjects) | AUC = 0.895 |
| Wang et al. [78] | CNN | DW-MRI (97 subjects) | AUC = 0.79 |
| Study | Aim | Method | Data | Performance |
|---|---|---|---|---|
| Mao et al. [84] | Discrimination of high/low grade HCC | Radiomics-based features and clinical biomarkers | CE-CT (297 subjects) | AUC = 0.8014 |
| Wu et al. [85] | Discrimination of high/low grade HCC | Radiomics-based features and clinical biomarkers | Multi-modal MRI (170 subjects) | AUC = 0.8 |
| Zhou et al. [87] | Discrimination of high/low grade HCC | CNN | DW-MRI (98 subjects) | Accuracy = 80% |
| Huang et al. [90] | Detection of DPHCC | Radiomics-based classification | CE-MRI (100 subjects) | Accuracy = 79.8% |
| Geng et al. [91] | Differentiation of four histopathology grades | Radiomics-based classification | SWI (53 subjects) | AUC = 0.8255 |
| Yang et al. [92] | Detection of CK-19 HCC | Radiomics-based classification | CE-MRI (257 subjects) | AUC = 0.758 |
| Wang et al. [93] | Detection of CK-19 HCC | Radiomics-based classification | CE-MRI (227 subjects) | AUC = 0.822 |
| Fan et al. [94] | Prediction of K-67 | Radiomics-based features | Multi-modal MRI (151 subjects) | AUC = 0.922 |
| Study | Aim | Method | Data | Performance |
|---|---|---|---|---|
| An et al. [101] | Prediction of MWA response | CNN | CE-MRI (141 subjects) | Qualitative evaluation |
| Hu et al. [102] | Prediction of MWA and RFA response | Radiomics-based features and clinical biomarkers | CE-MRI (160 subjects) | AUC = 0.835 |
| Liang et al. [103] | Prediction of RFA response | Clinical biomarkers | CT (83 subjects) | AUC = 0.69 |
| Liu et al. [104] | Prediction of RFA response | Radiomics-based deep learning | CE-US (419 subjects) | Qualitative evaluation |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fahmy, D.; Alksas, A.; Elnakib, A.; Mahmoud, A.; Kandil, H.; Khalil, A.; Ghazal, M.; van Bogaert, E.; Contractor, S.; El-Baz, A. The Role of Radiomics and AI Technologies in the Segmentation, Detection, and Management of Hepatocellular Carcinoma. Cancers 2022, 14, 6123. https://doi.org/10.3390/cancers14246123
Fahmy D, Alksas A, Elnakib A, Mahmoud A, Kandil H, Khalil A, Ghazal M, van Bogaert E, Contractor S, El-Baz A. The Role of Radiomics and AI Technologies in the Segmentation, Detection, and Management of Hepatocellular Carcinoma. Cancers. 2022; 14(24):6123. https://doi.org/10.3390/cancers14246123
Chicago/Turabian StyleFahmy, Dalia, Ahmed Alksas, Ahmed Elnakib, Ali Mahmoud, Heba Kandil, Ashraf Khalil, Mohammed Ghazal, Eric van Bogaert, Sohail Contractor, and Ayman El-Baz. 2022. "The Role of Radiomics and AI Technologies in the Segmentation, Detection, and Management of Hepatocellular Carcinoma" Cancers 14, no. 24: 6123. https://doi.org/10.3390/cancers14246123
APA StyleFahmy, D., Alksas, A., Elnakib, A., Mahmoud, A., Kandil, H., Khalil, A., Ghazal, M., van Bogaert, E., Contractor, S., & El-Baz, A. (2022). The Role of Radiomics and AI Technologies in the Segmentation, Detection, and Management of Hepatocellular Carcinoma. Cancers, 14(24), 6123. https://doi.org/10.3390/cancers14246123








