ACY1 Downregulation Enhances the Radiosensitivity of Cetuximab-Resistant Colorectal Cancer by Inactivating the Wnt/β-Catenin Signaling Pathway
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Patients and Clinical Tissue Samples
2.2. Cell Lines and Cell Culture
2.3. Cell Viability Assay
2.4. X-ray Irradiation
2.5. Colony Formation Assay
2.6. High-Throughput Sequencing
2.7. Bioinformatics Analysis
2.8. Immunohistochemistry
2.9. Wound-Healing Assay
2.10. Cell Invasion Assay
2.11. RNA Extraction and Real-Time PCR Assay Analysis
2.12. Cell Transfection
2.13. Western Blotting
2.14. Statistical Analyses
3. Results
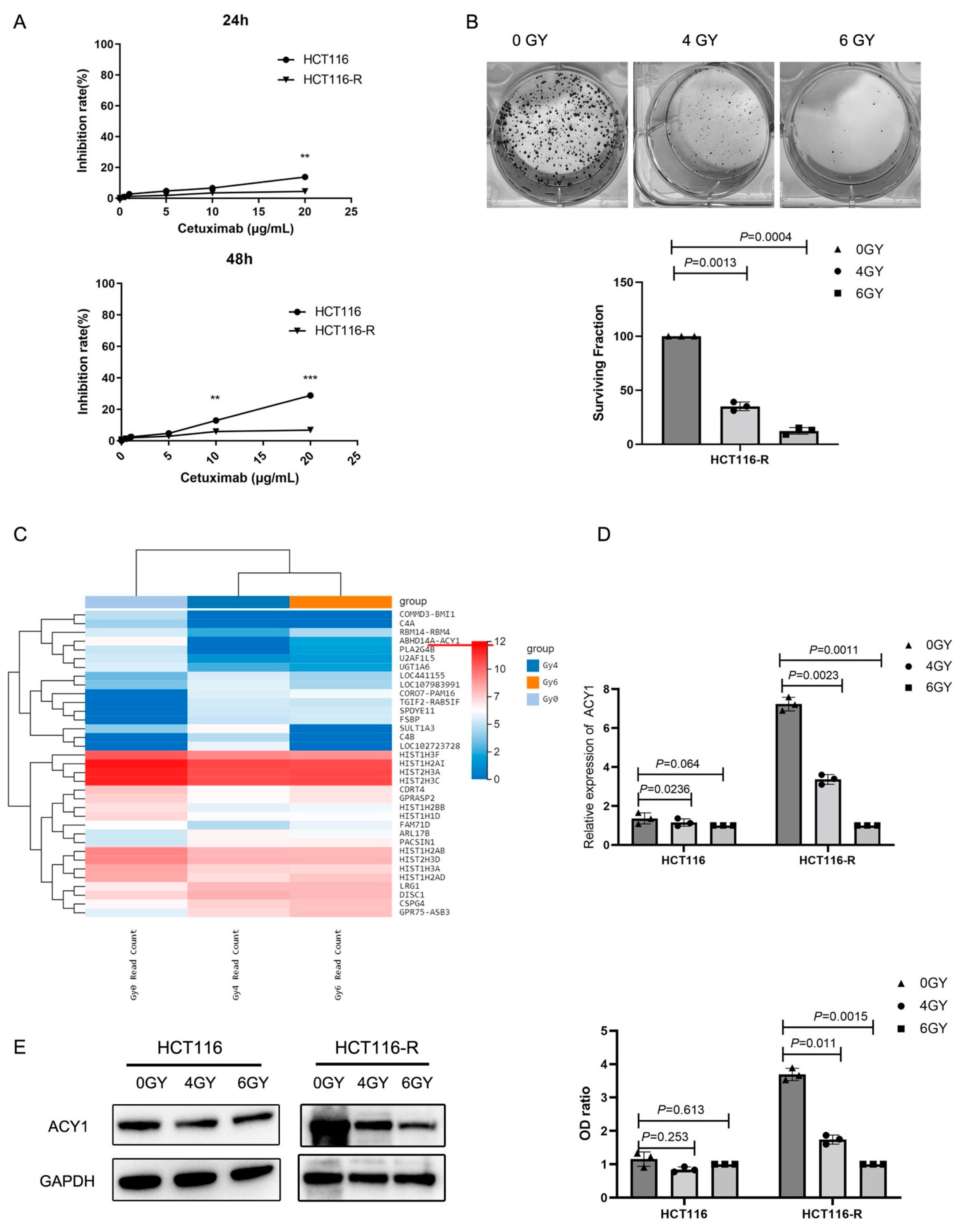
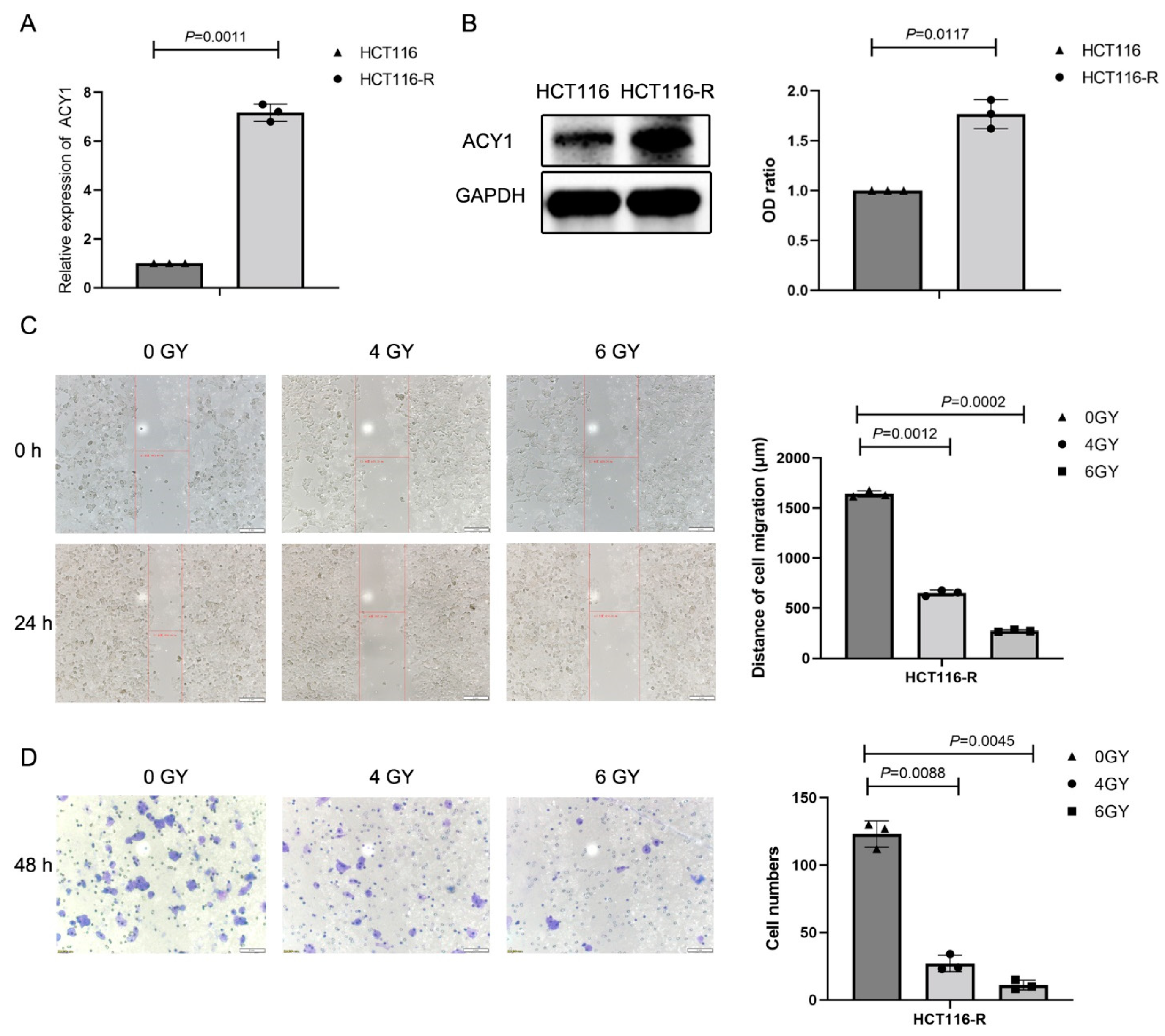
3.1. Irradiation Treatment Significantly Reduced ACY1 Expression in Cetuximab-Resistant CRC Cells
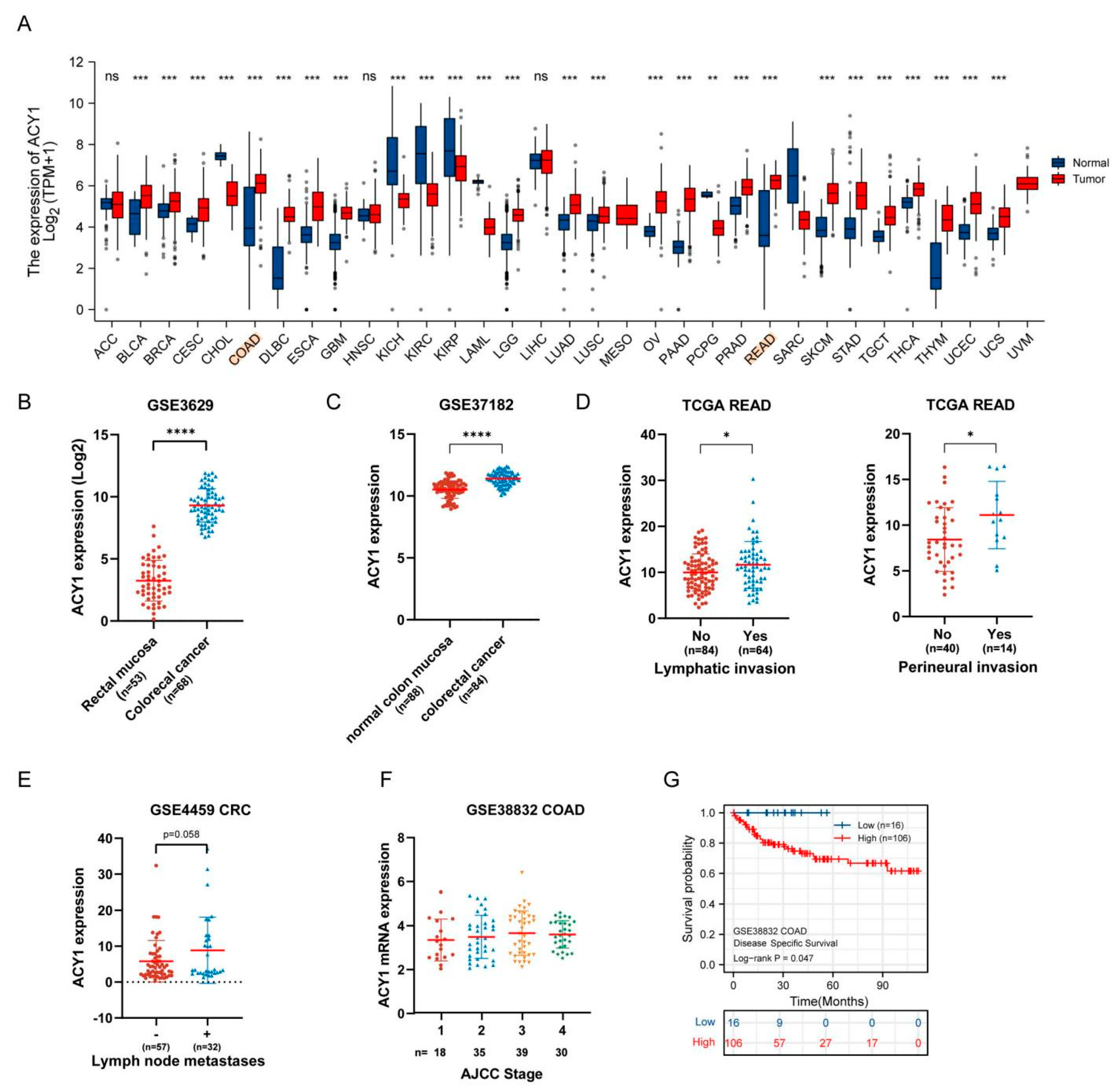
3.2. ACY1 Levels Were Associated with Tumor Metastasis, Prognosis, and Cetuximab Resistance in CRC
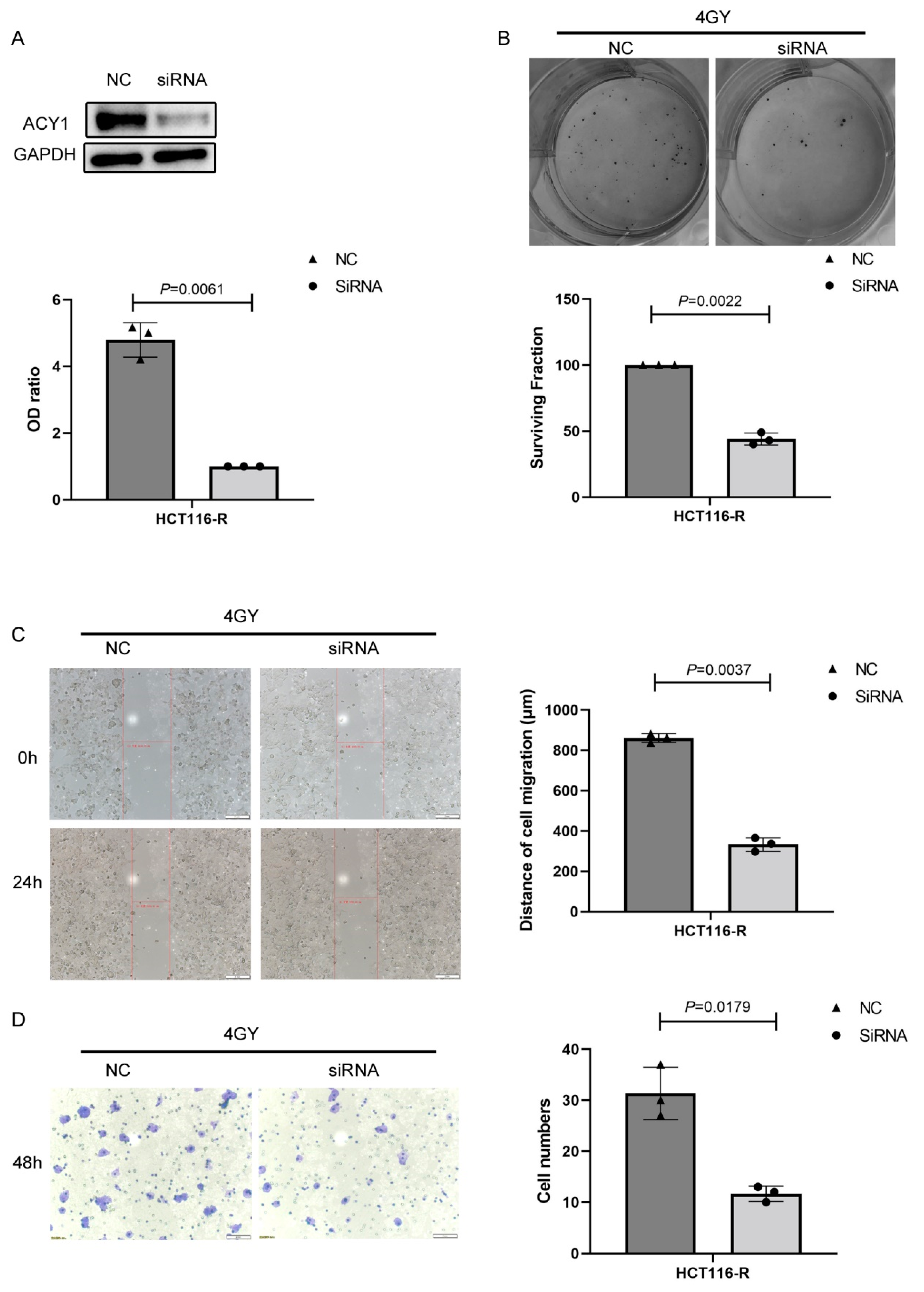
3.3. ACY1 Was Involved in Regulating the Radiosensitivity of Cetuximab-Resistant CRC
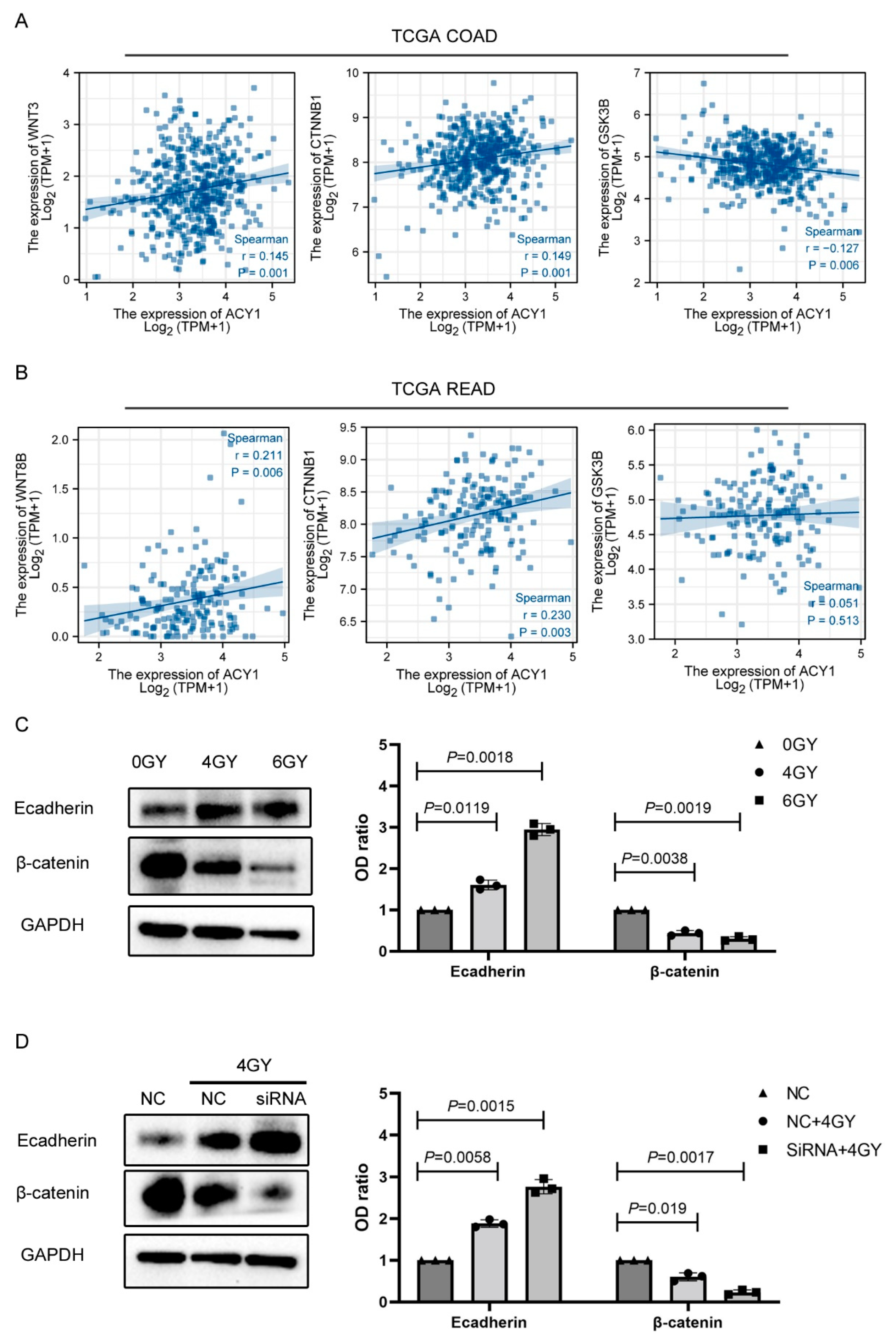
3.4. ACY1 Inactivated Wnt/β-Catenin Signaling to Regulate the Radiosensitivity of Cetuximab-Resistant CRC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wang, C.; Song, H.; Xu, Y.; Ji, G. RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers. Mol. Cancer 2019, 18, 8. [Google Scholar] [CrossRef] [PubMed]
- Bardhan, K.; Liu, K. Epigenetics and Colorectal Cancer Pathogenesis. Cancers 2013, 5, 676–713. [Google Scholar] [CrossRef] [PubMed]
- Van Cutsem, E.; Cervantes, A.; Nordlinger, B.; Arnold, D.; ESMO Guidelines Working Group. Metastatic colorectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2014, 25 (Suppl. S3), III1–III9. [Google Scholar] [CrossRef] [PubMed]
- Guren, M.G. The global challenge of colorectal cancer. Lancet Gastroenterol. Hepatol. 2019, 4, 894–895. [Google Scholar] [CrossRef]
- Khattak, M.A.; Martin, H.; Davidson, A.; Phillips, M. Role of First-Line Anti–Epidermal Growth Factor Receptor Therapy Compared with Anti–Vascular Endothelial Growth Factor Therapy in Advanced Colorectal Cancer: A Meta-Analysis of Randomized Clinical Trials. Clin. Color. Cancer 2015, 14, 81–90. [Google Scholar] [CrossRef]
- Zhou, B.; Wang, L.; Qiu, H.; Zhang, M.; Sun, L.; Peng, P.; Yu, Q.; Yuan, X. Mechanisms of resistance to anti-EGFR therapy in colorectal cancer. Oncotarget 2017, 8, 3980–4000. [Google Scholar] [CrossRef]
- De Roock, W.; Claes, B.; Bernasconi, D.; De Schutter, J.; Biesmans, B.; Fountzilas, G.; Kalogeras, K.T.; Kotoula, V.; Papamichael, D.; Laurent-Puig, P.; et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: A retrospective consortium analysis. Lancet Oncol. 2010, 11, 753–762. [Google Scholar] [CrossRef]
- Van Emburgh, B.O.; Sartore-Bianchi, A.; Di Nicolantonio, F.; Siena, S.; Bardelli, A. Acquired resistance to EGFR-targeted therapies in colorectal cancer. Mol. Oncol. 2014, 8, 1084–1094. [Google Scholar] [CrossRef]
- Marijnen, C.; Glimelius, B. The role of radiotherapy in rectal cancer. Eur. J. Cancer 2002, 38, 943–952. [Google Scholar] [CrossRef]
- Weber, G.F.; Rosenberg, R.; Murphy, J.E.; Meyer zum Büschenfelde, C.; Friess, H. Multimodal treatment strategies for locally advanced rectal cancer. Expert Rev. Anticancer. Ther. 2014, 12, 481–494. [Google Scholar] [CrossRef] [PubMed]
- Illum, H. Current Status of Radiosensitizing Agents for the Management of Rectal Cancer. Crit. Rev. Oncog. 2012, 17, 345–359. [Google Scholar] [CrossRef] [PubMed]
- Anders, M.W.; Dekant, W. Aminoacylases. Adv. Pharmacol. 1994, 27, 431–448. [Google Scholar]
- Polevoda, B.; Sherman, F. Nα-terminal Acetylation of Eukaryotic Proteins. J. Biol. Chem. 2000, 275, 36479–36482. [Google Scholar] [CrossRef]
- Sass, J.O.; Mohr, V.; Olbrich, H.; Engelke, U.; Horvath, J.; Fliegauf, M.; Loges, N.T.; Schweitzer-Krantz, S.; Moebus, R.; Weiler, P.; et al. Mutations in ACY1, the Gene Encoding Aminoacylase 1, Cause a Novel Inborn Error of Metabolism. Am. J. Hum. Genet. 2006, 78, 401–409. [Google Scholar] [CrossRef]
- Miller, Y.E.; Minna, J.D.; Gazdar, A.F. Lack of expression of aminoacylase-1 in small cell lung cancer. Evidence for inactivation of genes encoded by chromosome 3p. J. Clin. Investig. 1989, 83, 2120–2124. [Google Scholar] [CrossRef]
- Shi, H.; Hayes, M.T.; Kirana, C.; Miller, R.J.; Keating, J.P.; Stubbs, R.S. Overexpression of aminoacylase 1 is associated with colorectal cancer progression. Hum. Pathol. 2013, 44, 1089–1097. [Google Scholar] [CrossRef]
- Zhong, Y.; Onuki, J.; Yamasaki, T.; Ogawa, O.; Akatsuka, S.; Toyokuni, S. Genome-wide analysis identifies a tumor suppressor role for aminoacylase 1 in iron-induced rat renal cell carcinoma. Carcinogenesis 2009, 30, 158–164. [Google Scholar] [CrossRef]
- Balamurugan, T.S.T.; Huang, C.-H.; Chang, P.-C.; Huang, S.-T. Electrochemical Molecular Switch for the Selective Profiling of Cysteine in Live Cells and Whole Blood and for the Quantification of Aminoacylase-1. Anal. Chem. 2018, 90, 12631–12638. [Google Scholar] [CrossRef]
- Shi, H.; Hood, K.A.; Hayes, M.T.; Stubbs, R.S. Proteomic analysis of advanced colorectal cancer by laser capture microdissection and two-dimensional difference gel electrophoresis. J. Proteom. 2011, 75, 339–351. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef]
- Yaromina, A.; Krause, M.; Thames, H.; Rosner, A.; Krause, M.; Hessel, F.; Grenman, R.; Zips, D.; Baumann, M. Pre-treatment number of clonogenic cells and their radiosensitivity are major determinants of local tumour control after fractionated irradiation. Radiother. Oncol. 2007, 83, 304–310. [Google Scholar] [CrossRef]
- Ito, Y.; Yamada, Y.; Asada, K.; Ushijima, T.; Iwasa, S.; Kato, K.; Hamaguchi, T.; Shimada, Y. EGFR L2 domain mutation is not correlated with resistance to cetuximab in metastatic colorectal cancer patients. J. Cancer Res. Clin. Oncol. 2013, 139, 1391–1396. [Google Scholar] [CrossRef]
- Lièvre, A.; Bachet, J.-B.; Le Corre, D.; Boige, V.; Landi, B.; Emile, J.-F.; Côté, J.-F.; Tomasic, G.; Penna, C.; Ducreux, M.; et al. KRAS Mutation Status Is Predictive of Response to Cetuximab Therapy in Colorectal Cancer. Cancer Res. 2006, 66, 3992–3995. [Google Scholar] [CrossRef]
- Zhu, G.-X.; Gao, D.; Shao, Z.-Z.; Chen, L.; Ding, W.-J.; Yu, Q.-F. Wnt/β-catenin signaling: Causes and treatment targets of drug resistance in colorectal cancer (Review). Mol. Med. Rep. 2021, 23, 105. [Google Scholar] [CrossRef]
- Ji, Y.; Lv, J.; Sun, D.; Huang, Y. Therapeutic strategies targeting Wnt/β-catenin signaling for colorectal cancer (Review). Int. J. Mol. Med. 2022, 49, 1–17. [Google Scholar] [CrossRef]
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef]
- Dienstmann, R.; Vermeulen, L.; Guinney, J.; Kopetz, S.; Tejpar, S.; Tabernero, J. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat. Rev. Cancer 2017, 17, 79–92. [Google Scholar] [CrossRef]
- Wolf, A.M.D.; Fontham, E.T.H.; Church, T.R.; Flowers, C.R.; Guerra, C.E.; LaMonte, S.J.; Etzioni, R.; McKenna, M.T.; Oeffinger, K.C.; Shih, Y.-C.T.; et al. Colorectal cancer screening for average-risk adults: 2018 guideline update from the American Cancer Society. CA Cancer J. Clin. 2018, 68, 250–281. [Google Scholar] [CrossRef]
- Kopetz, S.; Chang, G.J.; Overman, M.J.; Eng, C.; Sargent, D.; Larson, D.W.; Grothey, A.; Vauthey, J.-N.; Nagorney, D.M.; McWilliams, R.R. Improved Survival in Metastatic Colorectal Cancer Is Associated with Adoption of Hepatic Resection and Improved Chemotherapy. J. Clin. Oncol. 2009, 27, 3677–3683. [Google Scholar] [CrossRef]
- Tabernero, J.; Grothey, A.; Van Cutsem, E.; Yaeger, R.; Wasan, H.; Yoshino, T.; Desai, J.; Ciardiello, F.; Loupakis, F.; Hong, Y.S.; et al. Encorafenib Plus Cetuximab as a New Standard of Care for Previously Treated BRAF V600E–Mutant Metastatic Colorectal Cancer: Updated Survival Results and Subgroup Analyses from the BEACON Study. J. Clin. Oncol. 2021, 39, 273–284. [Google Scholar] [CrossRef]
- Zhang, X.; Bi, K.; Tu, X.; Zhang, Q.; Cao, Q.; Liang, Y.; Zeng, P.; Wang, L.; Liu, T.; Fang, W.; et al. Interleukin-33 as an early predictor of cetuximab treatment efficacy in patients with colorectal cancer. Cancer Med. 2021, 10, 8338–8351. [Google Scholar] [CrossRef]
- Kim, S.-A.; Park, H.; Kim, K.-J.; Kim, J.-W.; Sung, J.H.; Nam, M.; Lee, J.H.; Jung, E.H.; Suh, K.J.; Lee, J.Y.; et al. Cetuximab resistance induced by hepatocyte growth factor is overcome by MET inhibition in KRAS, NRAS, and BRAF wild-type colorectal cancers. J. Cancer Res. Clin. Oncol. 2022, 148, 2995–3005. [Google Scholar] [CrossRef]
- Roh, M.S.; Colangelo, L.H.; O’Connell, M.J.; Yothers, G.; Deutsch, M.; Allegra, C.J.; Kahlenberg, M.S.; Baez-Diaz, L.; Ursiny, C.S.; Petrelli, N.J.; et al. Preoperative Multimodality Therapy Improves Disease-Free Survival in Patients with Carcinoma of the Rectum: NSABP R-03. J. Clin. Oncol. 2009, 27, 5124–5130. [Google Scholar] [CrossRef]
- Martin, S.T.; Heneghan, H.; Winter, D.C. Systematic review and meta-analysis of outcomes following pathological complete response to neoadjuvant chemoradiotherapy for rectal cancer. Br. J. Surg. 2012, 99, 918–928. [Google Scholar] [CrossRef]
- Song, C.; Chung, J.-H.; Kang, S.-B.; Kim, D.-W.; Oh, H.-K.; Lee, H.S.; Kim, J.W.; Lee, K.-W.; Kim, J.H.; Kim, J.-S. Impact of Tumor Regression Grade as a Major Prognostic Factor in Locally Advanced Rectal Cancer after Neoadjuvant Chemoradiotherapy: A Proposal for a Modified Staging System. Cancers 2018, 10, 319. [Google Scholar] [CrossRef]
- Tian, R.-F.; Li, X.-F.; Xu, C.; Wu, H.; Liu, L.; Wang, L.-H.; He, D.; Cao, K.; Cao, P.-G.; Ma, J.K.; et al. SiRNA targeting PFK1 inhibits proliferation and migration and enhances radiosensitivity by suppressing glycolysis in colorectal cancer. Am. J. Transl. Res. 2020, 12, 4923–4940. [Google Scholar]
- Kong, L.; Wei, Q.; Hu, X.; Chen, L.; Li, J. miR-193a-3p Promotes Radio-Resistance of Nasopharyngeal Cancer Cells by Targeting SRSF2 Gene and Hypoxia Signaling Pathway. Med. Sci. Monit. Basic Res. 2019, 25, 53–62. [Google Scholar] [CrossRef]
- Huang, D.; Zhu, X.; Wang, Y.; Yu, H.; Pu, Y. Long non-coding RNA FAM133B-2 represses the radio-resistance of nasopharyngeal cancer cells by targeting miR-34a-5p/CDK6 axis. Aging 2020, 12, 16936–16950. [Google Scholar] [CrossRef]
- Smith, M.P.W.; Zougman, A.; Cairns, D.A.; Wilson, M.; Wind, T.; Wood, S.L.; Thompson, D.; Messenger, M.P.; Mooney, A.; Selby, P.J.; et al. Serum aminoacylase-1 is a novel biomarker with potential prognostic utility for long-term outcome in patients with delayed graft function following renal transplantation. Kidney Int. 2013, 84, 1214–1225. [Google Scholar] [CrossRef]
- Wei, X.; Li, J.; Xie, H.; Ling, Q.; Wang, J.; Lu, D.; Zhou, L.; Xu, X.; Zheng, S. Proteomics-based identification of the tumor suppressor role of aminoacylase 1 in hepatocellular carcinoma. Cancer Lett. 2014, 351, 117–125. [Google Scholar] [CrossRef]
- Yu, B.; Liu, X.; Cao, X.; Zhang, M.; Chang, H. Study of the expression and function of ACY1 in patients with colorectal cancer. Oncol. Lett. 2017, 13, 2459–2464. [Google Scholar] [CrossRef]
- Chen, Z.; Gao, J.; Sun, J.; Wu, Z.; Wang, B. Aminoacylase 1 (ACY-1) Mediates the Proliferation and Migration of Neuroblastoma Cells in Humans Through the ERK/Transforming Growth Factor β (TGF-β) Signaling Pathways. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2020, 27, e928813-1–e928813-8. [Google Scholar] [CrossRef]
- Xu, Z.; Hu, Y.; Yu, Z. Effect of the ACY-1 gene on HER2 and TRAIL expression in rectal carcinoma. Exp. Ther. Med. 2021, 22, 817. [Google Scholar] [CrossRef]
- Chen, H.; Wang, W.; Xiao, C.; Xia, D.; Li, F.; Liu, S. ACY1 regulating PTEN/PI3K/AKT signaling in the promotion of non-small cell lung cancer progression. Ann. Transl. Med. 2021, 9, 1378. [Google Scholar] [CrossRef]
- Liu, R.; Deng, P.; Zhang, Y.; Wang, Y.; Peng, C. Circ_0082182 promotes oncogenesis and metastasis of colorectal cancer in vitro and in vivo by sponging miR-411 and miR-1205 to activate the Wnt/β-catenin pathway. World J. Surg. Oncol. 2021, 19, 51. [Google Scholar] [CrossRef]
- Sun, G.; Shang, Z.; Liu, W. SPP1 Regulates Radiotherapy Sensitivity of Gastric Adenocarcinoma via the Wnt/Beta-Catenin Pathway. J. Oncol. 2021, 2021, 1642852. [Google Scholar] [CrossRef]
- Huang, Y.; Sheng, H.; Xiao, Y.; Hu, W.; Zhang, Z.; Chen, Y.; Zhu, Z.; Wu, D.; Cao, C.; Sun, J. Wnt/β-catenin inhibitor ICG-001 enhances the antitumor efficacy of radiotherapy by increasing radiation-induced DNA damage and improving tumor immune microenvironment in hepatocellular carcinoma. Radiother. Oncol. 2021, 162, 34–44. [Google Scholar] [CrossRef]
- Clevers, H.; Nusse, R. Wnt/β-catenin signaling and disease. Cell 2012, 149, 1192–1205. [Google Scholar] [CrossRef]
- Nelson, W.J.; Nusse, R. Convergence of Wnt, ß-Catenin, and Cadherin Pathways. Science 2004, 303, 1483–1487. [Google Scholar] [CrossRef]
- Li, T.-H.; Zhao, B.-B.; Qin, C.; Wang, Y.-Y.; Li, Z.-R.; Cao, H.-T.; Yang, X.-Y.; Zhou, X.-T.; Wang, W.-B. IFIT1 modulates the proliferation, migration and invasion of pancreatic cancer cells via Wnt/β-catenin signaling. Cell. Oncol. 2021, 44, 1425–1437. [Google Scholar] [CrossRef]
- Xu, J.-Y.; Liu, F.-Y.; Liu, S.-X.; Xie, L.-Z.; Li, J.; Ma, Y.-T.; Han, F.-J. Plant-Derived Chinese Medicine Monomers on Ovarian Cancer via the Wnt/β-Catenin Signaling Pathway: Review of Mechanisms and Prospects. J. Oncol. 2021, 2021, 6852867. [Google Scholar] [CrossRef]
- Pouya, F.D.; Rasmi, Y.; Camci, I.Y.; Tutar, Y.; Nemati, M. Performance of capecitabine in novel combination therapies in colorectal cancer. J. Chemother. 2021, 33, 375–389. [Google Scholar] [CrossRef]
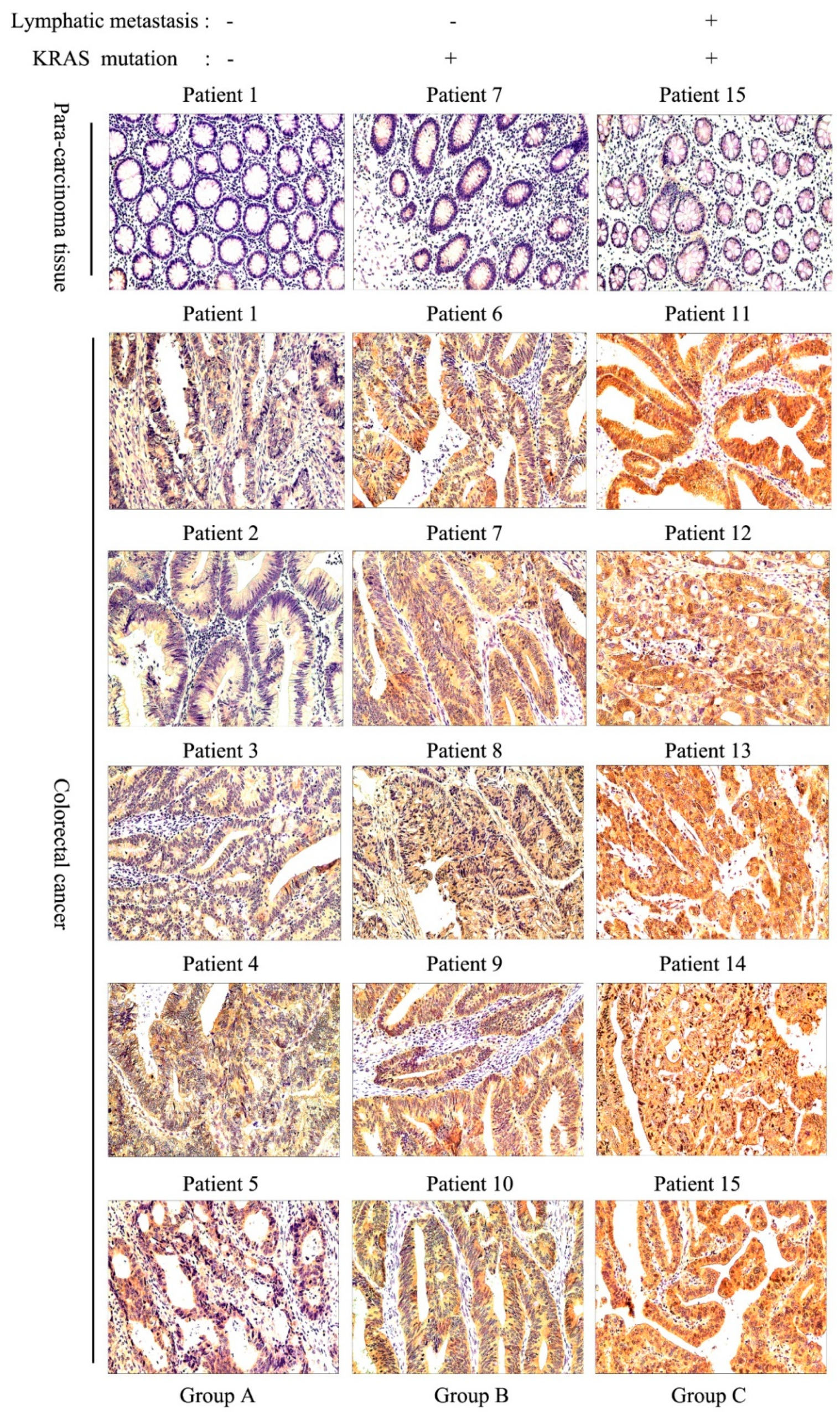


| Group | No. of Patient | Sex | Age | Lymphatic Metastasis | KRAS Mutation | TNM Stage | Pathological Grades |
|---|---|---|---|---|---|---|---|
| A | P1 | M | 63 | N | Wild type | II | Middle |
| P2 | F | 82 | N | Wild type | II | Middle | |
| P3 | M | 55 | N | Wild type | II | Middle-Low | |
| P4 | M | 67 | N | Wild type | II | Middle | |
| P5 | F | 43 | N | Wild type | I | Middle | |
| B | P6 | M | 67 | N | Mutant type | II | Middle |
| P7 | M | 48 | N | Mutant type | III | Middle-Low | |
| P8 | F | 49 | N | Mutant type | II | Middle-Low | |
| P9 | M | 67 | N | Mutant type | IV | Middle | |
| P10 | M | 75 | N | Mutant type | I | Middle | |
| C | P11 | M | 49 | P | Mutant type | III | Middle |
| P12 | M | 55 | P | Mutant type | IV | Middle | |
| P13 | M | 69 | P | Mutant type | IV | Low | |
| P14 | M | 61 | P | Mutant type | IV | Low | |
| P15 | F | 57 | P | Mutant type | III | Middle-Low |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shan, W.; Dai, C.; Zhang, H.; Han, D.; Yi, Q.; Xia, B. ACY1 Downregulation Enhances the Radiosensitivity of Cetuximab-Resistant Colorectal Cancer by Inactivating the Wnt/β-Catenin Signaling Pathway. Cancers 2022, 14, 5704. https://doi.org/10.3390/cancers14225704
Shan W, Dai C, Zhang H, Han D, Yi Q, Xia B. ACY1 Downregulation Enhances the Radiosensitivity of Cetuximab-Resistant Colorectal Cancer by Inactivating the Wnt/β-Catenin Signaling Pathway. Cancers. 2022; 14(22):5704. https://doi.org/10.3390/cancers14225704
Chicago/Turabian StyleShan, Wulin, Chunyang Dai, Huanhuan Zhang, Dan Han, Qiyi Yi, and Bairong Xia. 2022. "ACY1 Downregulation Enhances the Radiosensitivity of Cetuximab-Resistant Colorectal Cancer by Inactivating the Wnt/β-Catenin Signaling Pathway" Cancers 14, no. 22: 5704. https://doi.org/10.3390/cancers14225704
APA StyleShan, W., Dai, C., Zhang, H., Han, D., Yi, Q., & Xia, B. (2022). ACY1 Downregulation Enhances the Radiosensitivity of Cetuximab-Resistant Colorectal Cancer by Inactivating the Wnt/β-Catenin Signaling Pathway. Cancers, 14(22), 5704. https://doi.org/10.3390/cancers14225704






