Machine Learning in the Classification of Pediatric Posterior Fossa Tumors: A Systematic Review
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Inclusion and Exclusion Criteria
2.2. Data Extraction
2.3. Gold Standard Comparison
3. Results
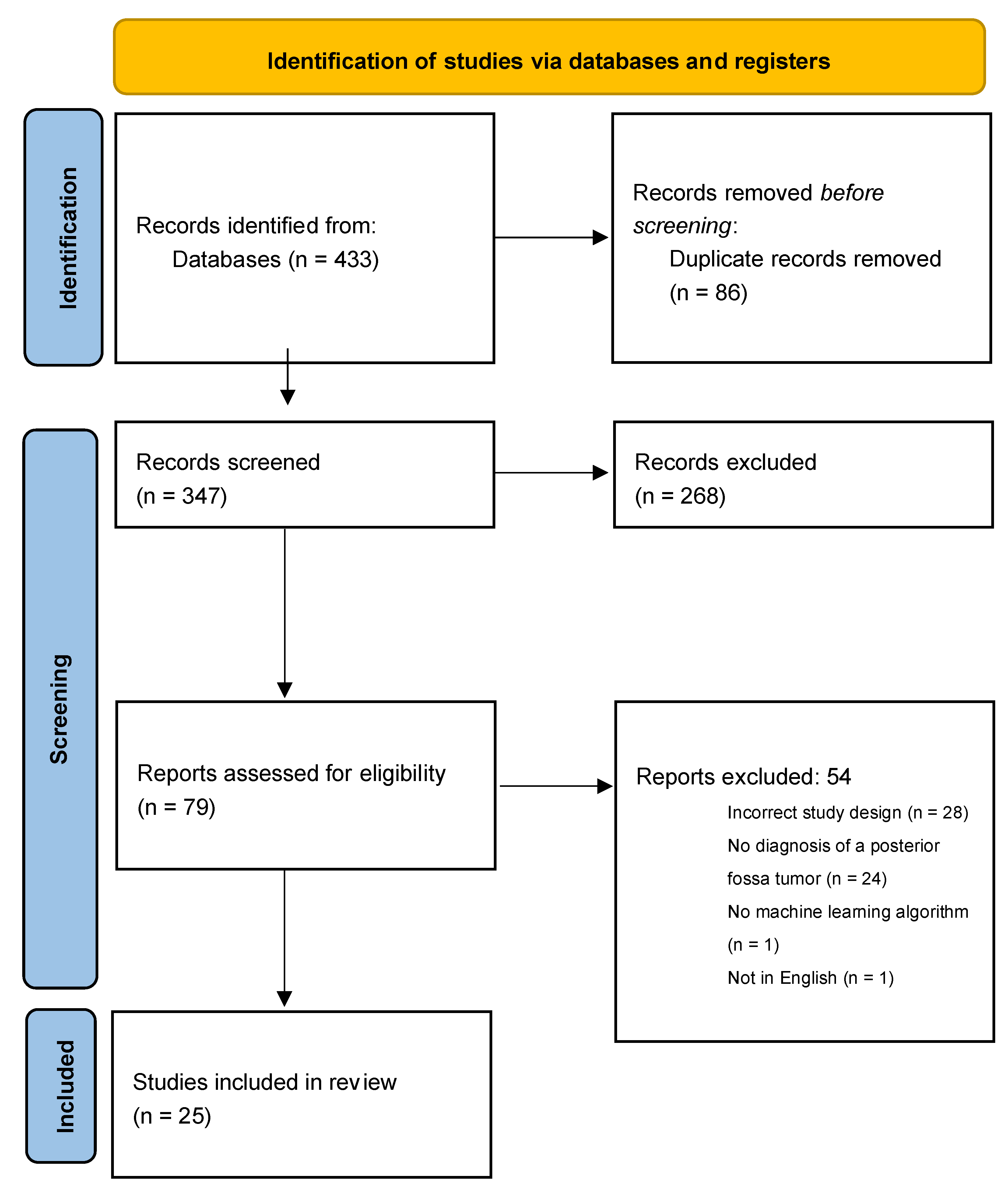
3.1. Search Results
3.2. Algorithm Study Parameters and Design
3.3. High-Yield Features
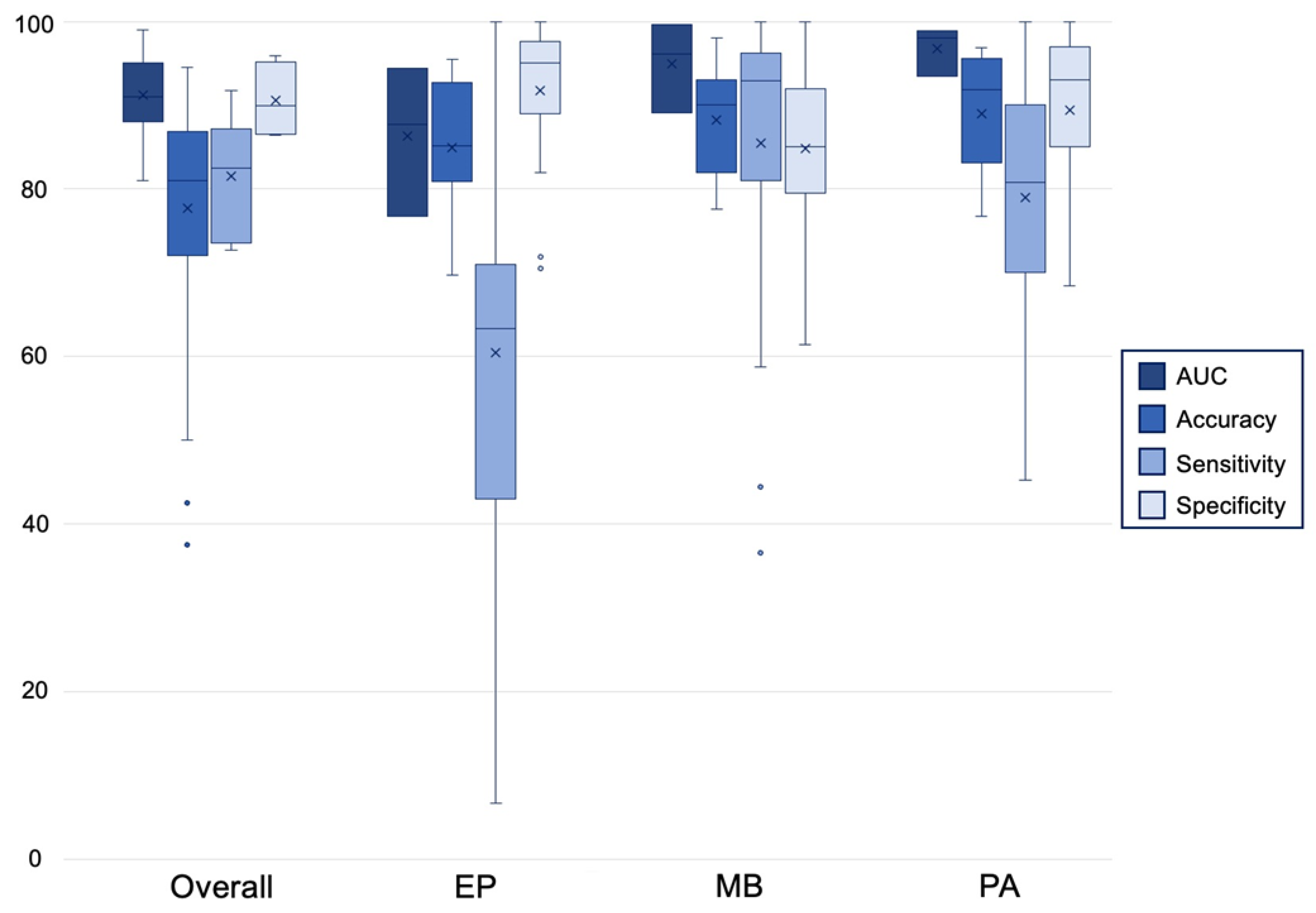
3.4. Algorithm Performance
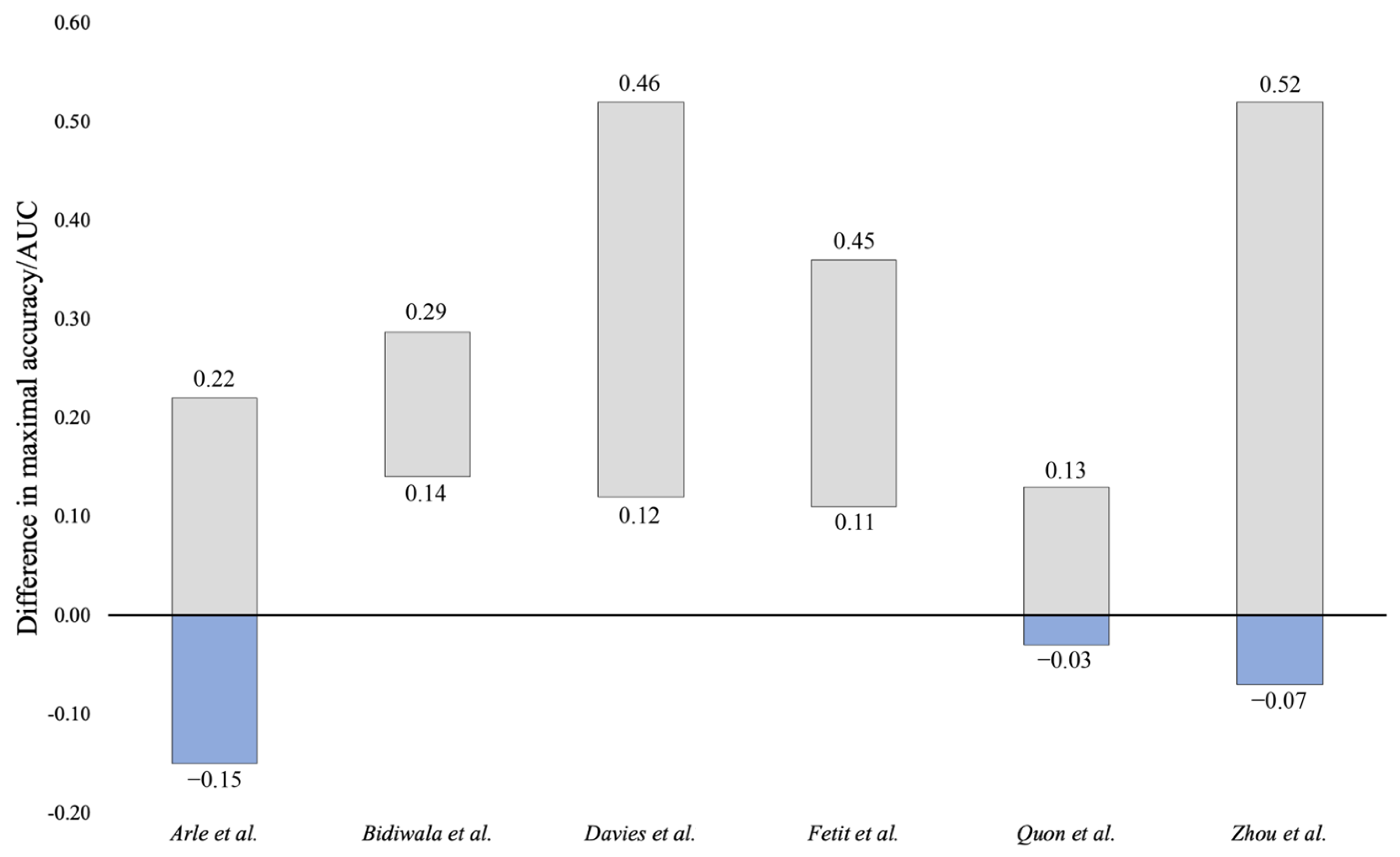
3.5. Comparison to Neuroradiologist
3.6. Observed Limitations
4. Discussion
4.1. Algorithm Selection
4.2. Objective of Machine Learning Application
4.3. Translation to Clinical Practice
4.4. Algorithm Limitations
4.5. Posterior Fossa Algorithm Recommendations of Best Practice
4.6. Limitations & Future Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- O′Brien, D.F.; Caird, J.; Kennedy, M.; Roberts, G.A.; Marks, J.C.; Allcutt, D.A. Posterior fossa tumours in childhood: Evaluation of presenting clinical features. Ir. Med. J. 2001, 94, 52–53. [Google Scholar] [PubMed]
- Bright, C.; Reulen, R.; Fidler-Benaoudia, M.; Guha, J.; Henson, K.; Wong, K.; Kelly, J.; Frobisher, C.; Winter, D.; Hawkins, M. Cerebrovascular complications in 208,769 5-year survivors of cancer diagnosed aged 15-39 years using hospital episode statistics: The population-based Teenage and Young Adult Cancer Survivor Study (TYACSS). Eur. J. Cancer Care 2015, 24, 9. [Google Scholar]
- Lannering, B.; Marky, I.; Nordborg, C. Brain tumors in childhood and adolescence in west Sweden 1970-1984. Epidemiology and survival. Cancer 1990, 66, 604–609. [Google Scholar] [CrossRef]
- Prasad, K.S.V.; Ravi, D.; Pallikonda, V.; Raman, B.V.S. Clinicopathological Study of Pediatric Posterior Fossa Tumors. J. Pediatr. Neurosci. 2017, 12, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Shay, V.; Fattal-Valevski, A.; Beni-Adani, L.; Constantini, S. Diagnostic delay of pediatric brain tumors in Israel: A retrospective risk factor analysis. Childs Nerv. Syst. 2012, 28, 93–100. [Google Scholar] [CrossRef]
- Culleton, S.; McKenna, B.; Dixon, L.; Taranath, A.; Oztekin, O.; Prasad, C.; Siddiqui, A.; Mankad, K. Imaging pitfalls in paediatric posterior fossa neoplastic and non-neoplastic lesions. Clin. Radiol. 2021, 76, e319–e391. [Google Scholar] [CrossRef]
- Kerleroux, B.; Cottier, J.P.; Janot, K.; Listrat, A.; Sirinelli, D.; Morel, B. Posterior fossa tumors in children: Radiological tips & tricks in the age of genomic tumor classification and advance MR technology. J. Neuroradiol. 2020, 47, 46–53. [Google Scholar] [CrossRef]
- Hwang, E.J.; Park, S.; Jin, K.N.; Kim, J.I.; Choi, S.Y.; Lee, J.H.; Goo, J.M.; Aum, J.; Yim, J.J.; Cohen, J.G.; et al. Development and Validation of a Deep Learning-Based Automated Detection Algorithm for Major Thoracic Diseases on Chest Radiographs. JAMA Netw. Open 2019, 2, e191095. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, J.Y.; Kim, G.H.; Kang, D.; Kim, I.J.; Seo, J.; Andrews, J.R.; Park, C.M. Clinical Validation of a Deep Learning Algorithm for Detection of Pneumonia on Chest Radiographs in Emergency Department Patients with Acute Febrile Respiratory Illness. J. Clin. Med. 2020, 9, 1981. [Google Scholar] [CrossRef]
- Pringle, C.; Kilday, J.-P.; Kamaly-Asl, I.; Stivaros, S.M. The role of artificial intelligence in paediatric neuroradiology. Pediatr. Radiol. 2022, 52, 2159–2172. [Google Scholar] [CrossRef]
- Abdelaziz Ismael, S.A.; Mohammed, A.; Hefny, H. An enhanced deep learning approach for brain cancer MRI images classification using residual networks. Artif. Intell. Med. 2020, 102, 101779. [Google Scholar] [CrossRef] [PubMed]
- Buchlak, Q.D.; Esmaili, N.; Leveque, J.C.; Bennett, C.; Farrokhi, F.; Piccardi, M. Machine learning applications to neuroimaging for glioma detection and classification: An artificial intelligence augmented systematic review. J. Clin. Neurosci. 2021, 89, 177–198. [Google Scholar] [CrossRef] [PubMed]
- Nakamoto, T.; Takahashi, W.; Haga, A.; Takahashi, S.; Kiryu, S.; Nawa, K.; Ohta, T.; Ozaki, S.; Nozawa, Y.; Tanaka, S.; et al. Prediction of malignant glioma grades using contrast-enhanced T1-weighted and T2-weighted magnetic resonance images based on a radiomic analysis. Sci. Rep. 2019, 9, 19411. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.; Wang, L.; Ji, B.; Lei, Y.; Ali, A.; Liu, T.; Curran, W.J.; Mao, H.; Yang, X. Machine-learning based classification of glioblastoma using delta-radiomic features derived from dynamic susceptibility contrast enhanced magnetic resonance images: Introduction. Quant. Imaging Med. Surg. 2019, 9, 1201–1213. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Arle, J.E.; Morriss, C.; Wang, Z.J.; Zimmerman, R.A.; Phillips, P.G.; Sutton, L.N. Prediction of posterior fossa tumor type in children by means of magnetic resonance image properties, spectroscopy, and neural networks. J. Neurosurg. 1997, 86, 755–761. [Google Scholar] [CrossRef]
- Bidiwala, S.; Pittman, T. Neural network classification of pediatric posterior fossa tumors using clinical and imaging data. Pediatr. Neurosurg. 2004, 40, 8–15. [Google Scholar] [CrossRef]
- Davies, N.P.; Rose, H.E.L.; Manias, K.A.; Natarajan, K.; Abernethy, L.J.; Oates, A.; Janjua, U.; Davies, P.; MacPherson, L.; Arvanitis, T.N.; et al. Added value of magnetic resonance spectroscopy for diagnosing childhood cerebellar tumours. NMR Biomed. 2022, 35, e4630. [Google Scholar] [CrossRef]
- Dong, J.; Li, L.; Liang, S.; Zhao, S.; Zhang, B.; Meng, Y.; Zhang, Y.; Li, S. Differentiation Between Ependymoma and Medulloblastoma in Children with Radiomics Approach. Acad. Radiol. 2021, 28, 318–327. [Google Scholar] [CrossRef]
- Dong, J.; Li, S.; Li, L.; Liang, S.; Zhang, B.; Meng, Y.; Zhang, X.; Zhang, Y.; Zhao, S. Differentiation of paediatric posterior fossa tumours by the multiregional and multiparametric MRI radiomics approach: A study on the selection of optimal multiple sequences and multiregions. Br. J. Radiol. 2022, 95, 20201302. [Google Scholar] [CrossRef]
- Fetit, A.E.; Novak, J.; Peet, A.C.; Arvanitits, T.N. Three-dimensional textural features of conventional MRI improve diagnostic classification of childhood brain tumours. NMR Biomed. 2015, 28, 1174–1184. [Google Scholar] [CrossRef] [PubMed]
- Grist, J.T.; Withey, S.; MacPherson, L.; Oates, A.; Powell, S.; Novak, J.; Abernethy, L.; Pizer, B.; Grundy, R.; Bailey, S.; et al. Distinguishing between paediatric brain tumour types using multi-parametric magnetic resonance imaging and machine learning: A multi-site study. NeuroImage Clin. 2020, 25, 102172. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez Gutierrez, D.; Awwad, A.; Meijer, L.; Manita, M.; Jaspan, T.; Dineen, R.A.; Grundy, R.G.; Auer, D.P. Metrics and textural features of MRI diffusion to improve classification of pediatric posterior fossa tumors. AJNR Am. J. Neuroradiol. 2014, 35, 1009–1015. [Google Scholar] [CrossRef]
- Li, M.; Wang, H.; Shang, Z.; Yang, Z.; Zhang, Y.; Wan, H. Ependymoma and pilocytic astrocytoma: Differentiation using radiomics approach based on machine learning. J. Clin. Neurosci. 2020, 78, 175–180. [Google Scholar] [CrossRef]
- Li, M.M.; Shang, Z.G.; Yang, Z.L.; Zhang, Y.; Wan, H. Machine learning methods for MRI biomarkers analysis of pediatric posterior fossa tumors. Biocybern. Biomed. Eng. 2019, 39, 765–774. [Google Scholar] [CrossRef]
- Novak, J.; Zarinabad, N.; Rose, H.; Arvanitis, T.; MacPherson, L.; Pinkey, B.; Oates, A.; Hales, P.; Grundy, R.; Auer, D.; et al. Classification of paediatric brain tumours by diffusion weighted imaging and machine learning. Sci. Rep. 2021, 11, 2987. [Google Scholar] [CrossRef]
- Orphanidou-Vlachou, E.; Vlachos, N.; Davies, N.P.; Arvanitis, T.N.; Grundy, R.G.; Peet, A.C. Texture analysis of T1- and T2-weighted MR images and use of probabilistic neural network to discriminate posterior fossa tumours in children. NMR Biomed. 2014, 27, 632–639. [Google Scholar] [CrossRef]
- Payabvash, S.; Aboian, M.; Tihan, T.; Cha, S. Machine Learning Decision Tree Models for Differentiation of Posterior Fossa Tumors Using Diffusion Histogram Analysis and Structural MRI Findings. Front. Oncol. 2020, 10, 71. [Google Scholar] [CrossRef]
- Wang, S.; Wang, G.; Zhang, W.; He, J.; Sun, W.; Yang, M.; Sun, Y.; Peet, A. MRI-based whole-tumor radiomics to classify the types of pediatric posterior fossa brain tumor. Neurochirurgie 2022. [Google Scholar] [CrossRef]
- Zarinabad, N.; Abernethy, L.J.; Avula, S.; Davies, N.P.; Rodriguez Gutierrez, D.; Jaspan, T.; MacPherson, L.; Mitra, D.; Rose, H.E.L.; Wilson, M.; et al. Application of pattern recognition techniques for classification of pediatric brain tumors by in vivo 3T (1) H-MR spectroscopy-A multi-center study. Magn. Reson. Med. 2018, 79, 2359–2366. [Google Scholar] [CrossRef]
- Quon, J.L.; Bala, W.; Chen, L.C.; Wright, J.; Kim, L.H.; Han, M.; Shpanskaya, K.; Lee, E.H.; Tong, E.; Iv, M.; et al. Deep Learning for Pediatric Posterior Fossa Tumor Detection and Classification: A Multi-Institutional Study. AJNR Am. J. Neuroradiol. 2020, 41, 1718–1725. [Google Scholar] [CrossRef]
- Zarinabad, N.; Wilson, M.; Gill, S.K.; Manias, K.A.; Davies, N.P.; Peet, A.C. Multiclass imbalance learning: Improving classification of pediatric brain tumors from magnetic resonance spectroscopy. Magn. Reson. Med. 2017, 77, 2114–2124. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Tam, L.; Wright, J.; Mohammadzadeh, M.; Han, M.; Chen, E.; Wagner, M.; Nemalka, J.; Lai, H.; Eghbal, A.; et al. Radiomics Can Distinguish Pediatric Supratentorial Embryonal Tumors, High-Grade Gliomas, and Ependymomas. AJNR Am. J. Neuroradiol. 2022, 43, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wong, S.W.; Lummus, S.; Han, M.; Radmanesh, A.; Ahmadian, S.S.; Prolo, L.M.; Lai, H.; Eghbal, A.; Oztekin, O.; et al. Radiomic Phenotypes Distinguish Atypical Teratoid/Rhabdoid Tumors from Medulloblastoma. AJNR Am. J. Neuroradiol. 2021, 42, 1702–1708. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Wong, S.W.; Wright, J.N.; Toescu, S.; Mohammadzadeh, M.; Han, M.; Lummus, S.; Wagner, M.W.; Yecies, D.; Lai, H.; et al. Machine Assist for Pediatric Posterior Fossa Tumor Diagnosis: A Multinational Study. Neurosurgery 2021, 89, 892–900. [Google Scholar] [CrossRef]
- Zhao, D.; Grist, J.T.; Rose, H.E.L.; Davies, N.P.; Wilson, M.; MacPherson, L.; Abernethy, L.J.; Avula, S.; Pizer, B.; Gutierrez, D.R.; et al. Metabolite selection for machine learning in childhood brain tumour classification. NMR Biomed. 2022, 35, e4673. [Google Scholar] [CrossRef]
- Zhou, H.; Hu, R.; Tang, O.; Hu, C.; Tang, L.; Chang, K.; Shen, Q.; Wu, J.; Zou, B.; Xiao, B.; et al. Automatic Machine Learning to Differentiate Pediatric Posterior Fossa Tumors on Routine MR Imaging. AJNR Am. J. Neuroradiol. 2020, 41, 1279–1285. [Google Scholar] [CrossRef]
- Danielsson, A.; Nemes, S.; Tisell, M.; Lannering, B.; Nordborg, C.; Sabel, M.; Carén, H. MethPed: A DNA methylation classifier tool for the identification of pediatric brain tumor subtypes. Clin. Epigenetics 2015, 7, 62. [Google Scholar] [CrossRef]
- Hollon, T.C.; Lewis, S.; Pandian, B.; Niknafs, Y.S.; Garrard, M.R.; Garton, H.; Maher, C.O.; McFadden, K.; Snuderl, M.; Lieberman, A.P.; et al. Rapid Intraoperative Diagnosis of Pediatric Brain Tumors Using Stimulated Raman Histology. Cancer Res. 2018, 78, 278–289. [Google Scholar] [CrossRef]
- Leslie, D.G.; Kast, R.E.; Poulik, J.M.; Rabah, R.; Sood, S.; Auner, G.W.; Klein, M.D. Identification of pediatric brain neoplasms using Raman spectroscopy. Pediatr. Neurosurg. 2012, 48, 109–117. [Google Scholar] [CrossRef]
- Zhang, G.P. Neural networks for classification: A survey. IEEE Trans. Syst. Man Cybern. Part C 2000, 30, 451–462. [Google Scholar] [CrossRef]
- Zhang, S. Challenges in KNN Classification. IEEE Trans. Knowl. Data Eng. 2022, 34, 4663–4675. [Google Scholar] [CrossRef]
- Hengl, T.; Nussbaum, M.; Wright, M.; Heuvelink, G.; Graeler, B. Random forest as a generic framework for predictive modeling of spatial and spatio-temporal variables. PeerJ 2018, 6, e5518. [Google Scholar] [CrossRef] [PubMed]
- Specht, D.F. Probabilistic neural networks. Neural. Netw. 1990, 3, 109–118. [Google Scholar] [CrossRef]
- Othman, M.F.; Basri, M.A.M. Probabilistic Neural Network for Brain Tumor Classification. In Proceedings of the 2011 Second International Conference on Intelligent Systems, Modelling and Simulation, Phnom Penh, Cambodia, 25–27 January 2011; pp. 136–138. [Google Scholar]
- Kaviani, P.; Dhotre, S. Short Survey on Naive Bayes Algorithm. Int. J. Adv. Res. Comput. Sci. Manag. 2017, 4, 143–147. [Google Scholar]
- Abdullah, N.; Ngah, U.K.; Aziz, S.A. Image classification of brain MRI using support vector machine. In Proceedings of the 2011 IEEE International Conference on Imaging Systems and Techniques, 17–18 May 2011; pp. 242–247. [Google Scholar]
- Drucker, H.; Donghui, W.; Vapnik, V.N. Support vector machines for spam categorization. IEEE Trans. Neural Netw. 1999, 10, 1048–1054. [Google Scholar] [CrossRef]
- Gholami, R.; Fakhari, N. Chapter 27–Support Vector Machine: Principles, Parameters, and Applications. In Handbook of Neural Computation; Samui, P., Sekhar, S., Balas, V.E., Eds.; Academic Press: Cambridge, UK, 2017; pp. 515–535. [Google Scholar]
- Abujudeh, H.H.; Boland, G.W.; Kaewlai, R.; Rabiner, P.; Halpern, E.F.; Gazelle, G.S.; Thrall, J.H. Abdominal and pelvic computed tomography (CT) interpretation: Discrepancy rates among experienced radiologists. Eur. Radiol. 2010, 20, 1952–1957. [Google Scholar] [CrossRef]
- Yin, P.; Mao, N.; Zhao, C.; Wu, J.; Sun, C.; Chen, L.; Hong, N. Comparison of radiomics machine-learning classifiers and feature selection for differentiation of sacral chordoma and sacral giant cell tumour based on 3D computed tomography features. Eur. Radiol. 2019, 29, 1841–1847. [Google Scholar] [CrossRef]
- Kim, Y.; Cho, H.H.; Kim, S.T.; Park, H.; Nam, D.; Kong, D.S. Radiomics features to distinguish glioblastoma from primary central nervous system lymphoma on multi-parametric MRI. Neuroradiology 2018, 60, 1297–1305. [Google Scholar] [CrossRef]
- Suh, H.B.; Choi, Y.S.; Bae, S.; Ahn, S.S.; Chang, J.H.; Kang, S.G.; Kim, E.H.; Kim, S.H.; Lee, S.K. Primary central nervous system lymphoma and atypical glioblastoma: Differentiation using radiomics approach. Eur. Radiol. 2018, 28, 3832–3839. [Google Scholar] [CrossRef]
- Aerts, H.J.W.L.; Velazquez, E.R.; Leijenaar, R.T.H.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef] [PubMed]
- Muzumdar, D.; Ventureyra, E.C.G. Treatment of posterior fossa tumors in children. Expert Rev. Neurother. 2010, 10, 525–546. [Google Scholar] [CrossRef] [PubMed]
- McDermott, M.B.A.; Wang, S.; Marinsek, N.; Ranganath, R.; Foschini, L.; Ghassemi, M. Reproducibility in machine learning for health research: Still a ways to go. Sci. Transl. Med. 2021, 13, eabb1655. [Google Scholar] [CrossRef] [PubMed]
- Gordillo, N.; Montseny, E.; Sobrevilla, P. State of the art survey on MRI brain tumor segmentation. Magn. Reson. Imaging 2013, 31, 1426–1438. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhao, Z.; Wu, W.; Lin, Y.; Wang, M. Automatic glioma segmentation based on adaptive superpixel. BMC Med. Imaging 2019, 19, 73. [Google Scholar] [CrossRef] [PubMed]
- Traverso, A.; Wee, L.; Dekker, A.; Gillies, R. Repeatability and Reproducibility of Radiomic Features: A Systematic Review. Int. J. Radiat. Oncol. Biol. Phys. 2018, 102, 1143–1158. [Google Scholar] [CrossRef]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Gronau, Q.F.; Wagenmakers, E.J. Limitations of Bayesian Leave-One-Out Cross-Validation for Model Selection. Comput. Brain Behav. 2019, 2, 1–11. [Google Scholar] [CrossRef]
- Davis, F.G.; McCarthy, B.J. Epidemiology of brain tumors. Curr. Opin. Neurol. 2000, 13, 635–640. [Google Scholar] [CrossRef]
- Bej, S.; Davtyan, N.; Wolfien, M.; Nassar, M.; Wolkenhauer, O. LoRAS: An oversampling approach for imbalanced datasets. Mach. Learn. 2021, 110, 279–301. [Google Scholar] [CrossRef]
- Hashimoto, D.A.; Witkowski, E.; Gao, L.; Meireles, O.; Rosman, G. Artificial Intelligence in Anesthesiology: Current Techniques, Clinical Applications, and Limitations. Anesthesiology 2020, 132, 379–394. [Google Scholar] [CrossRef] [PubMed]
- Kassner, A.; Thornhill, R.E. Texture analysis: A review of neurologic MR imaging applications. AJNR Am. J. Neuroradiol. 2010, 31, 809–816. [Google Scholar] [CrossRef] [PubMed]
- Collewet, G.; Strzelecki, M.; Mariette, F. Influence of MRI acquisition protocols and image intensity normalization methods on texture classification. Magn. Reson. Imaging 2004, 22, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Lotan, E.; Jain, R.; Razavian, N.; Fatterpekar, G.M.; Lui, Y.W. State of the Art: Machine Learning Applications in Glioma Imaging. Am. J. Roentgenol. 2018, 212, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med. Imaging 2015, 34, 1993–2024. [Google Scholar] [CrossRef]
- Azizi, S.; Mustafa, B.; Ryan, F.; Beaver, Z.; Freyberg, J.; Deaton, J.; Loh, A.; Karthikesalingam, A.; Kornblith, S.; Chen, T.; et al. Big Self-Supervised Models Advance Medical Image Classification. In Proceedings of the 2021 IEEE/CVF International Conference on Computer Vision (ICCV), Montreal, QC, Canada, 10–17 October 2021; pp. 3458–3468. [Google Scholar]
- Lin, Y.; Li, Q.; Yang, B.; Yan, Z.; Tan, H.; Chen, Z. Improving speech recognition models with small samples for air traffic control systems. Neurocomputing 2021, 445, 287–297. [Google Scholar] [CrossRef]
- Yan, K.; Wang, X.; Lu, L.; Summers, R. DeepLesion: Automated mining of large-scale lesion annotations and universal lesion detection with deep learning. J. Med. Imaging 2018, 5, 036501. [Google Scholar] [CrossRef]
- Summers, R.M. NIH Clinical Center Provides One of the Largest Publicly Available Chest X-ray Datasets to Scientific Community. Available online: https://www.nih.gov/news-events/news-releases/nih-clinical-center-provides-one-largest-publicly-available-chest-x-ray-datasets-scientific-community (accessed on 22 September 2022).
- Murdoch, W.J.; Singh, C.; Kumbier, K.; Abbasi-Asl, R.; Yu, B. Definitions, methods, and applications in interpretable machine learning. Proc. Natl. Acad. Sci. USA 2019, 116, 22071–22080. [Google Scholar] [CrossRef]
- Chen, H.; Gomez, C.; Huang, C.-M.; Unberath, M. Explainable medical imaging AI needs human-centered design: Guidelines and evidence from a systematic review. NPJ Digit. Med. 2022, 5, 156. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Erion, G.; Chen, H.; DeGrave, A.; Prutkin, J.M.; Nair, B.; Katz, R.; Himmelfarb, J.; Bansal, N.; Lee, S.-I. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell. 2020, 2, 56–67. [Google Scholar] [CrossRef]
- Kompa, B.; Snoek, J.; Beam, A.L. Second opinion needed: Communicating uncertainty in medical machine learning. NPJ Digit. Med. 2021, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H. Artificial intelligence in radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef] [PubMed]
| Paper | Tumor Type | Imaging/Assay Used | Prospective vs. Retrospective | Study Population | # of Sites | Ground Truth | Training Set | Validation Set | Image Segmentation Method | Normalization Used | Feature Selection Used | Number of Features Extracted | Texture Analysis Employed | Deep Learning Architecture |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Radiographic Algorithms | ||||||||||||||
| Arle et al., 1997 [16] | AS, EP, PNET | NC-T2MR, MR-spectroscopy | Prospective | 10 AS, 7 EP, 16 PNET | 1 | Histologic diagnosis | 150 * | 9 | Manual | No | No | 20 | No | NN |
| Bidiwala et al., 2004 [17] | EP, MB, PA, | CE-T1MR, CE-T2MR | Retrospective | 4 EP, 15 MB, 14 PA | 1 | Histologic diagnosis | 32 | 1 (× 33) # | Manual | Yes | No | 36 | No | NN |
| Davies et al., 2022 [18] | EP, MB, PA | NC-T1MR, NC-T2MR, DWI, MR-spectroscopy | Prospective | 7 EP, 32 MB, 28 PA | 1 | Histologic diagnosis | 34 | 33 | Manual | Yes | No | 19 | No | Multivariate classifier w/bootstrap cross-validation |
| Dong et al., 2021 [19] | EP, MB | CE-T1MR, DWI | Retrospective | 24 EP, 27 MB | 1 | Histologic diagnosis | ~46 (90% of cases) | ~5 (~10% of cases) | Semi-automatic | Yes | Yes | 188 | Yes | Adaptive boosting w/3 classifiers: kNN, RF, SVM |
| Dong et al., 2022 [20] | EP, MB, PA | NC-T1MR, NC-T2MR, CE-T1MR, FLAIR-MR, DWI | Retrospective | 32 EP, 67 MB, 37 PA | 1 | Histologic diagnosis | 106 | 30 | Semi-automatic | Yes | Yes | 11,958 | No | SVM |
| Fetit et al., 2015 [21] | EP, MB, PA | NC-T1MR, NC-T2MR | Retrospective | 7 EP, 21 MB, 20 PA | 1 | Histologic diagnosis | 47 | 1 (×48) # | Semi-automatic | Yes | Yes | 2D—454 3D—566 | Yes | 6 classifiers: NB, kNN, classification tree, SVM, ANN, LR |
| Grist et al., 2020 [22] | EP, MB, PA | NC-T1MR, NC-T2MR, CE-T1MR, FLAIR-MR, DWI, DSC-MR | Prospective | 10 EP, 17 MB, 22 PA | 4 | Histologic diagnosis | - | - | Manual | Yes | Yes | Not reported | No | 4 classifiers: NN, RF, SVM, kNN |
| Li et al., 2019 [25] | EP, MB | NC-T1MR, NC-T2MR | Retrospective | 58 patients, breakdown unspecified | 1 | Histologic diagnosis | ~41 (70%) | ~17 (30%) | Manual | Yes | Yes | 300 | Yes | Bagging and boosting w/9 classifiers: kNN, SVM, NN, classification and regression trees, RSM, ELM, NB, RF, partial LSR |
| Li et al., 2020 [24] | EP, PA | NC-T1MR, NC-T2MR | Retrospective | 45 patients, breakdown unspecified | 1 | Histologic diagnosis | ~32 (70%) | ~13 (30%) | Manual | No | Yes | 300 | Yes | SVM |
| Novak et al., 2021 [26] | ATRT, EP, LGT MB, PA | DWI | Retrospective | 4 ATRT, 26 EP, 3 LGT 55 MB, 36 PA | 5 | Histologic diagnosis | - | - | Manual | Yes | Yes | Not reported | No | 2 classifiers: NB, RF |
| Orphanidou-Vlachou et al., 2014 [27] | EP, MB, PA | NC-T1MR, NC-T2MR | Retrospective | 5 EP, 21 MB, 14 PA | 1 | Histologic diagnosis | - | - | Manual | Yes | Yes | 279 | Yes | 2 classifiers: LDA, PNN |
| Payabvash et al., 2020 [28] | AAS, ATRT, AXA, CPP, EP, GBM, GG, GNT, HB, LGG, lymphoma, MB metastases, PA, SEP | DWI | Retrospective | 7 AAS, 6 ATRT, 1 AXA, 4 CPP, 27 EP, 6 GBM, 1 GG, 2 GNT, 44 HB, 10 LGG, 8 lymphoma, 26 MB 65 metastases, 43 PA, 6 SEP | 1 | Histologic diagnosis | 199 | 49 | Manual | Yes | No | 24 | No | 4 classifiers: NB, RF, SVM, NN |
| Quon et al., 2020 [31] | DMG, EP, MB, PA | NC-T1MR, NC-T2MR, DWI | Retrospective | 122 DMG, 88 EP, 272 MB, 135 PA | 5 | Histologic diagnosis | 527 (scans) | 212 (scans) | N/A | Yes | No | Not reported | No | Modified ResNet architecture |
| Rodriguez et al., 2014 [23] | EP, MB, PA | NC-T1MR, NC-T2MR, DWI | Retrospective | 7 EP, 17 MB, 16 PA | Multiple | Histologic diagnosis | - | - | Manual | Yes | Yes | 183 | Yes | SVM |
| Wang et al., 2022 [29] | EP, MB, PA | NC-T1MR, NC-T2MR, DWI | Retrospective | 13 EP, 59 MB, 27 PA | 1 | Histologic diagnosis | 70 | 20 | Manual | Yes | Yes | 315 | Yes | RF |
| Zarinabad et al., 2017 [32] | EP, MB, PA | NC-T1MR, NC-T2MR, MR-spectroscopy | Retrospective | 10 EP, 38 MB, 42 PA | 1 | Histologic diagnosis | - | - | Automatic w/manual review | No | Yes | 17 | No | Adaptive boosting w/4 classifiers: NB, SVM, ANN, LDA |
| Zarinabad et al., 2018 [30] | EP, MB, PA | MR-spectroscopy | Retrospective | 4 EP, 17 MB, 20 PA | 4 | Histologic diagnosis | 37 | 4 | Manual | No | Yes | 19 | No | 3 classifiers: LDA, SVM, RF |
| Zhang et al., 2021 [34] | ATRT, MB | CE-T1MR, NC-T2MR | Retrospective | 48 ATRT, 96 MB | 7 | Histologic diagnosis | 108 | 36 | Manual | No | Yes | 1800 | Yes | Extreme gradient boosting w/5 classifiers: SVM, LR, kNN, RF, NN |
| Zhang et al., 2021 [35] | EP, MB, PA | CE-T1MR, CE-T2MR | Retrospective | 97 EP, 274 MB, 156 PA | Multiple | Histologic diagnosis | 395 | 132 | Manual | No | Yes | 1800 | No | Extreme gradient boosting w/5 classifiers: SVM, LR, kNN, RF, NN |
| Zhang et al., 2022 [33] | EP, HGG, SET | CE-T1MR, NC-T2MR | Retrospective | 54 EP, 127 HGG, 50 SET | 7 | Histologic diagnosis | 173 | 58 | Manual | Yes | Yes | 1800 | Yes | Extreme gradient boosting w/binary and single-stage multiclass classifier: SVM, LR, kNN, RF, NN |
| Zhao et al., 2022 [36] | EP, MB, PA | CE-T1MR, NC-T2MR, DWI, MR-spectroscopy | Prospective | 17 EP, 48 MB, 60 PA | 4 | Histologic diagnosis | - | 116 | Manual | Yes | Yes | 15 | No | 5 classifiers: NB, LDA, SVM, kNN, multinomial log-linear model fitting via NN |
| Zhou et al., 2020 [37] | EP, MB, PA | CE-T1MR, NC-T2MR, DWI | Retrospective | 70 EP, 111 MB, 107 PA | 4 | Histologic diagnosis | 202 | 86 | Manual | Yes | Yes | 3087 | Yes | Used tree-based pipeline optimization tool to find optimal architecture using 8 classifiers w/bagging and boosting: NN, decision tree, NB, RF, SVM, LDA, kNN, generalized linear models |
| Molecular Algorithms | ||||||||||||||
| Danielsson et al., 2015 [38] | EP, ETMR, DIPG, GBM, MB, PA | Illumina 450K methylation array data | Retrospective | 48 EP, 10 ETMR, 28 DIPG, 178 GBM, 238 MB, 58 PA | Multiple | Histologic diagnosis | 472 | 18, 28 separately | N/A | No | Yes | 900 | No | 3 classifiers: RF, LDA, stochastic generalized boosted models |
| Hollon et al., 2018 [39] | AS, chordoma, CPP, DMG, EP, ET, germinoma, GG, HB, MB, PA | Microscope slides | Prospective | 33 patients, breakdown unspecified | 1 | Histologic diagnosis | 25 | - | N/A | Yes | No | 13 | No | RF |
| Leslie et al., 2012 [40] | AS, EP, GG, MB, ODG, other glioma | Microscope slides | Prospective | 23 patients, breakdown unspecified | 1 | Histologic diagnosis | - | - | N/A | Yes | Yes | Variable by tumor type | No | SVM |
| Classifier Algorithm | Description |
|---|---|
| K-nearest neighbor | Determines the probability a datapoint will fall into a group based on its distance from the group’s members |
| Support vector machine | Assigns datapoints to one of two or more categories based on their locations on a space where the distance between the categories is maximized |
| Neural network | Infers the category of input data through layers of weighted non-linear or linear operations |
| Extreme learning machine | A feedforward neural network method with faster convergence |
| Classification tree | Divides datapoints into categories based on the homogeneity of independent variables |
| Regression tree | Divides data by iteratively partitioning independent variables to minimize mean square error |
| Random forest | An ensemble method that aggregates outputs of regression trees or classification trees |
| Naïve Bayes | Applies Bayes’ theorem to classify datapoints by independently considering the value of each independent variable |
| Partial least square regression | Identifies a subset of independent variables as significant predictors and then runs a regression with these predictors |
| Linear discriminant analysis | Identifies a linear combination of independent variables that divides datapoints into categories |
| Study | AUC | Accuracy | Sensitivity | Specificity |
|---|---|---|---|---|
| Discrimination of EP vs. MB | ||||
| Dong et al., 2021 [19] | 0.75–0.91 | 68.6–86.3 | - | - |
| Li et al., 2019 [25] | - | 74.6–85.4 | - | - |
| Zhang et al., 2021 [35] | 0.92 | 87.2 | 91.9 | 70.0 |
| Discrimination of EP vs. PA | ||||
| Li et al., 2020 [24] | 0.87–0.88 | 87.0–88.0 | 90.0–93.0 | 80.0–83.0 |
| Discrimination of EP vs. MB vs. PA | ||||
| Bidiwala et al., 2004 [17] | - | - | 72.7–85.7 | 86.4–92.9 |
| Dong et al., 2022 [20] | 0.94–0.98 | 80.0–84.9 | 80.0–84.9 | - |
| Fetit et al., 2015 [21] | 0.81–0.99 | 71.0–92.0 | - | - |
| Grist et al., 2020 [22] | - | 50.0–85.0 | - | - |
| Novak et al., 2021 [26] | - | 84.6–86.3 | - | - |
| Orphanidou-Vlachou et al., 2014 [27] | - | 37.5–93.8 | - | - |
| Rodriguez et al., 2014 [23] | - | 75.2–91.4 | - | - |
| Wang et al., 2022 [29] | - | 93.8 | - | - |
| Zarinabad et al., 2018 [30] | - | 81.0–86.0 | - | - |
| Zarinabad et al., 2017 [32] | - | 80.0–93.0 | - | - |
| Zhang et al., 2021 [35] | 0.90 | 82.6–94.5 | 73.9–91.8 | 86.9–95.9 |
| Zhao et al., 2022 [36] | - | 84.0–88.0 | - | - |
| Zhou et al., 2020 [37] | 0.91–0.92 | 74.0–83.0 | - | - |
| Algorithm | Accuracy (Mean +/− SD) | |||
|---|---|---|---|---|
| Overall | EP | MB | PA | |
| PNN | 89.7 +/− 3.8 | - | - | - |
| Naïve Bayes | 85.7 +/− 2.5 | 87.4 +/− 6.3 | 88.9 +/− 4.3 | 90.7 +/− 3.5 |
| LR | 82.5 +/− 7.5 | 85.4 +/− 11.2 | 85.5 +/− 9.5 | 88.6 +/− 8.4 |
| ANN | 82.5 +/− 13.4 | 91.5 +/− 4.9 | 88.5 +/− 10.6 | 86.5 +/− 13.4 |
| Classification tree | 79.0 +/− 5.7 | 90.0 +/− 7.1 | 87.5 +/− 3.5 | 82.0 +/− 4.2 |
| SVM | 78.2 +/− 10.7 | 84.3 +/− 7.1 | 88.7 +/− 5.9 | 90.5 +/− 7.0 |
| RF | 77.7 +/− 12.3 | 81.6 +/− 12.0 | 93.6 +/− 1.3 | 95.8 +/− 5.8 |
| kNN | 69.4 +/− 13.1 | 86.2 +/− 6.2 | 87.5 +/− 7.3 | 85.5 +/− 6.4 |
| LDA | 60.5 +/− 21.4 | - | - | - |
| Limitation | N (%) |
|---|---|
| Retrospective data collection | 19 (76%) |
| Small training or validation sets | 18 (72%) |
| Unequal distribution of tumor types in training cohorts | 17 (68%) |
| Methods lacking sufficient detail | 9 (36%) |
| Performance varies significantly by tumor type | 9 (36%) |
| Institutional differences in imaging/molecular acquisition | 8 (32%) |
| No inclusion of relevant clinical variables | 6 (24%) |
| Training and validation completed on the same dataset | 4 (16%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yearley, A.G.; Blitz, S.E.; Patel, R.V.; Chan, A.; Baird, L.C.; Friedman, G.K.; Arnaout, O.; Smith, T.R.; Bernstock, J.D. Machine Learning in the Classification of Pediatric Posterior Fossa Tumors: A Systematic Review. Cancers 2022, 14, 5608. https://doi.org/10.3390/cancers14225608
Yearley AG, Blitz SE, Patel RV, Chan A, Baird LC, Friedman GK, Arnaout O, Smith TR, Bernstock JD. Machine Learning in the Classification of Pediatric Posterior Fossa Tumors: A Systematic Review. Cancers. 2022; 14(22):5608. https://doi.org/10.3390/cancers14225608
Chicago/Turabian StyleYearley, Alexander G., Sarah E. Blitz, Ruchit V. Patel, Alvin Chan, Lissa C. Baird, Gregory K. Friedman, Omar Arnaout, Timothy R. Smith, and Joshua D. Bernstock. 2022. "Machine Learning in the Classification of Pediatric Posterior Fossa Tumors: A Systematic Review" Cancers 14, no. 22: 5608. https://doi.org/10.3390/cancers14225608
APA StyleYearley, A. G., Blitz, S. E., Patel, R. V., Chan, A., Baird, L. C., Friedman, G. K., Arnaout, O., Smith, T. R., & Bernstock, J. D. (2022). Machine Learning in the Classification of Pediatric Posterior Fossa Tumors: A Systematic Review. Cancers, 14(22), 5608. https://doi.org/10.3390/cancers14225608






