Next Generation Sequencing of Reactive Stroma and Residual Breast Cancer Cells in Tumor Bed after Neoadjuvant Chemotherapy
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient’s Cohort
2.2. Histological Characteristics in Breast Specimens after Neoadjuvant Treatment
2.3. Next Generation Sequencing (NGS)
3. Results
3.1. Tumor Infiltrating Lymphocytes (TILs)
3.2. PD-L1 Status
3.3. Statistical Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Burstein, H.J.; Curigliano, G.; Thurlimann, B.; Weber, W.P.; Poortmans, P.; Regan, M.M.; Senn, H.J.; Winer, E.P.; Gnant, M.; Panelists of the St Gallen Consensus Conference. Customizing local and systemic therapies for women with early breast cancer: The St. Gallen International Consensus Guidelines for treatment of early breast cancer 2021. Ann. Oncol. 2021, 32, 1216–1235. [Google Scholar] [CrossRef] [PubMed]
- Goldhirsch, A.; Wood, W.C.; Coates, A.S.; Gelber, R.D.; Thurlimann, B.; Senn, H.J.; Panel, M. Strategies for subtypes—Dealing with the diversity of breast cancer: Highlights of the St. Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann. Oncol. 2011, 22, 1736–1747. [Google Scholar] [CrossRef] [PubMed]
- Hasebe, T.; Iwasaki, M.; Akashi-Tanaka, S.; Hojo, T.; Shimizu, C.; Andoh, M.; Fujiwara, Y.; Shibata, T.; Sasajima, Y.; Kinoshita, T.; et al. Atypical tumor-stromal fibroblasts in invasive ductal carcinomas of the breast treated with neoadjuvant therapy. Hum. Pathol. 2011, 42, 998–1006. [Google Scholar] [CrossRef] [PubMed]
- Mieog, J.S.; van der Hage, J.A.; van de Velde, C.J. Preoperative chemotherapy for women with operable breast cancer. Cochrane Database Syst. Rev. 2007, 2007, CD005002. [Google Scholar] [CrossRef]
- Rubens, R.D.; Sexton, S.; Tong, D.; Winter, P.J.; Knight, R.K.; Hayward, J.L. Combined chemotherapy and radiotherapy for locally advanced breast cancer. Eur. J. Cancer 1980, 16, 351–356. [Google Scholar] [CrossRef]
- von Minckwitz, G.; Blohmer, J.U.; Costa, S.D.; Denkert, C.; Eidtmann, H.; Eiermann, W.; Gerber, B.; Hanusch, C.; Hilfrich, J.; Huober, J.; et al. Response-guided neoadjuvant chemotherapy for breast cancer. J. Clin. Oncol. 2013, 31, 3623–3630. [Google Scholar] [CrossRef]
- von Minckwitz, G.; Untch, M.; Blohmer, J.U.; Costa, S.D.; Eidtmann, H.; Fasching, P.A.; Gerber, B.; Eiermann, W.; Hilfrich, J.; Huober, J.; et al. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J. Clin. Oncol. 2012, 30, 1796–1804. [Google Scholar] [CrossRef]
- Zarotti, C.; Papassotiropoulos, B.; Elfgen, C.; Dedes, K.; Vorburger, D.; Pestalozzi, B.; Trojan, A.; Varga, Z. Biomarker dynamics and prognosis in breast cancer after neoadjuvant chemotherapy. Sci. Rep. 2022, 12, 91. [Google Scholar] [CrossRef]
- Haque, W.; Verma, V.; Hatch, S.; Suzanne Klimberg, V.; Brian Butler, E.; Teh, B.S. Response rates and pathologic complete response by breast cancer molecular subtype following neoadjuvant chemotherapy. Breast Cancer Res. Treat 2018, 170, 559–567. [Google Scholar] [CrossRef]
- Cortazar, P.; Zhang, L.; Untch, M.; Mehta, K.; Costantino, J.P.; Wolmark, N.; Bonnefoi, H.; Cameron, D.; Gianni, L.; Valagussa, P.; et al. Pathological complete response and long-term clinical benefit in breast cancer: The CTNeoBC pooled analysis. Lancet 2014, 384, 164–172. [Google Scholar] [CrossRef]
- Marchio, C.; Maletta, F.; Annaratone, L.; Sapino, A. The Perfect Pathology Report After Neoadjuvant Therapy. J. Natl. Cancer Inst. Monogr. 2015, 2015, 47–50. [Google Scholar] [CrossRef][Green Version]
- Zombori, T.; Cserni, G. Patterns of Regression in Breast Cancer after Primary Systemic Treatment. Pathol. Oncol. Res. 2019, 25, 1153–1161. [Google Scholar] [CrossRef]
- Laws, A.; Pastorello, R.; Dey, T.; Grossmith, S.; King, C.; McGrath, M.; Schnitt, S.J.; Mittendorf, E.A.; King, T. Impact of the Histologic Pattern of Residual Tumor After Neoadjuvant Chemotherapy on Recurrence and Survival in Stage I–III Breast Cancer. Ann. Surg. Oncol. 2022, 29, 7726–7736. [Google Scholar] [CrossRef]
- Hoffmann, L.G.; Sarian, L.O.; Vassallo, J.; de Paiva Silva, G.R.; Ramalho, S.O.B.; Ferracini, A.C.; da Silva Araujo, K.; Jales, R.M.; Figueira, D.E.; Derchain, S. Evaluation of PD-L1 and tumor infiltrating lymphocytes in paired pretreatment biopsies and post neoadjuvant chemotherapy surgical specimens of breast carcinoma. Sci. Rep. 2021, 11, 22478. [Google Scholar] [CrossRef]
- Pelekanou, V.; Barlow, W.E.; Nahleh, Z.A.; Wasserman, B.; Lo, Y.C.; von Wahlde, M.K.; Hayes, D.; Hortobagyi, G.N.; Gralow, J.; Tripathy, D.; et al. Tumor-Infiltrating Lymphocytes and PD-L1 Expression in Pre- and Posttreatment Breast Cancers in the SWOG S0800 Phase II Neoadjuvant Chemotherapy Trial. Mol. Cancer Ther. 2018, 17, 1324–1331. [Google Scholar] [CrossRef]
- Riemenschnitter, C.; Teleki, I.; Tischler, V.; Guo, W.; Varga, Z. Stability and prognostic value of Slug, Sox9 and Sox10 expression in breast cancers treated with neoadjuvant chemotherapy. Springerplus 2013, 2, 695. [Google Scholar] [CrossRef][Green Version]
- Roswall, P.; Bocci, M.; Bartoschek, M.; Li, H.; Kristiansen, G.; Jansson, S.; Lehn, S.; Sjolund, J.; Reid, S.; Larsson, C.; et al. Microenvironmental control of breast cancer subtype elicited through paracrine platelet-derived growth factor-CC signaling. Nat. Med. 2018, 24, 463–473. [Google Scholar] [CrossRef]
- Sobottka, B.; Pestalozzi, B.; Fink, D.; Moch, H.; Varga, Z. Similar lymphocytic infiltration pattern in primary breast cancer and their corresponding distant metastases. Oncoimmunology 2016, 5, e1153208. [Google Scholar] [CrossRef]
- Sonnenblick, A.; Salmon-Divon, M.; Salgado, R.; Dvash, E.; Ponde, N.; Zahavi, T.; Salmon, A.; Loibl, S.; Denkert, C.; Joensuu, H.; et al. Reactive stroma and trastuzumab resistance in HER2-positive early breast cancer. Int. J. Cancer 2020, 147, 266–276. [Google Scholar] [CrossRef]
- Van Bockstal, M.R.; Noel, F.; Guiot, Y.; Duhoux, F.P.; Mazzeo, F.; Van Marcke, C.; Fellah, L.; Ledoux, B.; Berliere, M.; Galant, C. Predictive markers for pathological complete response after neo-adjuvant chemotherapy in triple-negative breast cancer. Ann. Diagn. Pathol. 2020, 49, 151634. [Google Scholar] [CrossRef]
- Wang, Y.; Brodsky, A.S.; Xiong, J.; Lopresti, M.L.; Yang, D.; Resnick, M.B. Stromal Clusterin Expression Predicts Therapeutic Response to Neoadjuvant Chemotherapy in Triple Negative Breast Cancer. Clin. Breast Cancer 2018, 18, e373–e379. [Google Scholar] [CrossRef] [PubMed]
- WHO Classification of Tumours. Breast Tumours; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Lanjewar, S.; Patil, P.; Fineberg, S. Pathologic reporting practices for breast cancer specimens after neoadjuvant chemotherapy—A survey of pathologists in academic institutions across the United States. Mod. Pathol. 2020, 33, 91–98. [Google Scholar] [CrossRef]
- Kantor, O.; Laws, A.; Pastorello, R.G.; King, C.; Wong, S.; Dey, T.; Schnitt, S.; King, T.A.; Mittendorf, E.A. Comparison of Breast Cancer Staging Systems After Neoadjuvant Chemotherapy. Ann. Surg. Oncol. 2021, 28, 7347–7355. [Google Scholar] [CrossRef] [PubMed]
- Sejben, A.; Koszo, R.; Kahan, Z.; Cserni, G.; Zombori, T. Examination of Tumor Regression Grading Systems in Breast Cancer Patients Who Received Neoadjuvant Therapy. Pathol. Oncol. Res. 2020, 26, 2747–2754. [Google Scholar] [CrossRef] [PubMed]
- Varga, Z.; Caduff, R.; Pestalozzi, B. Stability of the HER2 gene after primary chemotherapy in advanced breast cancer. Virchows Arch. 2005, 446, 136–141. [Google Scholar] [CrossRef]
- Hagenaars, S.C.; de Groot, S.; Cohen, D.; Dekker, T.J.A.; Charehbili, A.; Meershoek-Klein Kranenbarg, E.; Duijm-de Carpentier, M.; Pijl, H.; Putter, H.; Tollenaar, R.; et al. Tumor-stroma ratio is associated with Miller-Payne score and pathological response to neoadjuvant chemotherapy in HER2-negative early breast cancer. Int. J. Cancer 2021, 149, 1181–1188. [Google Scholar] [CrossRef]
- Arole, V.; Nitta, H.; Wei, L.; Shen, T.; Parwani, A.V.; Li, Z. M2 tumor-associated macrophages play important role in predicting response to neoadjuvant chemotherapy in triple-negative breast carcinoma. Breast Cancer Res. Treat 2021, 188, 37–42. [Google Scholar] [CrossRef]
- Buldakov, M.; Zavyalova, M.; Krakhmal, N.; Telegina, N.; Vtorushin, S.; Mitrofanova, I.; Riabov, V.; Yin, S.; Song, B.; Cherdyntseva, N.; et al. CD68+, but not stabilin-1+ tumor associated macrophages in gaps of ductal tumor structures negatively correlate with the lymphatic metastasis in human breast cancer. Immunobiology 2017, 222, 31–38. [Google Scholar] [CrossRef][Green Version]
- de Jong, V.M.T.; Wang, Y.; Ter Hoeve, N.D.; Opdam, M.; Stathonikos, N.; Jozwiak, K.; Hauptmann, M.; Cornelissen, S.; Vreuls, W.; Rosenberg, E.H.; et al. Prognostic Value of Stromal Tumor-Infiltrating Lymphocytes in Young, Node-Negative, Triple-Negative Breast Cancer Patients Who Did Not Receive (neo) Adjuvant Systemic Therapy. J. Clin. Oncol. 2022, 40, 2361–2374. [Google Scholar] [CrossRef]
- Farmer, P.; Bonnefoi, H.; Anderle, P.; Cameron, D.; Wirapati, P.; Becette, V.; Andre, S.; Piccart, M.; Campone, M.; Brain, E.; et al. A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat. Med. 2009, 15, 68–74. [Google Scholar] [CrossRef]

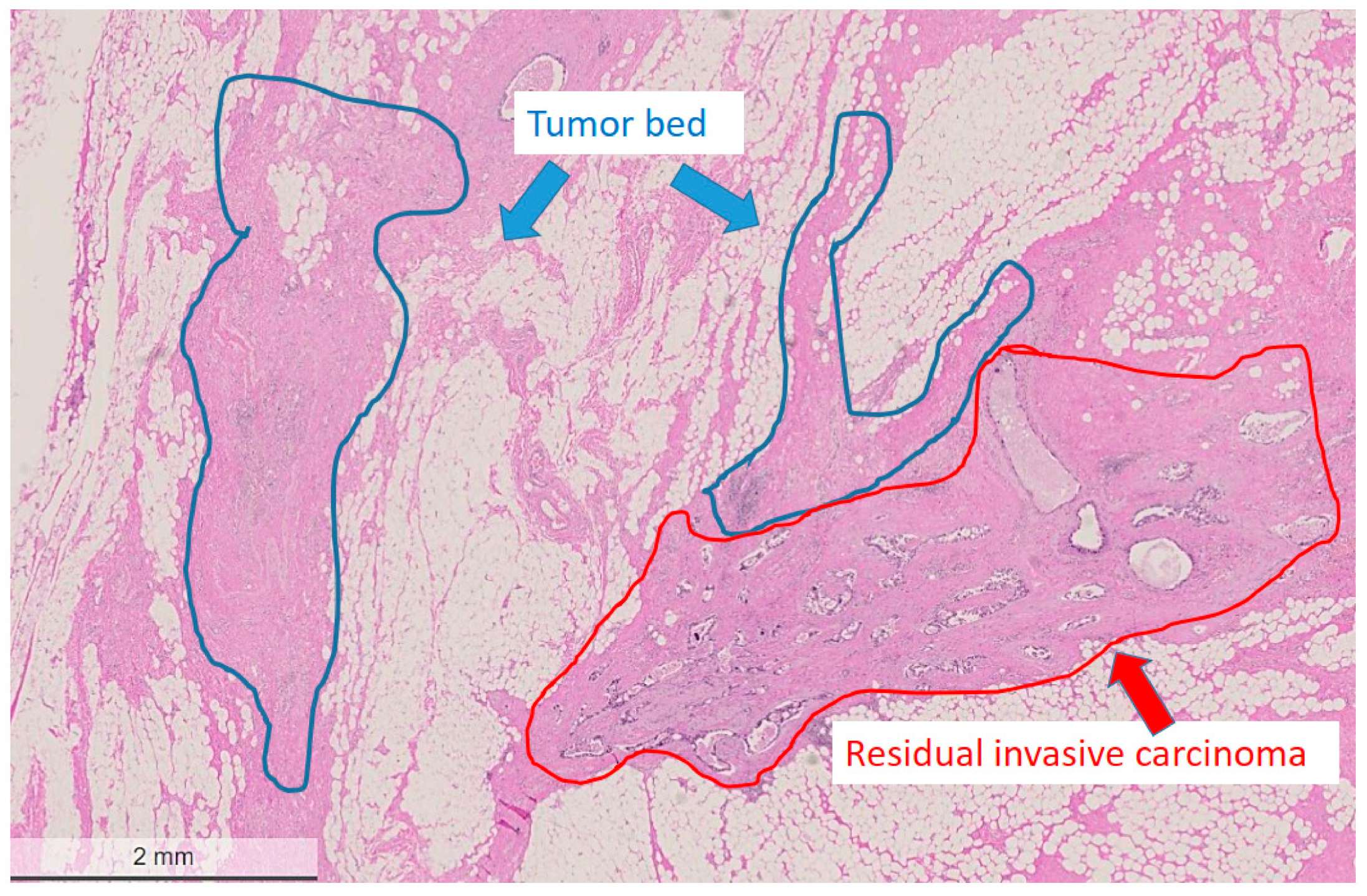
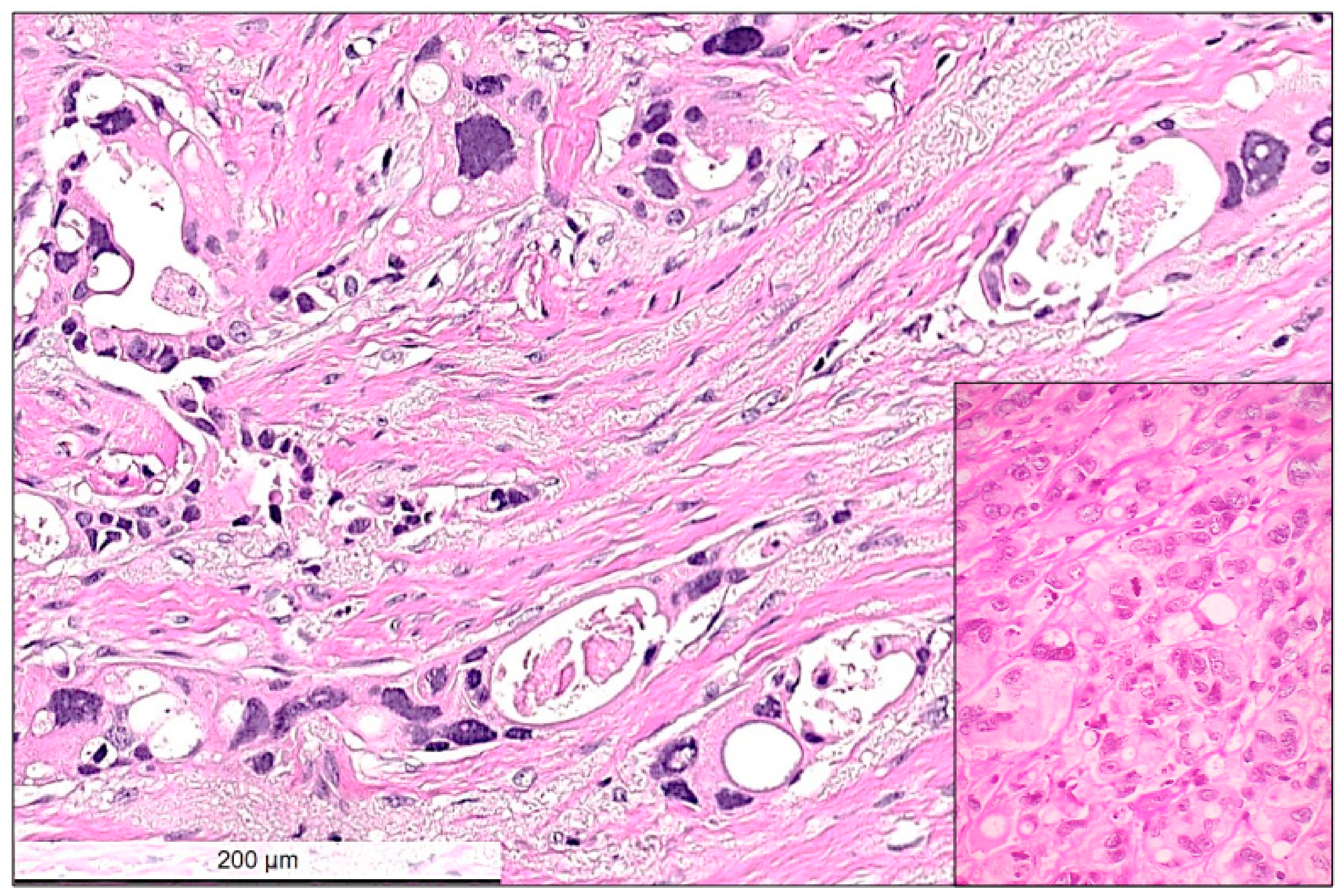
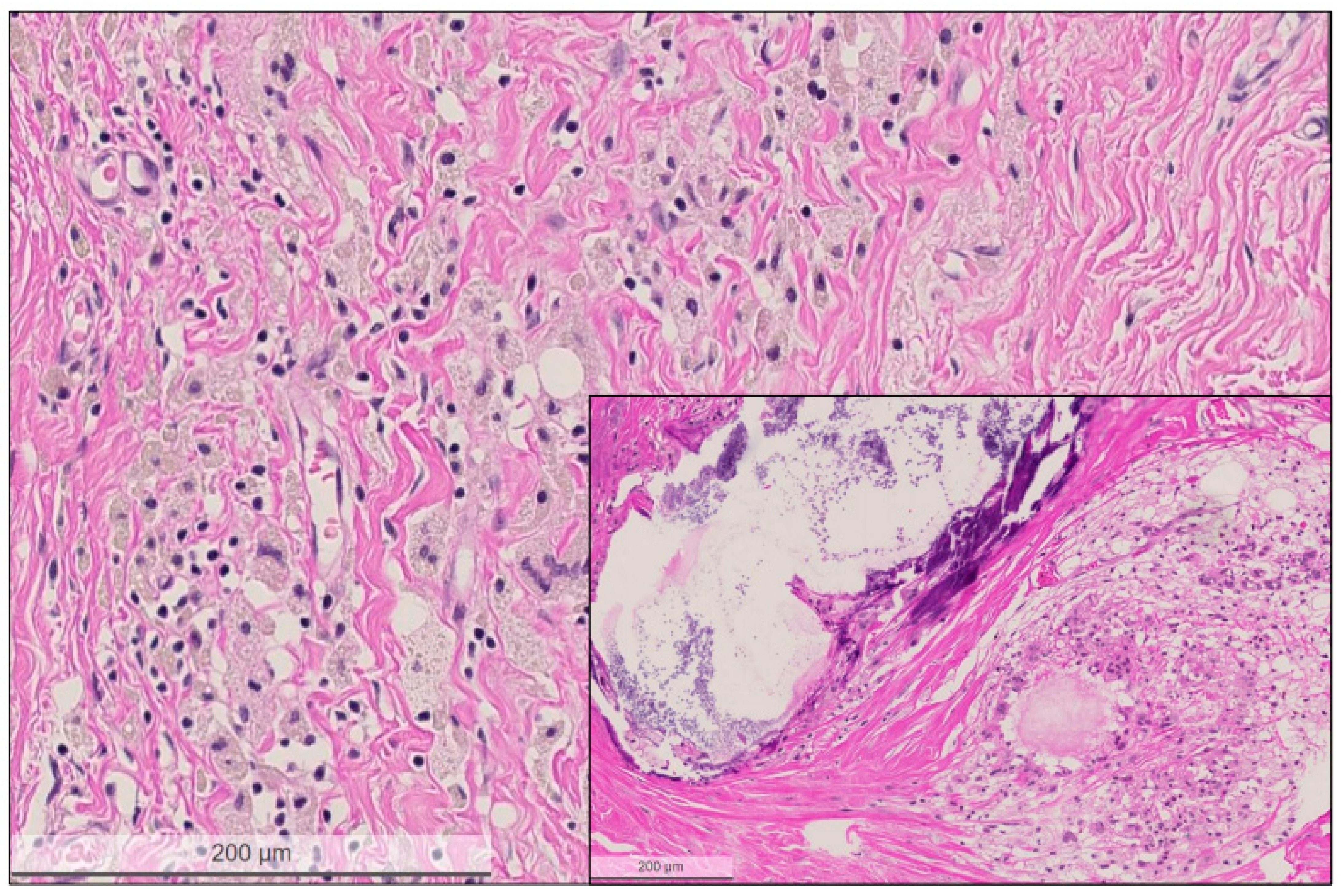
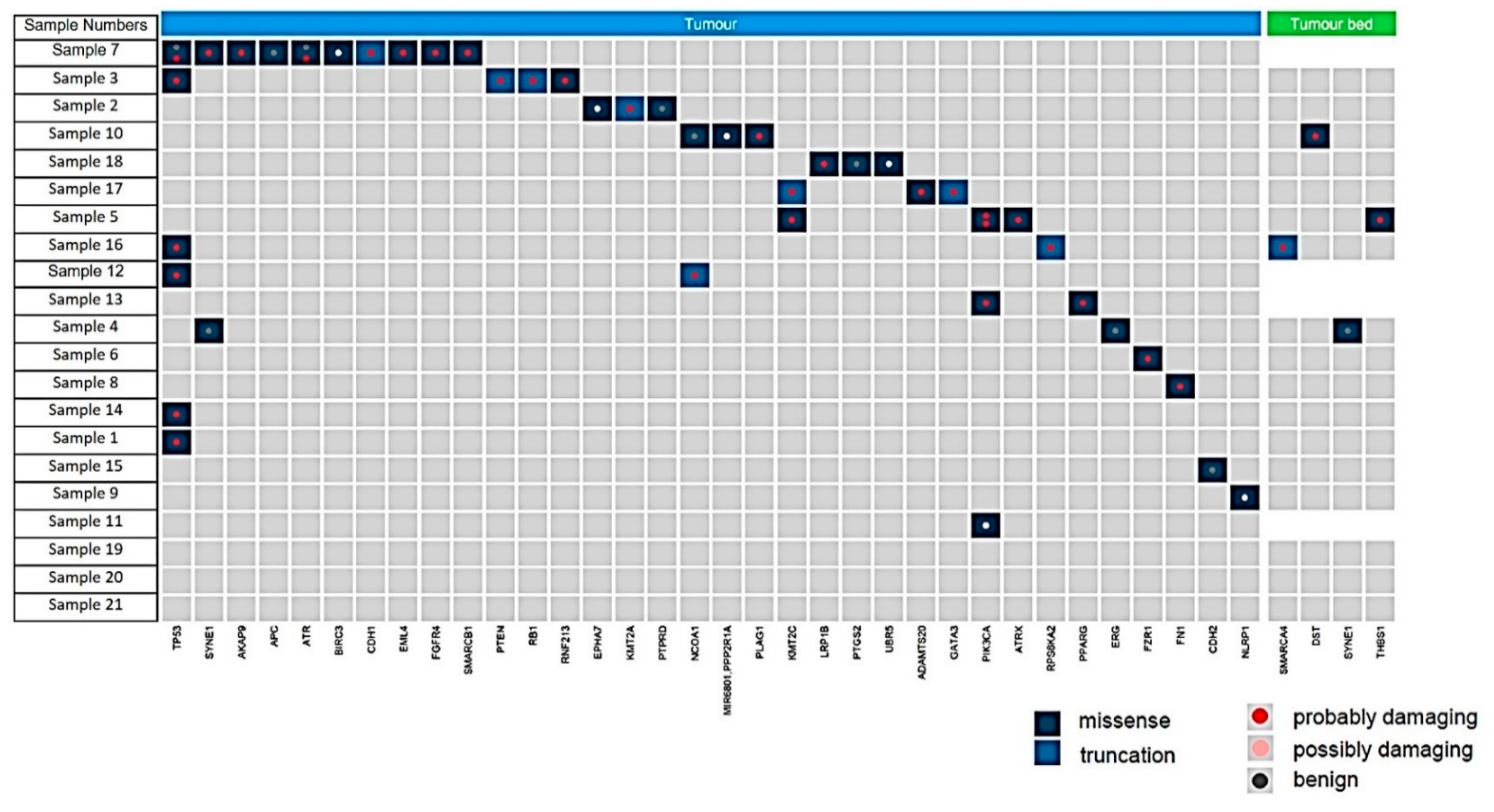
| Patient’s Number | Age at Diagnosis [Years] | cT,cN | ypT | ypN | ER/PR /Her2 Status | PD-L1 Status | TILs | Response | Follow-Up [Years] | Mutations in Tumour Cells | Mutations in Tumour Bed |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 43 | T2/N1 | pTis | N1 | triple negative | NA | NA | CR | 2 years dead | TP53 | no mutations |
| 2 | 74 | T4b/N1/M1 | pT4d | N2 | ERpos /PRpos /Her2neg | neg | + | NR | 1 year dead | EPHA7, KMT2A | no mutations |
| 3 | 50 | T2/N1 | pT2 | N0 | triple negative | neg | + | NR | 13 years alive | PTEN, TP53, RNF213, RB1 | no mutations |
| 4 | 62 | T3/N1 | pT3 | N0 | ERpos /PRneg /Her2 neg | pos IC 2% | + | NR | 11 years dead | no mutations | no mutations |
| 5 | 40 | T2/N1/M1 | pT1c | - | ERpos /PRpos /Her2neg | neg | 0 | NR | 10 years dead | PIK3CA, KMT2A, ATRX | no mutations |
| 6 | 45 | T4/N0 | pT2 | N0 | ERpos /PRpos /Her2pos | neg | 0 | NR | 11 years alive | FZR1 | no mutations |
| 7 | 76 | T2/N1 | pT2 | N1 | ERpos /PRpos /Her2neg | pos IC 5% | +++ | NR | 11 years dead | EML4, ATR, FGFR4, SYBE1, AKAP9, BIRC3, CDH1, TP53 | no mutations |
| 8 | 45 | T2/N0 | pT1b | - | ERneg /PRneg /Her2pos | neg | 0 | NR | 8 years dead | FN1 | no mutations |
| 9 | 56 | T2/N1 | - | N1 | triple negative | pos IC 2% | +++ | NR | 6 years dead | NLRP1 | no mutations |
| 10 | 82 | T3/N2 | pT3 | N2 | triple negative | pos IC 5% | +++ | NR | 9 years dead | PLAG1, MR6801, PP2R1A | no mutations |
| 11 | 38 | T2(m)/N2 | pT1c | N2 | ERpos /PRpos /Her2neg | neg | 0 | NR | 12 years alive | PIK3CA | no mutations |
| 12 | 54 | T2/N1 | pT2 | N1 | ERpos /PRpos /Her2neg | NA | NA | NR | lost to follow-up | TP53, NCOA1 | no mutations |
| 13 | 67 | T1/N0 | pT2 | N0 | ERpos /PRpos /Her2neg | neg | 0 | NR | 9 years alive | PPARG. PIK3CA | no mutations |
| 19 | 50 | T2/N0 | pT2 | N0 | ERpos /PRpos /Her2pos | NA | NA | NR | lost to follow-up | no mutations | no mutations |
| 20 | 39 | T3/N1/M1 | pT3 | N1 | ERpos /PRpos /Her2neg | NA | NA | NR | 4 years dead | no mutations | no mutations |
| 21 | 66 | T2/N1 | pT3 | N0 | ERpos /PRpos /Her2neg | NA | NA | NR | 12 years alive | no mutations | no mutations |
| 14 | 68 | T2/N1 | pT2 | N1 | ERpos /PRpos /Her2pos | neg | ++ | PR | 7 years dead | TP53 | no mutations |
| 15 | 43 | T3/N1 | pT3 | N1 | ERpos /PRpos /Her2neg | neg | + | PR | 9 years alive | no mutations | no mutations |
| 16 | 43 | T4/N1/M1 | pT3 | N2 | ERpos /PRpos /Her2neg | neg | + | PR | 4 years dead | RPS6KA2 | no mutations |
| 17 | 72 | T2/N0 | pT2 | N0 | ERpos /PRpos /Her2neg | neg | + | PR | lost to follow-up | KMT2C, GATA3, ADAMTS20 | no mutations |
| 18 | 37 | T1(m)/N1 | pT1a | N1 | ERpos /PRpos /Her2neg | neg | 0 | PR | lost to follow-up | LRP1B, UBR5 | no mutations |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Varga, Z.; Christiansen, A.; Lukamowicz-Rajska, M.; Batavia, A.A.; von Teichman, A.; Schraml, P.; Moch, H. Next Generation Sequencing of Reactive Stroma and Residual Breast Cancer Cells in Tumor Bed after Neoadjuvant Chemotherapy. Cancers 2022, 14, 5609. https://doi.org/10.3390/cancers14225609
Varga Z, Christiansen A, Lukamowicz-Rajska M, Batavia AA, von Teichman A, Schraml P, Moch H. Next Generation Sequencing of Reactive Stroma and Residual Breast Cancer Cells in Tumor Bed after Neoadjuvant Chemotherapy. Cancers. 2022; 14(22):5609. https://doi.org/10.3390/cancers14225609
Chicago/Turabian StyleVarga, Zsuzsanna, Ailsa Christiansen, Magdalena Lukamowicz-Rajska, Aashil A. Batavia, Adriana von Teichman, Peter Schraml, and Holger Moch. 2022. "Next Generation Sequencing of Reactive Stroma and Residual Breast Cancer Cells in Tumor Bed after Neoadjuvant Chemotherapy" Cancers 14, no. 22: 5609. https://doi.org/10.3390/cancers14225609
APA StyleVarga, Z., Christiansen, A., Lukamowicz-Rajska, M., Batavia, A. A., von Teichman, A., Schraml, P., & Moch, H. (2022). Next Generation Sequencing of Reactive Stroma and Residual Breast Cancer Cells in Tumor Bed after Neoadjuvant Chemotherapy. Cancers, 14(22), 5609. https://doi.org/10.3390/cancers14225609






