RNA Polymerase I Is Uniquely Vulnerable to the Small-Molecule Inhibitor BMH-21
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Purification of RNA Polymerases (Pols) I, II, and III
2.2. Multi-Nucleotide Addition Assay
2.3. Analysis of Multi-Nucleotide Addition Data
2.4. Elongation Complex (EC) Stability Assay
3. Results
3.1. BMH-21 Alters the Nucleotide Addition Mechanism of Pol I
3.2. Pol II Is Unaffected by BMH-21
3.3. BMH-21 Has a Modest Inhibitory Effect on Pol III
3.4. BMH-21 Does Not Destabilize Pol I, II, or III Elongation Complexes
4. Discussion
4.1. BMH-21 Significantly Impairs Pol I Transcription Elongation
4.2. BMH-21’s Modest Inhibitory Effect on Pol III May Be Therapeutically Advantageous
4.3. Intrinsic Biochemical Properties of the Pols Render Them More or Less Sensitive to BMH-21
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Pol I | RNA polymerase I |
| Pol II | RNA polymerase II |
| Pol III | RNA polymerase III |
| rDNA | ribosomal DNA |
| rRNA | ribosomal RNA |
| mRNA | messenger RNA |
| EC | elongation complex |
| DNAt | DNA template |
| DNAnt | DNA non-template |
| MENOTR | Multi-start Evolutionary Nonlinear OpTimizeR |
| tRNA | transfer RNA |
References
- Roeder, R.G.; Rutter, W.J. Multiple Forms of DNA-dependent RNA Polymerase in Eukaryotic Organisms. Nature 1969, 224, 234–237. [Google Scholar] [CrossRef] [PubMed]
- Misiaszek, A.D.; Girbig, M.; Grötsch, H.; Baudin, F.; Murciano, B.; Lafita, A.; Müller, C.W. Cryo-EM structures of human RNA polymerase I. Nat. Struct. Mol. Biol. 2021, 28, 997–1008. [Google Scholar] [CrossRef] [PubMed]
- Bernecky, C.; Herzog, F.; Baumeister, W.; Plitzko, J.M.; Cramer, P. Structure of transcribing mammalian RNA polymerase II. Nature 2016, 529, 551–554. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Liu, W.; Chen, K.; Wu, Z.; Yang, H.; Xu, Y. Structure of the human RNA polymerase I elongation complex. Cell Discov. 2021, 7, 97. [Google Scholar] [CrossRef] [PubMed]
- Girbig, M.; Misiaszek, A.D.; Vorländer, M.K.; Lafita, A.; Grötsch, H.; Baudin, F.; Bateman, A.; Müller, C.W. Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states. Nat. Struct. Mol. Biol. 2021, 28, 210–219. [Google Scholar] [CrossRef]
- Ramsay, E.P.; Abascal-Palacios, G.; Daiß, J.L.; King, H.; Gouge, J.; Pilsl, M.; Beuron, F.; Morris, E.; Gunkel, P.; Engel, C.; et al. Structure of human RNA polymerase III. Nat. Commun. 2020, 11, 6409. [Google Scholar] [CrossRef]
- Wang, Q.; Li, S.; Wan, F.; Xu, Y.; Wu, Z.; Cao, M.; Lan, P.; Lei, M.; Wu, J. Structural insights into transcriptional regulation of human RNA polymerase III. Nat. Struct. Mol. Biol. 2021, 28, 220–227. [Google Scholar] [CrossRef]
- Li, L.; Yu, Z.; Zhao, D.; Ren, Y.; Hou, H.; Xu, Y. Structure of human RNA polymerase III elongation complex. Cell Res. 2021, 31, 791–800. [Google Scholar] [CrossRef]
- Girbig, M.; Misiaszek, A.D.; Müller, C.W. Structural insights into nuclear transcription by eukaryotic DNA-dependent RNA polymerases. Nat. Rev. Mol. Cell Biol. 2022, 23, 603–622. [Google Scholar] [CrossRef]
- Farley-Barnes, K.I.; McCann, K.L.; Ogawa, L.M.; Merkel, J.; Surovtseva, Y.V.; Baserga, S.J. Diverse Regulators of Human Ribosome Biogenesis Discovered by Changes in Nucleolar Number. Cell Rep. 2018, 22, 1923–1934. [Google Scholar] [CrossRef]
- Mayer, C.; Grummt, I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 2006, 25, 6384–6391. [Google Scholar] [CrossRef] [PubMed]
- Warner, J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999, 24, 437–440. [Google Scholar] [CrossRef]
- Russell, J.; Zomerdijk, J.C. RNA-polymerase-I-directed rDNA transcription, life and works. Trends Biochem. Sci. 2005, 30, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Henderson, A.S.; Warburton, D.; Atwood, K.C. Location of Ribosomal DNA in the Human Chromosome Complement. Proc. Natl. Acad. Sci. USA 1972, 69, 3394–3398. [Google Scholar] [CrossRef] [PubMed]
- Smirnov, E.; Chmúrčiaková, N.; Liška, F.; Bažantová, P.; Cmarko, D. Variability of Human rDNA. Cells 2021, 10, 196. [Google Scholar] [CrossRef] [PubMed]
- Aubert, M.; O’Donohue, M.F.; LeBaron, S.; Gleizes, P.-E. Pre-Ribosomal RNA Processing in Human Cells: From Mechanisms to Congenital Diseases. Biomolecules 2018, 8, 123. [Google Scholar] [CrossRef]
- Sørensen, P.D.; Frederiksen, S. Characterization of human 5S rRNA genes. Nucleic Acids Res. 1991, 19, 4147–4151. [Google Scholar] [CrossRef]
- Steffensen, D.M.; Duffey, P.; Prensky, W. Localisation of 5S ribosomal RNA genes on human chromosome 1. Nature 1974, 252, 741–743. [Google Scholar] [CrossRef]
- Khatter, H.; Myasnikov, A.G.; Natchiar, S.K.; Klaholz, B.P. Structure of the human 80S ribosome. Nature 2015, 520, 640–645. [Google Scholar] [CrossRef]
- Ruggero, D. Revisiting the Nucleolus: From Marker to Dynamic Integrator of Cancer Signaling. Sci. Signal. 2012, 5, pe38. [Google Scholar] [CrossRef]
- Ruggero, D.; Pandolfi, P.P. Does the ribosome translate cancer? Nat. Rev. Cancer 2003, 3, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Montanaro, L.; Treré, D.; Derenzini, M. Changes in ribosome biogenesis may induce cancer by down-regulating the cell tumor suppressor potential. Biochim. Biophys. Acta BBA Rev. Cancer 2012, 1825, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, R.; Schneekloth, J.J.S.; Panov, K.I.; Hannan, K.M.; Hannan, R.D. Targeting the RNA Polymerase I Transcription for Cancer Therapy Comes of Age. Cells 2020, 9, 266. [Google Scholar] [CrossRef]
- Burger, K.; Mühl, B.; Harasim, T.; Rohrmoser, M.; Malamoussi, A.; Orban, M.; Kellner, M.; Gruber-Eber, A.; Kremmer, E.; Hölzel, M.; et al. Chemotherapeutic Drugs Inhibit Ribosome Biogenesis at Various Levels. J. Biol. Chem. 2010, 285, 12416–12425. [Google Scholar] [CrossRef]
- Awad, D.; Prattes, M.; Kofler, L.; Rössler, I.; Loibl, M.; Pertl, M.; Zisser, G.; Wolinski, H.; Pertschy, B.; Bergler, H. Inhibiting eukaryotic ribosome biogenesis. BMC Biol. 2019, 17, 46. [Google Scholar] [CrossRef] [PubMed]
- Catez, F.; Venezia, N.D.; Marcel, V.; Zorbas, C.; Lafontaine, D.L.J.; Diaz, J.-J. Ribosome biogenesis: An emerging druggable pathway for cancer therapeutics. Biochem. Pharmacol. 2019, 159, 74–81. [Google Scholar] [CrossRef]
- Scull, C.E.; Zhang, Y.; Tower, N.; Rasmussen, L.; Padmalayam, I.; Hunter, R.; Zhai, L.; Bostwick, R.; Schneider, D.A. Discovery of novel inhibitors of ribosome biogenesis by innovative high throughput screening strategies. Biochem. J. 2019, 476, 2209–2219. [Google Scholar] [CrossRef]
- Sanchez-Martin, V.; Schneider, D.A.; Ortiz-Gonzalez, M.; Soriano-Lerma, A.; Linde-Rodriguez, A.; Perez-Carrasco, V.; Gutierrez-Fernandez, J.; Cuadros, M.; González, C.; Soriano, M.; et al. Targeting ribosomal G-quadruplexes with naphthalene-diimides as RNA polymerase I inhibitors for colorectal cancer treatment. Cell Chem. Biol. 2021, 28, 1590–1601. [Google Scholar] [CrossRef]
- Peltonen, K.D.; Colis, L.; Liu, H.; Trivedi, R.; Moubarek, M.S.; Moore, H.M.; Bai, B.; Rudek, M.A.; Bieberich, C.; Laiho, M. A Targeting Modality for Destruction of RNA Polymerase I that Possesses Anticancer Activity. Cancer Cell 2014, 25, 77–90. [Google Scholar] [CrossRef]
- Peltonen, K.; Colis, L.; Liu, H.; Jäämaa, S.; Zhang, Z.; Af Hällström, T.; Moore, H.M.; Sirajuddin, P.; Laiho, M. Small molecule BMH-compounds that inhibit RNA polymerase I and cause nucleolar stress. Mol. Cancer Ther. 2014, 13, 2537–2546. [Google Scholar] [CrossRef]
- Harris, C.C. The carcinogenicity of anticancer drugs: A hazard in man. Cancer 1976, 37 (Suppl. 2), 1014–1023. [Google Scholar] [CrossRef]
- Sanij, E.; Hannan, K.M.; Xuan, J.; Yan, S.; Ahern, J.E.; Trigos, A.S.; Brajanovski, N.; Son, J.; Chan, K.T.; Kondrashova, O.; et al. CX-5461 activates the DNA damage response and demonstrates therapeutic efficacy in high-grade serous ovarian cancer. Nat. Commun. 2020, 11, 2641. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Di Antonio, M.; McKinney, S.; Mathew, V.; Ho, B.; O’Neil, N.J.; Santos, N.D.; Silvester, J.; Wei, V.; Garcia, J.; et al. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat. Commun. 2017, 8, 14432. [Google Scholar] [CrossRef] [PubMed]
- Hannan, R.D.; Drygin, D.; Pearson, R. Targeting RNA polymerase I transcription and the nucleolus for cancer therapy. Expert Opin. Ther. Targets 2013, 17, 873–878. [Google Scholar] [CrossRef] [PubMed]
- Drygin, D.; Rice, W.G.; Grummt, I. The RNA Polymerase I Transcription Machinery: An Emerging Target for the Treatment of Cancer. Annu. Rev. Pharmacol. Toxicol. 2010, 50, 131–156. [Google Scholar] [CrossRef] [PubMed]
- Laferté, A.; Favry, E.; Sentenac, A.; Riva, M.; Carles, C.; Chédin, S. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev. 2006, 20, 2030–2040. [Google Scholar] [CrossRef]
- Chédin, S.; Laferté, A.; Hoang, T.; Lafontaine, D.L.; Riva, M.; Carles, C. Is ribosome synthesis controlled by pol I transcription? Cell Cycle 2007, 6, 11–15. [Google Scholar] [CrossRef]
- Haddach, M.; Schwaebe, M.K.; Michaux, J.; Nagasawa, J.; O’Brien, S.E.; Whitten, J.P.; Pierre, F.; Kerdoncuff, P.; Darjania, L.; Stansfield, R.; et al. Discovery of CX-5461, the First Direct and Selective Inhibitor of RNA Polymerase I, for Cancer Therapeutics. ACS Med. Chem. Lett. 2012, 3, 602–606. [Google Scholar] [CrossRef]
- Drygin, D.; Lin, A.; Bliesath, J.; Ho, C.B.; O’Brien, S.E.; Proffitt, C.; Omori, M.; Haddach, M.; Schwaebe, M.K.; Siddiqui-Jain, A.; et al. Targeting RNA Polymerase I with an Oral Small Molecule CX-5461 Inhibits Ribosomal RNA Synthesis and Solid Tumor Growth. Cancer Res. 2011, 71, 1418–1430. [Google Scholar] [CrossRef]
- Gorski, J.J.; Pathak, S.; Panov, K.; Kasciukovic, T.; Panova, T.; Russell, J.; Zomerdijk, J.C.B.M. A novel TBP-associated factor of SL1 functions in RNA polymerase I transcription. EMBO J. 2007, 26, 1560–1568. [Google Scholar] [CrossRef]
- Russell, J.; Zomerdijk, J.C. The RNA polymerase I transcription machinery. Biochem. Soc. Symp. 2006, 73, 203–216. [Google Scholar] [CrossRef]
- Bruno, P.M.; Lu, M.; Dennis, K.A.; Inam, H.; Moore, C.J.; Sheehe, J.; Elledge, S.J.; Hemann, M.T.; Pritchard, J.R. The primary mechanism of cytotoxicity of the chemotherapeutic agent CX-5461 is topoisomerase II poisoning. Proc. Natl. Acad. Sci. USA 2020, 117, 4053–4060. [Google Scholar] [CrossRef]
- Pan, M.; Wright, W.C.; Chapple, R.H.; Zubair, A.; Sandhu, M.; Batchelder, J.E.; Huddle, B.C.; Low, J.; Blankenship, K.B.; Wang, Y.; et al. The chemotherapeutic CX-5461 primarily targets TOP2B and exhibits selective activity in high-risk neuroblastoma. Nat. Commun. 2021, 12, 6468. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Hurley, L.H. A first-in-class clinical G-quadruplex-targeting drug. The bench-to-bedside translation of the fluoroquinolone QQ58 to CX-5461 (Pidnarulex). Bioorganic Med. Chem. Lett. 2022, 77, 129016. [Google Scholar] [CrossRef] [PubMed]
- Peltonen, K.D.; Colis, L.; Liu, H.; Jäämaa, S.; Moore, H.M.; Enbäck, J.; Laakkonen, P.; Vaahtokari, A.; Jones, R.J.; Hällström, T.M.A.; et al. Identification of Novel p53 Pathway Activating Small-Molecule Compounds Reveals Unexpected Similarities with Known Therapeutic Agents. PLoS ONE 2010, 5, e12996. [Google Scholar] [CrossRef] [PubMed]
- Colis, L.; Peltonen, K.; Sirajuddin, P.; Liu, H.; Sanders, S.; Ernst, G.; Barrow, J.C.; Laiho, M. DNA intercalator BMH-21 inhibits RNA polymerase I independent of DNA damage response. Oncotarget 2014, 5, 4361–4369. [Google Scholar] [CrossRef]
- Wei, T.; Najmi, S.M.; Liu, H.; Peltonen, K.D.; Kučerová, A.; Schneider, D.; Laiho, M. Small-Molecule Targeting of RNA Polymerase I Activates a Conserved Transcription Elongation Checkpoint. Cell Rep. 2018, 23, 404–414. [Google Scholar] [CrossRef]
- Jacobs, R.Q.; Huffines, A.K.; Laiho, M.; Schneider, D.A. The small-molecule BMH-21 directly inhibits transcription elongation and DNA occupancy of RNA polymerase I in vivo and in vitro. J. Biol. Chem. 2021, 298, 101450. [Google Scholar] [CrossRef]
- Jacobs, R.Q.; Ingram, Z.M.; Lucius, A.L.; Schneider, D.A. Defining the divergent enzymatic properties of RNA polymerases I and II. J. Biol. Chem. 2021, 296, 100051. [Google Scholar] [CrossRef]
- Jacobs, R.Q.; Carter, Z.I.; Lucius, A.L.; Schneider, D. Uncovering the mechanisms of transcription elongation by eukaryotic RNA polymerases I, II, and III. iScience 2022, 25, 105306. [Google Scholar] [CrossRef]
- Ingram, Z.M.; Schneider, D.A.; Lucius, A.L. Transient-state kinetic analysis of multi-nucleotide addition catalyzed by RNA polymerase I. Biophys. J. 2021, 120, 4378–4390. [Google Scholar] [CrossRef] [PubMed]
- Appling, F.D.; Lucius, A.L.; Schneider, D.A. Transient-State Kinetic Analysis of the RNA Polymerase I Nucleotide Incorporation Mechanism. Biophys. J. 2015, 109, 2382–2393. [Google Scholar] [CrossRef] [PubMed]
- Ingram, Z.M.; Scull, N.W.; Schneider, D.S.; Lucius, A.L. Multi-start Evolutionary Nonlinear OpTimizeR (MENOTR): A hybrid parameter optimization toolbox. Biophys. Chem. 2021, 279, 106682. [Google Scholar] [CrossRef]
- Appling, F.D.; Scull, C.E.; Lucius, A.L.; Schneider, D.A. The A12.2 Subunit Is an Intrinsic Destabilizer of the RNA Polymerase I Elongation Complex. Biophys. J. 2018, 114, 2507–2515. [Google Scholar] [CrossRef] [PubMed]
- Scull, C.E.; Clarke, A.M.; Lucius, A.L.; Schneider, D.A. Downstream sequence-dependent RNA cleavage and pausing by RNA polymerase I. J. Biol. Chem. 2020, 295, 1288–1299. [Google Scholar] [CrossRef]
- White, R.J. RNA polymerases I and III, non-coding RNAs and cancer. Trends Genet. 2008, 24, 622–629. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, S.J.; White, R.J. Regulation of RNA Polymerase III Transcription During Mammalian Cell Growth. Cell Cycle 2007, 6, 2323–2326. [Google Scholar] [CrossRef]
- Brown, T.R.P.; Scott, P.H.; Stein, T.; Winter, A.G.; White, R.J. RNA Polymerase III Transcription: Its Control by Tumor Suppressors and Its Deregulation by Transforming Agents. Gene Expr. 2001, 9, 15–28. [Google Scholar] [CrossRef][Green Version]
- Cabarcas, S.; Schramm, L. RNA polymerase III transcription in cancer: The BRF2 connection. Mol. Cancer 2011, 10, 47. [Google Scholar] [CrossRef]
- Pavon-Eternod, M.; Gomes, S.; Geslain, R.; Dai, Q.; Rosner, M.R.; Pan, T. tRNA over-expression in breast cancer and functional consequences. Nucleic Acids Res. 2009, 37, 7268–7280. [Google Scholar] [CrossRef]
- Winter, A.G.; Sourvinos, G.; Allison, S.J.; Tosh, K.; Scott, P.H.; Spandidos, D.A.; White, R.J. RNA polymerase III transcription factor TFIIIC2 is overexpressed in ovarian tumors. Proc. Natl. Acad. Sci. USA 2000, 97, 12619–12624. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.A.S.; Dubeau, L.; Johnson, D.L. Enhanced RNA Polymerase III-dependent Transcription Is Required for Oncogenic Transformation*. J. Biol. Chem. 2008, 283, 19184–19191. [Google Scholar] [CrossRef] [PubMed]





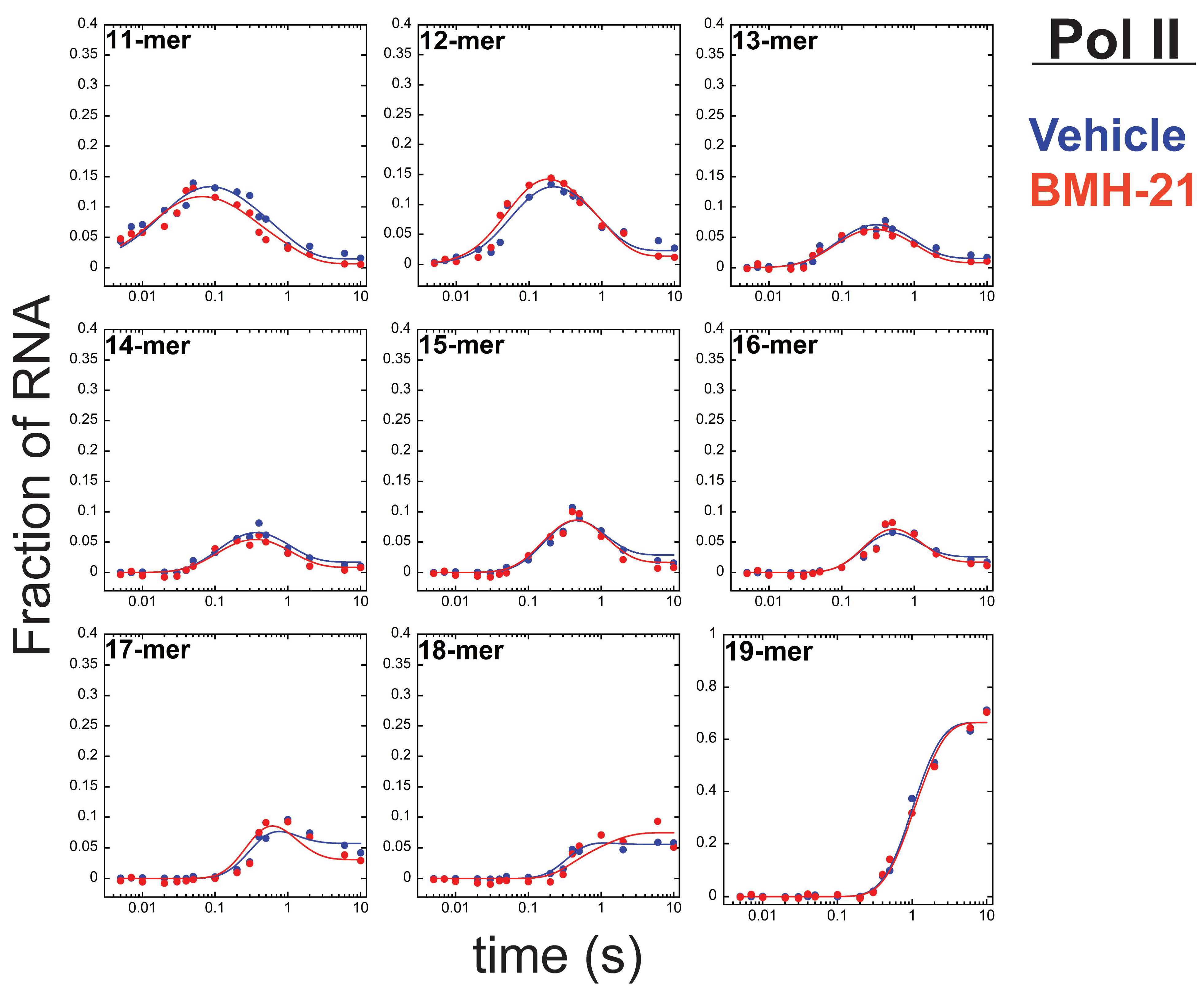
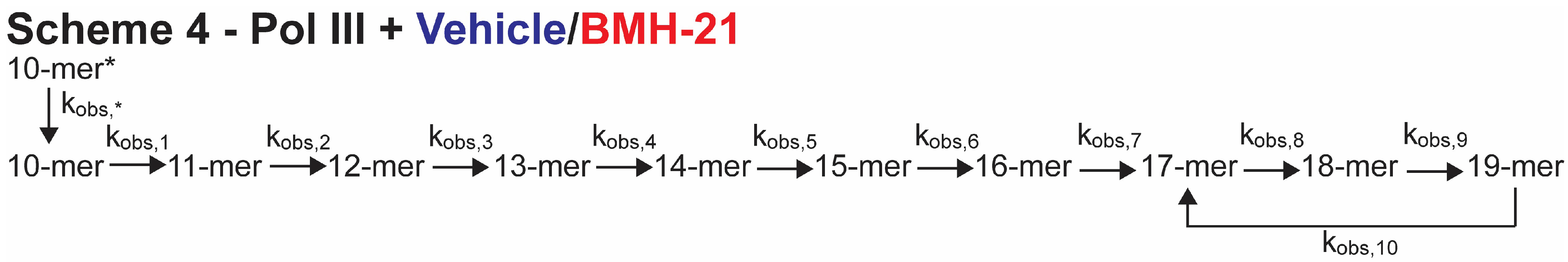
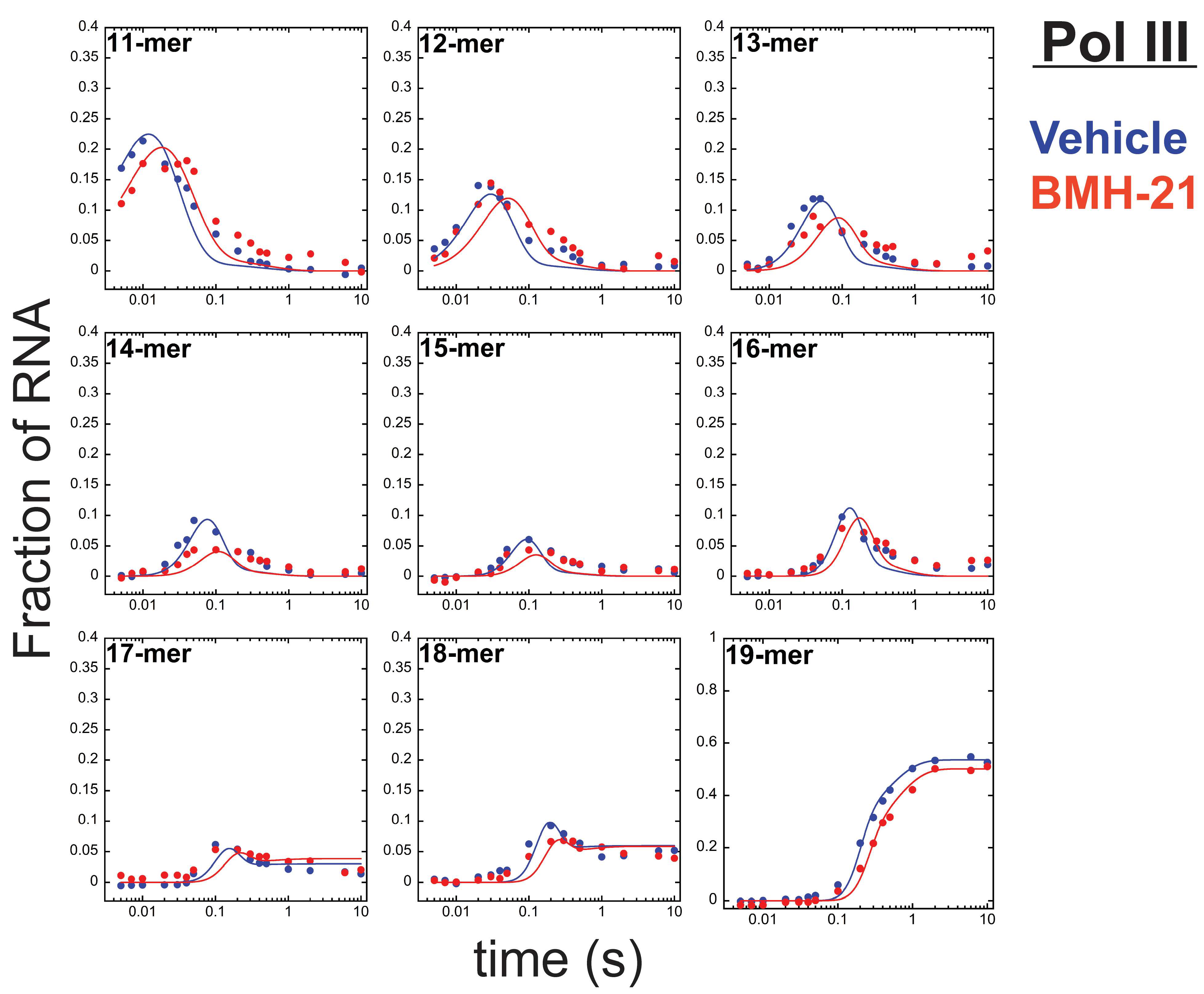
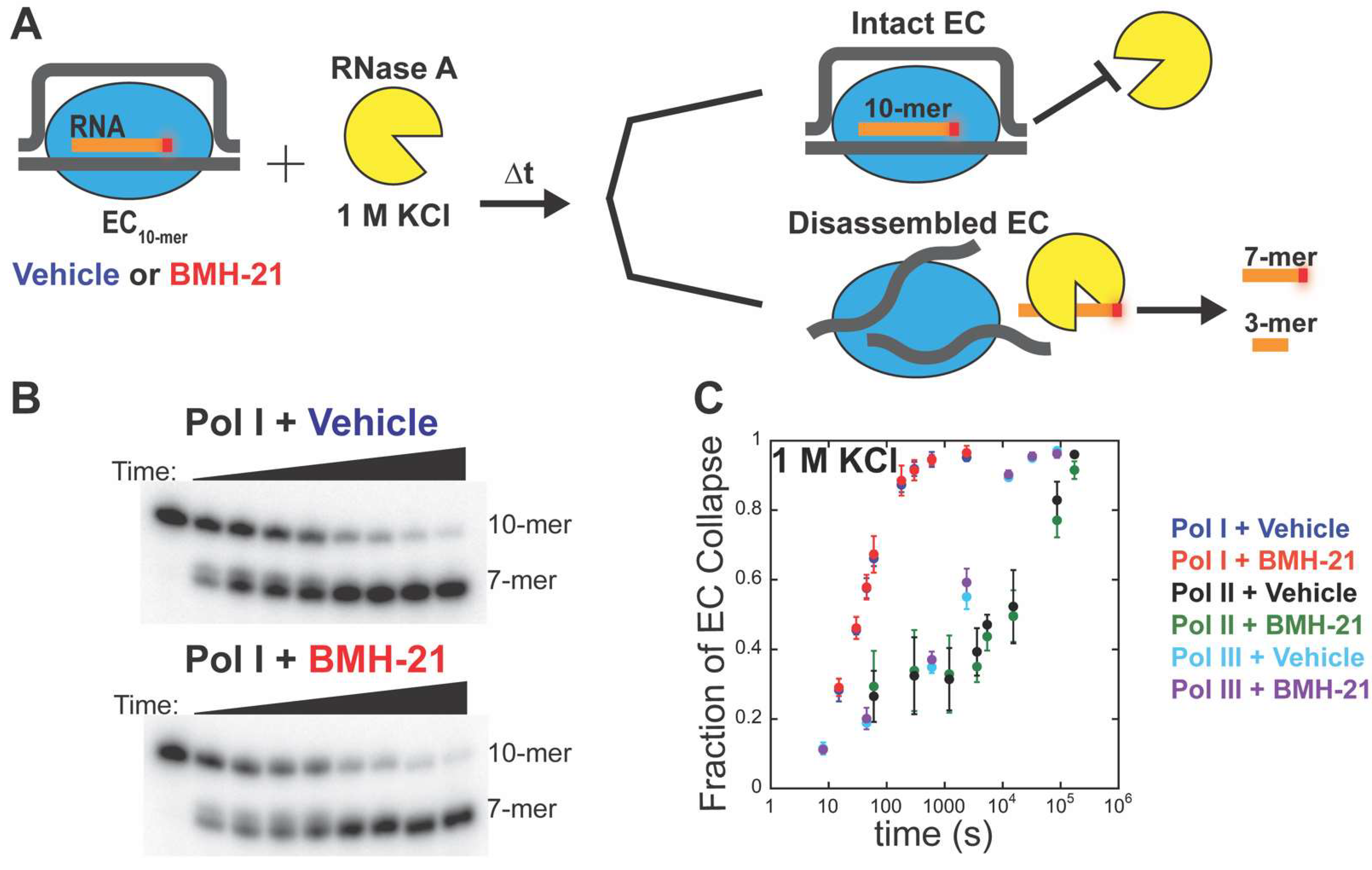
| Kinetic Parameter (s−1) | Vehicle a | BMH-21 (1 µM) b |
|---|---|---|
| kobs,1 | 80 ± 4 | 60 ± 20 |
| kobs,2 | 120 ± 30 | 90 ± 20 |
| kobs,3 | 66 ± 6 | 30 ± 7 |
| kobs,4 | 26 ± 3 | 13 ± 3 |
| kobs,5 | 53 ± 9 | 40 ± 20 |
| kobs,6 | 63 ± 5 | 40 ± 20 |
| kobs,7 | 17 ± 4 | 13 ± 3 |
| kobs,8 | 48 ± 5 | 70 ± 50 |
| kobs,9 | 52 ± 3 | 60 ± 40 |
| kobs,10 | 8 ± 2 | 8 ± 7 |
| kobs,1,on | - | 2.5 ± 0.8 |
| kobs,1,off | - | 25 ± 5 |
| kobs,2,on | - | 0.9 ± 0.4 |
| kobs,2,off | - | 17 ± 5 |
| Kinetic Parameter | Vehicle a | BMH-21 (1 µM) b |
|---|---|---|
| kobs,1,F/kobs,1,R | 0.43 ± 0.06 | 0.5 ± 0.1 |
| kobs,2,F/kobs,2,R | 1.9 ± 0.6 | 5 ± 3 |
| kobs,3,F/kobs,3,R | 0.8 ± 0.2 | 1.0 ± 0.5 |
| kobs,4,F/kobs,4,R | 1.4 ± 0.5 | 1.6 ± 0.5 |
| kobs,5,F/kobs,5,R | 1.8 ± 0.3 | 2.6 ± 0.7 |
| kobs,6,F/kobs,6,R | 0.92 ± 0.04 | 1.5 ± 0.6 |
| kobs,7,F/kobs,7,R | 2.0 ± 0.2 | 2.6 ± 0.7 |
| kobs,8,F/kobs,8,R | 1.0 ± 0.3 | 1.7 ± 0.7 |
| kobs,9,F/kobs,9,R | 20 ± 20 | 20 ± 10 |
| Kinetic Parameter (s−1) | Vehicle a | BMH-21 (1 µM) b |
|---|---|---|
| kobs,* | 1.8 ± 0.6 | 2.1 ± 0.5 |
| kobs,1 | 138 ± 7 | 104 ± 3 |
| kobs,2 | 46.3 ± 0.5 | 30 ± 4 |
| kobs,3 | 50 ± 1 | 25 ± 3 |
| kobs,4 | 40 ± 2 | 31 ± 5 |
| kobs,5 | 44 ± 2 | 54 ± 5 |
| kobs,6 | 60 ± 1 | 54 ± 9 |
| kobs,7 | 26 ± 1 | 20 ± 2 |
| kobs,8 | 54 ± 3 | 46 ± 4 |
| kobs,9 | 31 ± 3 | 30 ± 3 |
| kobs,10 | 3.5 ± 0.5 | 3.3 ± 0.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jacobs, R.Q.; Fuller, K.B.; Cooper, S.L.; Carter, Z.I.; Laiho, M.; Lucius, A.L.; Schneider, D.A. RNA Polymerase I Is Uniquely Vulnerable to the Small-Molecule Inhibitor BMH-21. Cancers 2022, 14, 5544. https://doi.org/10.3390/cancers14225544
Jacobs RQ, Fuller KB, Cooper SL, Carter ZI, Laiho M, Lucius AL, Schneider DA. RNA Polymerase I Is Uniquely Vulnerable to the Small-Molecule Inhibitor BMH-21. Cancers. 2022; 14(22):5544. https://doi.org/10.3390/cancers14225544
Chicago/Turabian StyleJacobs, Ruth Q., Kaila B. Fuller, Stephanie L. Cooper, Zachariah I. Carter, Marikki Laiho, Aaron L. Lucius, and David A. Schneider. 2022. "RNA Polymerase I Is Uniquely Vulnerable to the Small-Molecule Inhibitor BMH-21" Cancers 14, no. 22: 5544. https://doi.org/10.3390/cancers14225544
APA StyleJacobs, R. Q., Fuller, K. B., Cooper, S. L., Carter, Z. I., Laiho, M., Lucius, A. L., & Schneider, D. A. (2022). RNA Polymerase I Is Uniquely Vulnerable to the Small-Molecule Inhibitor BMH-21. Cancers, 14(22), 5544. https://doi.org/10.3390/cancers14225544





