Multi-Omics Approaches in Colorectal Cancer Screening and Diagnosis, Recent Updates and Future Perspectives
Abstract
Simple Summary
Abstract
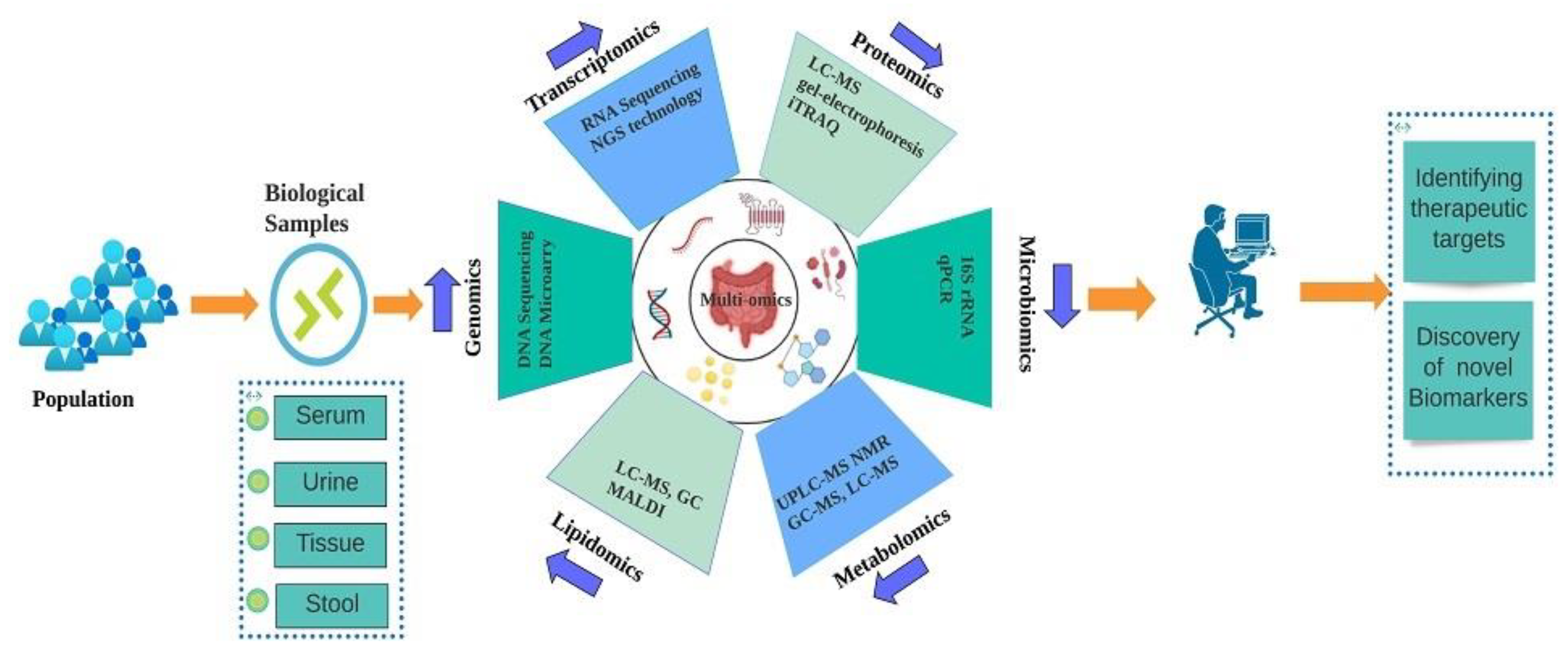
1. Introduction
2. Genomics of CRC
3. Transcriptomics of CRC
4. Proteomics of CRC
5. Microbiomics of CRC
6. Metabolomics of CRC
7. Lipidomics of CRC
8. Future Perspectives and Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CBX8 | Chromobox 8 |
| CD96 | CD96 Molecule |
| 8MTUS1 | Microtubule Associated Scaffold Protein 1 |
| SDC2 | Syndecan 2 |
| NDRG4 | NDRG Family Member 4 |
| SOX21 | SRY-Box Transcription Factor 21 |
| BDNF | Brain-Derived Neurotrophic Factor |
| PTGS2 | Prostaglandin–Endoperoxide Synthase 2 |
| GSK3B | Glycogen Synthase Kinase 3 Beta |
| CTNNB1 | Catenin Beta 1 |
| HPGD | 15-Hydroxyprostaglandin Dehydrogenase |
| YWHAB | Tyrosine 3–Monooxygenase/Tryptophan 5–Monooxygenase Activation Protein Beta |
| MCM4, | Minichromosome Maintenance Complex Component 4 |
| FBXO46 | F-Box Protein 46 |
| DPP7/2 | Dipeptidyl Peptidase 7 |
| SDC2 | Syndecan 2 |
| TFPI2 | Tissue Factor Pathway Inhibitor 2 |
| SNORD15B | Small Nucleolar RNA, C/D Box 15B |
| SNORA5C | Small Nucleolar RNA, H/ACA Box 5C |
| GALR1 | Galanin Receptor 1 |
| LRRC19 | Leucine-rich repeat-containing protein 19 |
| GPR55 | G protein-coupled receptor 55 |
| CCAT2 | Colon Cancer Associated Transcript 2 |
| CCAT1 | Colon Cancer Associated Transcript 1 |
| H19 | H19 Imprinted Maternally Expressed Transcript |
| MALAT1 | Metastasis Associated Lung Adenocarcinoma Transcript 1 |
| MEG3 | Maternally Expressed 3 |
| HULC | Hepatocellular Carcinoma Up-Regulated Long Non-Coding RNA |
| HOTAIR | HOX Transcript Antisense RNA |
| PCAT1 | Prostate Cancer Associated Transcript 1 |
| PTENP1 | Phosphatase And Tensin Homolog Pseudogene 1 |
| TUSC7 | Tumour Suppressor Candidate 7 |
| CHD 9 | Chromodomain Helicase DNA Binding Protein 9 |
| ACTBL2 | Actin Beta Like 2 |
| CDK3, | Cyclin Dependent Kinase 3 |
| CDK5 | Cyclin Dependent Kinase 5 |
| CDK8 | Cyclin-dependent kinase 8 |
| STK4 or MST1 | serine/threonine kinase 4 or Macrophage Stimulating 1 |
| MRC1 | Mannose Receptor C-Type 1 |
| S100A90 | S100 Calcium Binding Protein A9 |
| CEACAM-7 | CEA Cell Adhesion Molecule 7 |
| CEA | Carcinoembryonic antigen |
| SPG20 | spastic paraplegia 20 |
| STK31 | Serine/Threonine Kinase 31 |
| TPM3 | Tropomyosin 3 |
| FJX1 | Four-Jointed Box Kinase 1 |
| NOP14 | Nucleolar protein 14 |
| SPARCL1 | Secreted protein acidic and rich in cysteine-like 1 |
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, T.; Arnold, D.; Taniguchi, H.; Pentheroudakis, G.; Yamazaki, K.; Xu, R.H.; Kim, T.W.; Ismail, F.; Tan, I.B.; Yeh, K.H.; et al. Pan-Asian adapted ESMO consensus guidelines for the management of patients with metastatic colorectal cancer: A JSMO-ESMO initiative endorsed by CSCO, KACO, MOS, SSO and TOS. Ann. Oncol. 2018, 29, 44–70. [Google Scholar] [CrossRef]
- Benson, A.B.; Venook, A.P.; Al-Hawary, M.M.; Arain, M.A.; Chen, Y.J.; Ciombor, K.K.; Cohen, S.; Cooper, H.S.; Deming, D.; Farkas, L.; et al. Colon Cancer, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2021, 19, 329–359. [Google Scholar] [CrossRef] [PubMed]
- Iyer, P.; Deng, M.; Handorf, E.A.; Nakhoda, S.; Dotan, E. Assessing Oncologists’ Adoption of Biomarker Testing in Metastatic Colorectal Cancer Using Real World Data. JNCI Cancer Spectr. 2022, pkac065. [Google Scholar] [CrossRef]
- Amatu, A.; Sartore-Bianchi, A.; Bencardino, K.; Pizzutilo, E.G.; Tosi, F.; Siena, S. Tropomyosin receptor kinase (TRK) biology and the role of NTRK gene fusions in cancer. Ann. Oncol. 2019, 30 (Suppl. S8), viii5–viii15. [Google Scholar] [CrossRef]
- Marabelle, A.; Fakih, M.; Lopez, J.; Shah, M.; Shapira-Frommer, R.; Nakagawa, K.; Chung, H.C.; Kindler, H.L.; Lopez-Martin, J.A.; Miller, W.H., Jr.; et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: Prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 2020, 21, 1353–1365. [Google Scholar] [CrossRef]
- Argilés, G.; Tabernero, J.; Labianca, R.; Hochhauser, D.; Salazar, R.; Iveson, T.; Laurent-Puig, P.; Quirke, P.; Yoshino, T.; Taieb, J.; et al. Localised colon cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2020, 31, 1291–1305. [Google Scholar] [CrossRef]
- Testing for Colorectal Cancer. How Is Colorectal Cancer Diagnosed? Available online: https://www.cancer.gov/types/colorectal/screening-fact-sheet (accessed on 12 September 2022).
- Elsafi, S.H.; Alqahtani, N.I.; Zakary, N.Y.; Al Zahrani, E.M. The sensitivity, specificity, predictive values, and likelihood ratios of fecal occult blood test for the detection of colorectal cancer in hospital settings. Clin. Exp. Gastroenterol. 2015, 8, 279–284. [Google Scholar] [CrossRef]
- Chan, S.C.H.; Liang, J.Q. Advances in tests for colorectal cancer screening and diagnosis. Expert Rev. Mol. Diagn. 2022, 22, 449–460. [Google Scholar] [CrossRef]
- Tan, K.C.; Ipcho, S.V.; Trengove, R.D.; Oliver, R.P.; Solomon, P.S. Assessing the impact of transcriptomics, proteomics and metabolomics on fungal phytopathology. Mol. Plant Pathol. 2009, 10, 703–715. [Google Scholar] [CrossRef] [PubMed]
- Dalal, N.; Jalandra, R.; Sharma, M.; Prakash, H.; Makharia, G.K.; Solanki, P.R.; Singh, R.; Kumar, A. Omics technologies for improved diagnosis and treatment of colorectal cancer: Technical advancement and major perspectives. Biomed. Pharmacother. 2020, 131, 110648. [Google Scholar] [CrossRef]
- Jahani-Sherafat, S.; Alebouyeh, M.; Moghim, S.; Ahmadi Amoli, H.; Ghasemian-Safaei, H. Role of gut microbiota in the pathogenesis of colorectal cancer; a review article. Gastroenterol. Hepatol. Bed Bench 2018, 11, 101–109. [Google Scholar] [PubMed]
- Peng, Y.; Nie, Y.; Yu, J.; Wong, C.C. Microbial Metabolites in Colorectal Cancer: Basic and Clinical Implications. Metabolites 2021, 11, 159. [Google Scholar] [CrossRef] [PubMed]
- Kontush, A.; Chapman, M.J. Lipidomics as a tool for the study of lipoprotein metabolism. Curr. Atheroscler. Rep. 2010, 12, 194–201. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.M.; Shui, G. Lipidomics as a principal tool for advancing biomedical research. J. Genet. Genom. 2013, 40, 375–390. [Google Scholar] [CrossRef]
- Mato-Abad, V.; Pazos, A.; Munteanu, C.R.; Liñares-Blanco, J.; Alvarez-Gonzalez, S.; Vázquez-Naya, J.M.; Pedreira, N.; Amigo, J.; Fernandez-Lozano, C. Bioinformatic tools for research in CRC. In Foundations of Colorectal Cancer; Elsevier: Amsterdam, The Netherlands, 2022; pp. 231–247. [Google Scholar]
- Lopez, G.; Boggio, F.; Ferrero, S.; Fusco, N.; Del Gobbo, A. Molecular and Immunohistochemical Markers with Prognostic and Predictive Significance in Liver Metastases from Colorectal Carcinoma. Int. J. Mol. Sci. 2018, 19, 3014. [Google Scholar] [CrossRef]
- Loomans-Kropp, H.A.; Umar, A. Increasing Incidence of Colorectal Cancer in Young Adults. J. Cancer Epidemiol. 2019, 2019, 9841295. [Google Scholar] [CrossRef]
- Casamassimi, A.; Federico, A.; Rienzo, M.; Esposito, S.; Ciccodicola, A. Transcriptome Profiling in Human Diseases: New Advances and Perspectives. Int. J. Mol. Sci. 2017, 18, 1652. [Google Scholar] [CrossRef]
- Wheeler, D.A.; Wang, L. From human genome to cancer genome: The first decade. Genome Res. 2013, 23, 1054–1062. [Google Scholar] [CrossRef]
- López-Lázaro, M. A new view of carcinogenesis and an alternative approach to cancer therapy. Mol. Med. 2010, 16, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Supplitt, S.; Karpinski, P.; Sasiadek, M.; Laczmanska, I. Current Achievements and Applications of Transcriptomics in Personalized Cancer Medicine. Int. J. Mol. Sci. 2021, 22, 1422. [Google Scholar] [CrossRef] [PubMed]
- Sardo, E.; Napolitano, S.; Della Corte, C.M.; Ciardiello, D.; Raucci, A.; Arrichiello, G.; Troiani, T.; Ciardiello, F.; Martinelli, E.; Martini, G. Multi-Omic Approaches in Colorectal Cancer beyond Genomic Data. J. Pers. Med. 2022, 12, 128. [Google Scholar] [CrossRef] [PubMed]
- Paleri, P. Revisiting National Security: Prospecting Governance for Human Well-Being; Springer Nature: Berlin/Heidelberg, Germany, 2022. [Google Scholar]
- Bunnik, E.M.; Le Roch, K.G. An Introduction to Functional Genomics and Systems Biology. Adv. Wound Care 2013, 2, 490–498. [Google Scholar] [CrossRef]
- Przybyla, L.; Gilbert, L.A. A new era in functional genomics screens. Nat. Rev. Genet. 2022, 23, 89–103. [Google Scholar] [CrossRef]
- Definition of genomics. NCI Dictionary of Cancer Terms—National Cancer Institute. Available online: https://www.cancer.gov/publications/dictionaries/cancer-terms/def/genomics (accessed on 12 September 2022).
- Bosch, L.J.; Carvalho, B.; Fijneman, R.J.; Jimenez, C.R.; Pinedo, H.M.; van Engeland, M.; Meijer, G.A. Molecular tests for colorectal cancer screening. Clin. Colorectal. Cancer 2011, 10, 8–23. [Google Scholar] [CrossRef]
- Escobar-Zepeda, A.; Vera-Ponce de León, A.; Sanchez-Flores, A. The Road to Metagenomics: From Microbiology to DNA Sequencing Technologies and Bioinformatics. Front. Genet. 2015, 6, 348. [Google Scholar] [CrossRef]
- Gan, X.; Wang, T.; Chen, Z.Y.; Zhang, K.H. Blood-derived molecular signatures as biomarker panels for the early detection of colorectal cancer. Mol. Biol. Rep. 2020, 47, 8159–8168. [Google Scholar] [CrossRef]
- Ghatak, S.; Mehrabi, S.F.; Mehdawi, L.M.; Satapathy, S.R.; Sjölander, A. Identification of a Novel Five-Gene Signature as a Prognostic and Diagnostic Biomarker in Colorectal Cancers. Int. J. Mol. Sci. 2022, 23, 793. [Google Scholar] [CrossRef]
- Essa, H.Y.S.; Kusaf, G.; Yuruker, O.; Kalkan, R. Epigenetic Alteration in Colorectal Cancer: A Biomarker for Diagnostic and Therapeutic Application. Glob. Med. Genet. 2022, 9, 258–262. [Google Scholar] [CrossRef]
- Esteller, M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007, 8, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.P.; Rai, S.; Suyal, S.; Singh, S.K.; Singh, N.K.; Agarwal, A.; Srivastava, S. Genetic and epigenetic markers in colorectal cancer screening: Recent advances. Expert Rev. Mol. Diagn. 2017, 17, 665–685. [Google Scholar] [CrossRef]
- Zamani, M.; Hosseini, S.V.; Mokarram, P. Epigenetic biomarkers in colorectal cancer: Premises and prospects. Biomarkers 2018, 23, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Berdasco, M.; Esteller, M. Clinical epigenetics: Seizing opportunities for translation. Nat. Rev. Genet. 2019, 20, 109–127. [Google Scholar] [CrossRef] [PubMed]
- Galanopoulos, M.; Tsoukalas, N.; Papanikolaou, I.S.; Tolia, M.; Gazouli, M.; Mantzaris, G.J. Abnormal DNA methylation as a cell-free circulating DNA biomarker for colorectal cancer detection: A review of literature. World J. Gastrointest. Oncol. 2017, 9, 142–152. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, N.; Spencer, E.; Torkamani, A.; Nicholson, L. Liquid Biopsies for Cancer: Coming to a Patient near You. J. Clin. Med. 2017, 6, 3. [Google Scholar] [CrossRef]
- Belkhiri, A.; El-Rifai, W. 5-Methylcytosine hydroxylation-mediated LINE-1 hypomethylation: A novel mechanism of proto-oncogenes activation in colorectal cancer? Gut 2014, 63, 538–539. [Google Scholar] [CrossRef]
- Ichimura, N.; Shinjo, K.; An, B.; Shimizu, Y.; Yamao, K.; Ohka, F.; Katsushima, K.; Hatanaka, A.; Tojo, M.; Yamamoto, E.; et al. Aberrant TET1 Methylation Closely Associated with CpG Island Methylator Phenotype in Colorectal Cancer. Cancer Prev. Res. 2015, 8, 702–711. [Google Scholar] [CrossRef]
- Hur, K.; Cejas, P.; Feliu, J.; Moreno-Rubio, J.; Burgos, E.; Boland, C.R.; Goel, A. Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut 2014, 63, 635–646. [Google Scholar] [CrossRef]
- Zheng, R.; Gao, D.; He, T.; Zhang, M.; Zhang, X.; Linghu, E.; Wei, L.; Guo, M. Methylation of DIRAS1 promotes colorectal cancer progression and may serve as a marker for poor prognosis. Clin. Epigenetics 2017, 9, 50. [Google Scholar] [CrossRef]
- Wang, Y.; He, T.; Herman, J.G.; Linghu, E.; Yang, Y.; Fuks, F.; Zhou, F.; Song, L.; Guo, M. Methylation of ZNF331 is an independent prognostic marker of colorectal cancer and promotes colorectal cancer growth. Clin. Epigenetics 2017, 9, 115. [Google Scholar] [CrossRef]
- He, T.; Zhang, M.; Zheng, R.; Zheng, S.; Linghu, E.; Herman, J.G.; Guo, M. Methylation of SLFN11 is a marker of poor prognosis and cisplatin resistance in colorectal cancer. Epigenomics 2017, 9, 849–862. [Google Scholar] [CrossRef]
- Zhu, Y.; Lu, H.; Zhang, D.; Li, M.; Sun, X.; Wan, L.; Yu, D.; Tian, Y.; Jin, H.; Lin, A.; et al. Integrated analyses of multi-omics reveal global patterns of methylation and hydroxymethylation and screen the tumor suppressive roles of HADHB in colorectal cancer. Clin. Epigenetics 2018, 10, 30. [Google Scholar] [CrossRef] [PubMed]
- Vasanthakumar, A.; Godley, L.A. 5-hydroxymethylcytosine in cancer: Significance in diagnosis and therapy. Cancer Genet. 2015, 208, 167–177. [Google Scholar] [CrossRef]
- Wang, J.; Su, Y.; Tian, Y.; Ding, Y.; Wang, X. Characterization of DNA hydroxymethylation profile in cervical cancer. Artif. Cells Nanomed. Biotechnol. 2019, 47, 2706–2714. [Google Scholar] [CrossRef]
- Hlady, R.A.; Sathyanarayan, A.; Thompson, J.J.; Zhou, D.; Wu, Q.; Pham, K.; Lee, J.H.; Liu, C.; Robertson, K.D. Integrating the Epigenome to Identify Drivers of Hepatocellular Carcinoma. Hepatology 2019, 69, 639–652. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, S.; Pradhan, K.; Campbell, N.; Mazdo, J.; Vasantkumar, A.; Maqbool, S.; Bhagat, T.D.; Gupta, S.; Suzuki, M.; Yu, Y.; et al. Altered hydroxymethylation is seen at regulatory regions in pancreatic cancer and regulates oncogenic pathways. Genome Res. 2017, 27, 1830–1842. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.P.; Ottaviano, R.; Unterberger, E.B.; Lempiäinen, H.; Muller, A.; Terranova, R.; Illingworth, R.S.; Webb, S.; Kerr, A.R.; Lyall, M.J.; et al. Loss of Tet1-Associated 5-Hydroxymethylcytosine Is Concomitant with Aberrant Promoter Hypermethylation in Liver Cancer. Cancer Res. 2016, 76, 3097–3108. [Google Scholar] [CrossRef] [PubMed]
- Jiang, A.C.; Buckingham, L.; Barbanera, W.; Korang, A.Y.; Bishesari, F.; Melson, J. LINE-1 is preferentially hypomethylated within adenomatous polyps in the presence of synchronous colorectal cancer. Clin. Epigenetics 2017, 9, 25. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Tang, T.; Li, C.; Liu, X.; Zhou, L. CBX8 and CD96 Are Important Prognostic Biomarkers of Colorectal Cancer. Med. Sci. Monit. 2018, 24, 7820–7827. [Google Scholar] [CrossRef]
- Cheng, L.Y.; Huang, M.S.; Zhong, H.G.; Ru, H.M.; Mo, S.S.; Wei, C.Y.; Su, Z.J.; Mo, X.W.; Yan, L.H.; Tang, W.Z. MTUS1 is a promising diagnostic and prognostic biomarker for colorectal cancer. World J. Surg. Oncol. 2022, 20, 257. [Google Scholar] [CrossRef]
- Jin, S.; Ye, Q.; Hong, Y.; Dai, W.; Zhang, C.; Liu, W.; Guo, Y.; Zhu, D.; Zhang, Z.; Chen, S.; et al. A systematic evaluation of stool DNA preparation protocols for colorectal cancer screening via analysis of DNA methylation biomarkers. Clin. Chem. Lab. Med. 2020, 59, 91–99. [Google Scholar] [CrossRef]
- Moradi, K.; Babaei, E.; Hosseinpour Feizi, M.A.; Safaralizadeh, R.; Rezvani, N. Quantitative detection of SRY-Box 21 (SOX21) gene promoter methylation as a stool-based noninvasive biomarker for early diagnosis of colorectal cancer by MethyLight method. Indian J. Cancer 2021, 58, 217–224. [Google Scholar] [CrossRef]
- Ahluwalia, P.; Mondal, A.K.; Bloomer, C.; Fulzele, S.; Jones, K.; Ananth, S.; Gahlay, G.K.; Heneidi, S.; Rojiani, A.M.; Kota, V.; et al. Identification and Clinical Validation of a Novel 4 Gene-Signature with Prognostic Utility in Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 3818. [Google Scholar] [CrossRef]
- Zhang, W.; Yang, C.; Wang, S.; Xiang, Z.; Dou, R.; Lin, Z.; Zheng, J.; Xiong, B. SDC2 and TFPI2 Methylation in Stool Samples as an Integrated Biomarker for Early Detection of Colorectal Cancer. Cancer Manag. Res. 2021, 13, 3601–3617. [Google Scholar] [CrossRef]
- Shen, L.; Lu, W.; Huang, Y.; He, J.; Wang, Q.; Zheng, X.; Wang, Z. SNORD15B and SNORA5C: Novel Diagnostic and Prognostic Biomarkers for Colorectal Cancer. Biomed Res. Int. 2022, 2022, 8260800. [Google Scholar] [CrossRef]
- Gu, S.; Qian, S.; Lin, S.; Ye, D.; Li, Q.; Yang, J.; Ying, X.; Li, Z.; Tang, M.; Wang, J.; et al. Promoter hypermethylation of GALR1 acts as an early epigenetic susceptibility event in colorectal carcinogenesis. J. Hum. Genet. 2022, 67, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.J.; Liu, M.; Jiang, H.Y.; Yu, Y.W. Downregulation of LRRC19 Is Associated with Poor Prognosis in Colorectal Cancer. J. Oncol. 2022, 2022, 5848823. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh-Sedigh, M.; Mahmoodzadeh, H.; Fazeli, M.S.; Haddadi-Aghdam, M.; Teimoori-Toolabi, L. The potential of PIK3CA, KRAS, BRAF, and APC hotspot mutations as a non-invasive detection method for colorectal cancer. Mol. Cell. Probes 2022, 63, 101807. [Google Scholar] [CrossRef] [PubMed]
- Mantione, K.J.; Kream, R.M.; Kuzelova, H.; Ptacek, R.; Raboch, J.; Samuel, J.M.; Stefano, G.B. Comparing bioinformatic gene expression profiling methods: Microarray and RNA-Seq. Med. Sci. Monit. Basic Res. 2014, 20, 138–142. [Google Scholar] [CrossRef]
- Lowe, R.; Shirley, N.; Bleackley, M.; Dolan, S.; Shafee, T. Transcriptomics technologies. PLoS Comput. Biol. 2017, 13, e1005457. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Jacquier, A. The complex eukaryotic transcriptome: Unexpected pervasive transcription and novel small RNAs. Nat. Rev. Genet. 2009, 10, 833–844. [Google Scholar] [CrossRef]
- Popov, D.V.; Makhnovskii, P.A.; Shagimardanova, E.I.; Gazizova, G.R.; Lysenko, E.A.; Gusev, O.A.; Vinogradova, O.L. Contractile activity-specific transcriptome response to acute endurance exercise and training in human skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2019, 316, E605–E614. [Google Scholar] [CrossRef] [PubMed]
- Iwata, M.; Yuan, L.; Zhao, Q.; Tabei, Y.; Berenger, F.; Sawada, R.; Akiyoshi, S.; Hamano, M.; Yamanishi, Y. Predicting drug-induced transcriptome responses of a wide range of human cell lines by a novel tensor-train decomposition algorithm. Bioinformatics 2019, 35, i191–i199. [Google Scholar] [CrossRef]
- Jaeger, P.A.; Doherty, C.; Ideker, T. Modeling transcriptome dynamics in a complex world. Cell 2012, 151, 1161–1162. [Google Scholar] [CrossRef]
- Mallardo, M.; Poltronieri, P.; D’Urso, O.F. Non-protein coding RNA biomarkers and differential expression in cancers: A review. J. Exp. Clin. Cancer Res. 2008, 27, 19. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bashiardes, S.; Zilberman-Schapira, G.; Elinav, E. Use of Metatranscriptomics in Microbiome Research. Bioinform. Biol. Insights 2016, 10, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Slaby, O.; Svoboda, M.; Michalek, J.; Vyzula, R. MicroRNAs in colorectal cancer: Translation of molecular biology into clinical application. Mol. Cancer 2009, 8, 102. [Google Scholar] [CrossRef]
- Schee, K.; Lorenz, S.; Worren, M.M.; Günther, C.C.; Holden, M.; Hovig, E.; Fodstad, O.; Meza-Zepeda, L.A.; Flatmark, K. Deep Sequencing the MicroRNA Transcriptome in Colorectal Cancer. PLoS ONE 2013, 8, e66165. [Google Scholar] [CrossRef]
- Sun, Y.; Shen, S.; Tang, H.; Xiang, J.; Peng, Y.; Tang, A.; Li, N.; Zhou, W.; Wang, Z.; Zhang, D.; et al. miR-429 identified by dynamic transcriptome analysis is a new candidate biomarker for colorectal cancer prognosis. Omics 2014, 18, 54–64. [Google Scholar] [CrossRef] [PubMed]
- Chen, E.; Li, Q.; Wang, H.; Yang, F.; Min, L.; Yang, J. MiR-92a promotes tumorigenesis of colorectal cancer, a transcriptomic and functional based study. Biomed Pharmacother. 2018, 106, 1370–1377. [Google Scholar] [CrossRef] [PubMed]
- Durán-Vinet, B.; Araya-Castro, K.; Calderón, J.; Vergara, L.; Weber, H.; Retamales, J.; Araya-Castro, P.; Leal-Rojas, P. CRISPR/Cas13-Based Platforms for a Potential Next-Generation Diagnosis of Colorectal Cancer through Exosomes Micro-RNA Detection: A Review. Cancers 2021, 13, 4640. [Google Scholar] [CrossRef]
- Gallardo-Gómez, M.; De Chiara, L.; Álvarez-Chaver, P.; Cubiella, J. Colorectal cancer screening and diagnosis: Omics-based technologies for development of a non-invasive blood-based method. Expert Rev. Anticancer. Ther. 2021, 21, 723–738. [Google Scholar] [CrossRef]
- Chen, E.; Li, Q.; Wang, H.; Zhang, P.; Zhao, X.; Yang, F.; Yang, J. MiR-32 promotes tumorigenesis of colorectal cancer by targeting BMP5. Biomed. Pharmacother. 2018, 106, 1046–1051. [Google Scholar] [CrossRef]
- Yamada, A.; Yu, P.; Lin, W.; Okugawa, Y.; Boland, C.R.; Goel, A. A RNA-Sequencing approach for the identification of novel long non-coding RNA biomarkers in colorectal cancer. Sci. Rep. 2018, 8, 575. [Google Scholar] [CrossRef]
- Zhang, Z.; Jia, H.; Gu, T.; Hu, Q.; Yu, J.; Zang, D.; Song, N.; Wang, H. RNA sequencing and bioinformatics analysis of the long noncoding RNA-mRNA network in colorectal cancer. J. Cell. Biochem. 2018, 119, 9957–9966. [Google Scholar] [CrossRef]
- Jang, J.E.; Kim, H.P.; Han, S.W.; Jang, H.; Lee, S.H.; Song, S.H.; Bang, D.; Kim, T.Y. NFATC3-PLA2G15 Fusion Transcript Identified by RNA Sequencing Promotes Tumor Invasion and Proliferation in Colorectal Cancer Cell Lines. Cancer Res. Treat. 2019, 51, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, X.; Wu, F.; Huang, R.; Xue, F.; Liang, G.; Tao, M.; Cai, P.; Huang, Y. Transcriptome profiling of the cancer, adjacent non-tumor and distant normal tissues from a colorectal cancer patient by deep sequencing. PLoS ONE 2012, 7, e41001. [Google Scholar] [CrossRef]
- Kashima, Y.; Sakamoto, Y.; Kaneko, K.; Seki, M.; Suzuki, Y.; Suzuki, A. Single-cell sequencing techniques from individual to multiomics analyses. Exp. Mol. Med. 2020, 52, 1419–1427. [Google Scholar] [CrossRef]
- Haque, A.; Engel, J.; Teichmann, S.A.; Lönnberg, T. A practical guide to single-cell RNA-sequencing for biomedical research and clinical applications. Genome Med. 2017, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Hyeon, D.Y.; Hwang, D. Single-cell multiomics: Technologies and data analysis methods. Exp. Mol. Med. 2020, 52, 1428–1442. [Google Scholar] [CrossRef]
- Picelli, S. Single-cell RNA-sequencing: The future of genome biology is now. RNA Biol. 2017, 14, 637–650. [Google Scholar] [CrossRef]
- Pelka, K.; Hofree, M.; Chen, J.H.; Sarkizova, S.; Pirl, J.D.; Jorgji, V.; Bejnood, A.; Dionne, D.; Ge, W.H.; Xu, K.H.; et al. Spatially organized multicellular immune hubs in human colorectal cancer. Cell 2021, 184, 4734–4752.e4720. [Google Scholar] [CrossRef]
- Davis, R.T.; Blake, K.; Ma, D.; Gabra, M.B.I.; Hernandez, G.A.; Phung, A.T.; Yang, Y.; Maurer, D.; Lefebvre, A.; Alshetaiwi, H.; et al. Transcriptional diversity and bioenergetic shift in human breast cancer metastasis revealed by single-cell RNA sequencing. Nat. Cell Biol. 2020, 22, 310–320. [Google Scholar] [CrossRef]
- Shalek, A.K.; Benson, M. Single-cell analyses to tailor treatments. Sci. Transl. Med. 2017, 9, eaan4730. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.H.; Zhou, Z.G.; Chen, R.; Wang, M.J.; Zhou, B.; Li, Y.; Sun, X.F. Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancer. Tumour. Biol. 2013, 34, 2175–2181. [Google Scholar] [CrossRef]
- Wang, J.; Li, X.; Lu, L.; He, L.; Hu, H.; Xu, Z. Circular RNA hsa_circ_0000567 can be used as a promising diagnostic biomarker for human colorectal cancer. J. Clin. Lab. Anal. 2018, 32, e22379. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, D.; Zafari, Y.; Estaki, Z.; Mehrabi, M.; Moghbelinejad, S. Evaluation of plasma circ_0006282 as a novel diagnostic biomarker in colorectal cancer. J. Clin. Lab. Anal. 2022, 36, e24147. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Song, Y.; Wang, J.; Huang, J. Plasma circular RNA panel acts as a novel diagnostic biomarker for colorectal cancer detection. Am. J. Transl. Res. 2020, 12, 7395–7403. [Google Scholar]
- Iwasaki, H.; Shimura, T.; Kitagawa, M.; Yamada, T.; Nishigaki, R.; Fukusada, S.; Okuda, Y.; Katano, T.; Horike, S.I.; Kataoka, H. A Novel Urinary miRNA Biomarker for Early Detection of Colorectal Cancer. Cancers 2022, 14, 461. [Google Scholar] [CrossRef] [PubMed]
- Ismail, H.T.H.; AbdelMageed, M.; Lindmark, G.; Hammarström, M.L.; Hammarström, S.; Sitohy, B. Prognostic Significance of GPR55 mRNA Expression in Colon Cancer. Int. J. Mol. Sci. 2022, 23, 4556. [Google Scholar] [CrossRef] [PubMed]
- Kang, E.; Jung, S.C.; Nam, S.K.; Park, Y.; Seo, S.H.; Park, K.U.; Oh, H.K.; Kim, D.W.; Kang, S.B.; Lee, H.S. Tissue miR-200c-3p and circulating miR-1290 as potential prognostic biomarkers for colorectal cancer. Sci. Rep. 2022, 12, 2295. [Google Scholar] [CrossRef]
- Liu, X.; Xu, X.; Pan, B.; He, B.; Chen, X.; Zeng, K.; Xu, M.; Pan, Y.; Sun, H.; Xu, T.; et al. Circulating miR-1290 and miR-320d as Novel Diagnostic Biomarkers of Human Colorectal Cancer. J. Cancer 2019, 10, 43–50. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, M.; Shan, X.; Zhou, X.; Wang, T.; Zhang, J.; Tao, J.; Cheng, W.; Chen, G.; Li, J.; et al. A panel of seven-miRNA signature in plasma as potential biomarker for colorectal cancer diagnosis. Gene 2019, 687, 246–254. [Google Scholar] [CrossRef]
- Gharib, E.; Nazemalhosseini-Mojarad, E.; Baghdar, K.; Nayeri, Z.; Sadeghi, H.; Rezasoltani, S.; Jamshidi-Fard, A.; Larki, P.; Sadeghi, A.; Hashemi, M.; et al. Identification of a stool long non-coding RNAs panel as a potential biomarker for early detection of colorectal cancer. J. Clin. Lab. Anal. 2021, 35, e23601. [Google Scholar] [CrossRef]
- Ahmed, F.E.; Ahmed, N.C.; Gouda, M.M.; Vos, P.W.; Bonnerup, C. RT-qPCR for Fecal Mature MicroRNA Quantification and Validation. Methods Mol. Biol. 2018, 1765, 203–215. [Google Scholar] [CrossRef]
- Ghazanfar, S.; Fatima, I.; Aslam, M.; Musharraf, S.G.; Sherman, N.E.; Moskaluk, C.; Fox, J.W.; Akhtar, M.W.; Sadaf, S. Identification of actin beta-like 2 (ACTBL2) as novel, upregulated protein in colorectal cancer. J. Proteom. 2017, 152, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.J.; Zhi, X.; Wang, Y.; Zhang, Z.; Hao, Z.; Ye, R.; Tang, Z.; Qian, F.; Wang, Q.; Zhu, J. Comprehensive Proteomic Characterization of the Human Colorectal Carcinoma Reveals Signature Proteins and Perturbed Pathways. Sci. Rep. 2017, 7, 42436. [Google Scholar] [CrossRef]
- Yamamoto, T.; Kudo, M.; Peng, W.X.; Takata, H.; Takakura, H.; Teduka, K.; Fujii, T.; Mitamura, K.; Taga, A.; Uchida, E.; et al. Identification of aldolase A as a potential diagnostic biomarker for colorectal cancer based on proteomic analysis using formalin-fixed paraffin-embedded tissue. Tumour. Biol. 2016, 37, 13595–13606. [Google Scholar] [CrossRef] [PubMed]
- Torres, S.; Bartolomé, R.A.; Mendes, M.; Barderas, R.; Fernandez-Aceñero, M.J.; Peláez-García, A.; Peña, C.; Lopez-Lucendo, M.; Villar-Vázquez, R.; de Herreros, A.G.; et al. Proteome profiling of cancer-associated fibroblasts identifies novel proinflammatory signatures and prognostic markers for colorectal cancer. Clin. Cancer Res. 2013, 19, 6006–6019. [Google Scholar] [CrossRef]
- Alnabulsi, A.; Murray, G.I. Proteomics for early detection of colorectal cancer: Recent updates. Expert Rev. Proteom. 2018, 15, 55–63. [Google Scholar] [CrossRef]
- Ganepola, G.A.; Nizin, J.; Rutledge, J.R.; Chang, D.H. Use of blood-based biomarkers for early diagnosis and surveillance of colorectal cancer. World J. Gastrointest. Oncol. 2014, 6, 83–97. [Google Scholar] [CrossRef] [PubMed]
- Ivancic, M.M.; Megna, B.W.; Sverchkov, Y.; Craven, M.; Reichelderfer, M.; Pickhardt, P.J.; Sussman, M.R.; Kennedy, G.D. Noninvasive Detection of Colorectal Carcinomas Using Serum Protein Biomarkers. J. Surg. Res. 2020, 246, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, M.; Gies, A.; Weigl, K.; Tikk, K.; Benner, A.; Schrotz-King, P.; Borchers, C.H.; Brenner, H. Evaluation and Validation of Plasma Proteins Using Two Different Protein Detection Methods for Early Detection of Colorectal Cancer. Cancers 2019, 11, 1426. [Google Scholar] [CrossRef] [PubMed]
- Peltier, J.; Roperch, J.P.; Audebert, S.; Borg, J.P.; Camoin, L. Quantitative proteomic analysis exploring progression of colorectal cancer: Modulation of the serpin family. J. Proteom. 2016, 148, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Zajkowska, M.; Dulewicz, M.; Kulczyńska-Przybik, A.; Safiejko, K.; Juchimiuk, M.; Konopko, M.; Kozłowski, L.; Mroczko, B. The Significance of Selected C-C Motif Chemokine Ligands in Colorectal Cancer Patients. J. Clin. Med. 2022, 11, 1794. [Google Scholar] [CrossRef] [PubMed]
- Quesada-Calvo, F.; Massot, C.; Bertrand, V.; Longuespée, R.; Blétard, N.; Somja, J.; Mazzucchelli, G.; Smargiasso, N.; Baiwir, D.; De Pauw-Gillet, M.C.; et al. OLFM4, KNG1 and Sec24C identified by proteomics and immunohistochemistry as potential markers of early colorectal cancer stages. Clin. Proteom. 2017, 14, 9. [Google Scholar] [CrossRef]
- Chantaraamporn, J.; Champattanachai, V.; Khongmanee, A.; Verathamjamras, C.; Prasongsook, N.; Mingkwan, K.; Luevisadpibul, V.; Chutipongtanate, S.; Svasti, J. Glycoproteomic Analysis Reveals Aberrant Expression of Complement C9 and Fibronectin in the Plasma of Patients with Colorectal Cancer. Proteomes 2020, 8, 26. [Google Scholar] [CrossRef]
- Fan, N.J.; Chen, H.M.; Song, W.; Zhang, Z.Y.; Zhang, M.D.; Feng, L.Y.; Gao, C.F. Macrophage mannose receptor 1 and S100A9 were identified as serum diagnostic biomarkers for colorectal cancer through a label-free quantitative proteomic analysis. Cancer Biomark 2016, 16, 235–243. [Google Scholar] [CrossRef]
- Łukaszewicz-Zając, M.; Pączek, S.; Mroczko, P.; Kulczyńska-Przybik, A. The Significance of CXCL1 and CXCL8 as Well as Their Specific Receptors in Colorectal Cancer. Cancer Manag. Res. 2020, 12, 8435–8443. [Google Scholar] [CrossRef] [PubMed]
- Pączek, S.; Łukaszewicz-Zając, M.; Gryko, M.; Mroczko, P.; Kulczyńska-Przybik, A.; Mroczko, B. CXCL-8 in Preoperative Colorectal Cancer Patients: Significance for Diagnosis and Cancer Progression. Int. J. Mol. Sci. 2020, 21, 2040. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Peng, H.; Huang, X.X.; Xia, Y.B.; Hu, K.F.; Zhang, Z.M. Decreased expression of chromodomain helicase DNA-binding protein 9 is a novel independent prognostic biomarker for colorectal cancer. Braz. J. Med. Biol. Res. 2018, 51, e7588. [Google Scholar] [CrossRef]
- Wang, D.; Zhou, Y.; Hua, L.; Li, J.; Zhu, N.; Liu, Y. CDK3, CDK5 and CDK8 Proteins as Prognostic and Potential Biomarkers in Colorectal Cancer Patients. Int. J. Gen. Med. 2022, 15, 2233–2245. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Zhai, X.; Li, X.; Zhong, C.; Guo, C.; Yang, F.; Yuan, Y.; Zheng, S. Identification of MST1 as a potential early detection biomarker for colorectal cancer through a proteomic approach. Sci. Rep. 2017, 7, 14265. [Google Scholar] [CrossRef]
- Tu, C.; Mojica, W.; Straubinger, R.M.; Li, J.; Shen, S.; Qu, M.; Nie, L.; Roberts, R.; An, B.; Qu, J. Quantitative proteomic profiling of paired cancerous and normal colon epithelial cells isolated freshly from colorectal cancer patients. Proteom. Clin. Appl. 2017, 11, 1600155. [Google Scholar] [CrossRef]
- Li, H.; Zhao, S.; Jing, Z.; Li, J.; Shuanying, Y.; Zhang, N. Combination of D-dimer and carcinoembryonic antigen levels as a predictive and prognostic biomarker in advanced colorectal cancer patients. J. Cell. Biochem. 2018, 120, 8086–8092. [Google Scholar] [CrossRef] [PubMed]
- Hassan, N.A.; Idriss, N.K.; Gaber, N.; Ibrahim, A.; Tawfeek, M.A.; Mossad, E.; Mosa, A.A.; Ahmed, E.H.; Sayed, S.A.; Ahmed, H.A.; et al. Spastic Paraplegia 20 and Serine/Threonine Protein Kinase 31 Expression for the Detection of Colorectal Cancer. Cell. Physiol. Biochem. 2022, 56, 138–149. [Google Scholar] [CrossRef]
- Watany, M.M.; Elmashad, N.M.; Badawi, R.; Hawash, N. Serum FBLN1 and STK31 as biomarkers of colorectal cancer and their ability to noninvasively differentiate colorectal cancer from benign polyps. Clin. Chim. Acta 2018, 483, 151–155. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Huang, Y.; Li, Y.; Wang, Q.; Hao, Y.; Liu, L.; Yao, X.; Yao, X.; Wei, Y.; Sun, X.; et al. FJX1 as a candidate diagnostic and prognostic serum biomarker for colorectal cancer. Clin. Transl. Oncol. 2022, 24, 1964–1974. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Liao, W.; Huang, Y.; Huang, Y.; Luo, Y. Increased expression of NOP14 is associated with improved prognosis due to immune regulation in colorectal cancer. BMC Gastroenterol. 2022, 22, 207. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.P.; Wu, J.; Liu, Z.F.; Gao, J.W.; Li, S.Y. SPARCL1 Is a Novel Prognostic Biomarker and Correlates with Tumor Microenvironment in Colorectal Cancer. Biomed Res. Int. 2022, 2022, 1398268. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cheuk-Hay Lau, H.; Cheng, W.Y.; Yu, J. Gut microbiome in colorectal cancer: Clinical diagnosis and treatment. Genom. Proteom. Bioinform. 2022, in press. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.H.; Yu, J. Gut microbiota in colorectal cancer: Mechanisms of action and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 690–704. [Google Scholar] [CrossRef]
- Ternes, D.; Karta, J.; Tsenkova, M.; Wilmes, P.; Haan, S.; Letellier, E. Microbiome in Colorectal Cancer: How to Get from Meta-omics to Mechanism? Trends Microbiol. 2020, 28, 401–423. [Google Scholar] [CrossRef]
- Inamura, K. Colorectal Cancers: An Update on Their Molecular Pathology. Cancers 2018, 10, 26. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Helmink, B.A.; Spencer, C.N.; Reuben, A.; Wargo, J.A. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell 2018, 33, 570–580. [Google Scholar] [CrossRef]
- Grenham, S.; Clarke, G.; Cryan, J.F.; Dinan, T.G. Brain-gut-microbe communication in health and disease. Front. Physiol. 2011, 2, 94. [Google Scholar] [CrossRef]
- Holmes, E.; Li, J.V.; Marchesi, J.R.; Nicholson, J.K. Gut microbiota composition and activity in relation to host metabolic phenotype and disease risk. Cell Metab. 2012, 16, 559–564. [Google Scholar] [CrossRef]
- Clos-Garcia, M.; Garcia, K.; Alonso, C.; Iruarrizaga-Lejarreta, M.; D’Amato, M.; Crespo, A.; Iglesias, A.; Cubiella, J.; Bujanda, L.; Falcón-Pérez, J.M. Integrative Analysis of Fecal Metagenomics and Metabolomics in Colorectal Cancer. Cancers 2020, 12, 1142. [Google Scholar] [CrossRef]
- Zeller, G.; Tap, J.; Voigt, A.Y.; Sunagawa, S.; Kultima, J.R.; Costea, P.I.; Amiot, A.; Böhm, J.; Brunetti, F.; Habermann, N.; et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 2014, 10, 766. [Google Scholar] [CrossRef] [PubMed]
- Zackular, J.P.; Rogers, M.A.; Ruffin, M.T.t.; Schloss, P.D. The human gut microbiome as a screening tool for colorectal cancer. Cancer Prev. Res. 2014, 7, 1112–1121. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, D.; Yang, Z.; Dai, W.; Feng, X.; Liu, Y.; Jiang, Y.; Li, P.; Li, Y.; Tang, B.; et al. Establishing high-accuracy biomarkers for colorectal cancer by comparing fecal microbiomes in patients with healthy families. Gut Microbes 2020, 11, 918–929. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.Q.; Li, T.; Nakatsu, G.; Chen, Y.X.; Yau, T.O.; Chu, E.; Wong, S.; Szeto, C.H.; Ng, S.C.; Chan, F.K.L.; et al. A novel faecal Lachnoclostridium marker for the non-invasive diagnosis of colorectal adenoma and cancer. Gut 2020, 69, 1248–1257. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Li, L.; Xu, B.; Li, M.; Zeng, Q.; Xiao, H.; Xue, Y.; Wu, Y.; Wang, Y.; Liu, W.; et al. A Simple and Novel Fecal Biomarker for Colorectal Cancer: Ratio of Fusobacterium Nucleatum to Probiotics Populations, Based on Their Antagonistic Effect. Clin. Chem. 2018, 64, 1327–1337. [Google Scholar] [CrossRef]
- McQuade, J.L.; Daniel, C.R.; Helmink, B.A.; Wargo, J.A. Modulating the microbiome to improve therapeutic response in cancer. Lancet Oncol. 2019, 20, e77–e91. [Google Scholar] [CrossRef]
- Poore, G.D.; Kopylova, E.; Zhu, Q.; Carpenter, C.; Fraraccio, S.; Wandro, S.; Kosciolek, T.; Janssen, S.; Metcalf, J.; Song, S.J.; et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020, 579, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, A.H.; Wu, F.F.; Wang, X.J. Alterations in the Gut Microbiota and Their Metabolites in Colorectal Cancer: Recent Progress and Future Prospects. Front. Oncol. 2022, 12, 841552. [Google Scholar] [CrossRef]
- Xiao, Q.; Lu, W.; Kong, X.; Shao, Y.W.; Hu, Y.; Wang, A.; Bao, H.; Cao, R.; Liu, K.; Wang, X.; et al. Alterations of circulating bacterial DNA in colorectal cancer and adenoma: A proof-of-concept study. Cancer Lett. 2021, 499, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Q.; Zhang, A.H.; Miao, J.H.; Sun, H.; Yan, G.L.; Wu, F.F.; Wang, X.J. Gut microbiota as important modulator of metabolism in health and disease. RSC Adv. 2018, 8, 42380–42389. [Google Scholar] [CrossRef]
- Chen, F.; Dai, X.; Zhou, C.C.; Li, K.X.; Zhang, Y.J.; Lou, X.Y.; Zhu, Y.M.; Sun, Y.L.; Peng, B.X.; Cui, W. Integrated analysis of the faecal metagenome and serum metabolome reveals the role of gut microbiome-associated metabolites in the detection of colorectal cancer and adenoma. Gut 2022, 71, 1315–1325. [Google Scholar] [CrossRef] [PubMed]
- Jian, H.; Qinglian, Z. Correlation between plasma trimethylamine oxide levels and colorectal neoplastic lesions. J. Dig. Oncol. 2020, 12, 275–278. [Google Scholar]
- Haixia, Z. Serum Folate and Fecal Short-Chain Fatty Acid Levels and Expression of Tissue Nuclear Stem Factor and Proliferating Cell Nuclear Antigen in Patients with Colorectal Cancer; Third Military Medical University: Chongqing, China, 2011. [Google Scholar]
- Peinan, L. Study on the Correlation between Left and Right Colon Cancer and Bile Acid Metabolism; Nanjing Medical University: Nanjing, China, 2018. [Google Scholar]
- Yu, H. The Role of Intestinal Flora in the Pathogenesis of Colorectal Cancer and Its Clinical Diagnostic Value Based on Multi-Omics; Chinese People’s Liberation Army Naval Medical University: Beijing, China, 2019. [Google Scholar]
- Weir, T.L.; Manter, D.K.; Sheflin, A.M.; Barnett, B.A.; Heuberger, A.L.; Ryan, E.P. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS ONE 2013, 8, e70803. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.G.; Rao, S.; Weir, T.L.; O’Malia, J.; Bazan, M.; Brown, R.J.; Ryan, E.P. Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab. 2016, 4, 11. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Ma, C.; Bezabeh, T.; Wang, Z.; Liang, J.; Huang, Y.; Zhao, J.; Liu, X.; Ye, W.; Tang, W.; et al. (1) H NMR-based metabolomics reveal overlapping discriminatory metabolites and metabolic pathway disturbances between colorectal tumor tissues and fecal samples. Int. J. Cancer 2019, 145, 1679–1689. [Google Scholar] [CrossRef] [PubMed]
- Manna, S.K.; Tanaka, N.; Krausz, K.W.; Haznadar, M.; Xue, X.; Matsubara, T.; Bowman, E.D.; Fearon, E.R.; Harris, C.C.; Shah, Y.M.; et al. Biomarkers of coordinate metabolic reprogramming in colorectal tumors in mice and humans. Gastroenterology 2014, 146, 1313–1324. [Google Scholar] [CrossRef]
- Yang, Y.; Misra, B.B.; Liang, L.; Bi, D.; Weng, W.; Wu, W.; Cai, S.; Qin, H.; Goel, A.; Li, X.; et al. Integrated microbiome and metabolome analysis reveals a novel interplay between commensal bacteria and metabolites in colorectal cancer. Theranostics 2019, 9, 4101–4114. [Google Scholar] [CrossRef]
- Wong, S.H.; Kwong, T.N.Y.; Chow, T.C.; Luk, A.K.C.; Dai, R.Z.W.; Nakatsu, G.; Lam, T.Y.T.; Zhang, L.; Wu, J.C.Y.; Chan, F.K.L.; et al. Quantitation of faecal Fusobacterium improves faecal immunochemical test in detecting advanced colorectal neoplasia. Gut 2017, 66, 1441–1448. [Google Scholar] [CrossRef]
- Shah, M.S.; DeSantis, T.Z.; Weinmaier, T.; McMurdie, P.J.; Cope, J.L.; Altrichter, A.; Yamal, J.M.; Hollister, E.B. Leveraging sequence-based faecal microbial community survey data to identify a composite biomarker for colorectal cancer. Gut 2018, 67, 882–891. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Yang, M.; Wang, D.; Zhang, S.; Yan, S.; Zhu, Y.; Chen, W. Alteration of the abundance of Parvimonas micra in the gut along the adenoma-carcinoma sequence. Oncol. Lett. 2020, 20, 106. [Google Scholar] [CrossRef]
- Coker, O.O.; Liu, C.; Wu, W.K.K.; Wong, S.H.; Jia, W.; Sung, J.J.Y.; Yu, J. Altered gut metabolites and microbiota interactions are implicated in colorectal carcinogenesis and can be non-invasive diagnostic biomarkers. Microbiome 2022, 10, 35. [Google Scholar] [CrossRef]
- Wu, Y.; Jiao, N.; Zhu, R.; Zhang, Y.; Wu, D.; Wang, A.J.; Fang, S.; Tao, L.; Li, Y.; Cheng, S.; et al. Identification of microbial markers across populations in early detection of colorectal cancer. Nat. Commun. 2021, 12, 3063. [Google Scholar] [CrossRef] [PubMed]
- Eklöf, V.; Löfgren-Burström, A.; Zingmark, C.; Edin, S.; Larsson, P.; Karling, P.; Alexeyev, O.; Rutegård, J.; Wikberg, M.L.; Palmqvist, R. Cancer-associated fecal microbial markers in colorectal cancer detection. Int. J. Cancer 2017, 141, 2528–2536. [Google Scholar] [CrossRef] [PubMed]
- Song, E.M.; Byeon, J.S.; Lee, S.M.; Yoo, H.J.; Kim, S.J.; Lee, S.H.; Chang, K.; Hwang, S.W.; Yang, D.H.; Jeong, J.Y. Fecal Fatty Acid Profiling as a Potential New Screening Biomarker in Patients with Colorectal Cancer. Dig. Dis. Sci. 2018, 63, 1229–1236. [Google Scholar] [CrossRef]
- Nishiumi, S.; Kobayashi, T.; Kawana, S.; Unno, Y.; Sakai, T.; Okamoto, K.; Yamada, Y.; Sudo, K.; Yamaji, T.; Saito, Y.; et al. Investigations in the possibility of early detection of colorectal cancer by gas chromatography/triple-quadrupole mass spectrometry. Oncotarget 2017, 8, 17115–17126. [Google Scholar] [CrossRef]
- Ning, W.; Qiao, N.; Zhang, X.; Pei, D.; Wang, W. Metabolic profiling analysis for clinical urine of colorectal cancer. Asia Pac. J. Clin. Oncol. 2021, 17, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Troisi, J.; Cavallo, P.; Colucci, A.; Pierri, L.; Scala, G.; Symes, S.; Jones, C.; Richards, S. Metabolomics in genetic testing. Adv. Clin. Chem. 2020, 94, 85–153. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Sun, H.; Yan, G.; Wang, P.; Wang, X. Metabolomics for Biomarker Discovery: Moving to the Clinic. Biomed Res. Int. 2015, 2015, 354671. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Broadhurst, D.I.; Atherton, H.J.; Goodacre, R.; Griffin, J.L. Systems level studies of mammalian metabolomes: The roles of mass spectrometry and nuclear magnetic resonance spectroscopy. Chem. Soc. Rev. 2011, 40, 387–426. [Google Scholar] [CrossRef]
- Ivanisevic, J.; Want, E.J. From Samples to Insights into Metabolism: Uncovering Biologically Relevant Information in LC-HRMS Metabolomics Data. Metabolites 2019, 9, 308. [Google Scholar] [CrossRef]
- Vignoli, A.; Ghini, V.; Meoni, G.; Licari, C.; Takis, P.G.; Tenori, L.; Turano, P.; Luchinat, C. High-Throughput Metabolomics by 1D NMR. Angew Chem. Int. Ed. Engl. 2019, 58, 968–994. [Google Scholar] [CrossRef] [PubMed]
- Commisso, M.; Strazzer, P.; Toffali, K.; Stocchero, M.; Guzzo, F. Untargeted metabolomics: An emerging approach to determine the composition of herbal products. Comput. Struct. Biotechnol. J. 2013, 4, e201301007. [Google Scholar] [CrossRef]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef]
- Kim, E.R.; Kwon, H.N.; Nam, H.; Kim, J.J.; Park, S.; Kim, Y.H. Urine-NMR metabolomics for screening of advanced colorectal adenoma and early stage colorectal cancer. Sci. Rep. 2019, 9, 4786. [Google Scholar] [CrossRef] [PubMed]
- Daniluk, U.; Daniluk, J.; Kucharski, R.; Kowalczyk, T.; Pietrowska, K.; Samczuk, P.; Filimoniuk, A.; Kretowski, A.; Lebensztejn, D.; Ciborowski, M. Untargeted Metabolomics and Inflammatory Markers Profiling in Children With Crohn’s Disease and Ulcerative Colitis-A Preliminary Study. Inflamm. Bowel Dis. 2019, 25, 1120–1128. [Google Scholar] [CrossRef]
- Wang, H.; Tso, V.K.; Slupsky, C.M.; Fedorak, R.N. Metabolomics and detection of colorectal cancer in humans: A systematic review. Future Oncol. 2010, 6, 1395–1406. [Google Scholar] [CrossRef]
- Nannini, G.; Meoni, G.; Amedei, A.; Tenori, L. Metabolomics profile in gastrointestinal cancers: Update and future perspectives. World J. Gastroenterol. 2020, 26, 2514–2532. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Xiao, Y.; Shu, D.; Liang, X.; Hu, X.; Xie, Y.; Lin, D.; Li, H. Metabolomics Analysis in Serum from Patients with Colorectal Polyp and Colorectal Cancer by (1)H-NMR Spectrometry. Dis. Markers 2019, 2019, 3491852. [Google Scholar] [CrossRef]
- Erben, V.; Poschet, G.; Schrotz-King, P.; Brenner, H. Comparing Metabolomics Profiles in Various Types of Liquid Biopsies among Screening Participants with and without Advanced Colorectal Neoplasms. Diagnostics 2021, 11, 561. [Google Scholar] [CrossRef]
- Udo, R.; Katsumata, K.; Kuwabara, H.; Enomoto, M.; Ishizaki, T.; Sunamura, M.; Nagakawa, Y.; Soya, R.; Sugimoto, M.; Tsuchida, A. Urinary charged metabolite profiling of colorectal cancer using capillary electrophoresis-mass spectrometry. Sci. Rep. 2020, 10, 21057. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Sun, H.; Wang, X. Mass spectrometry-driven drug discovery for development of herbal medicine. Mass Spectrom. Rev. 2018, 37, 307–320. [Google Scholar] [CrossRef]
- Yin, F.T.; Zhou, X.H.; Kang, S.Y.; Li, X.H.; Li, J.; Ullah, I.; Zhang, A.H.; Sun, H.; Wang, X.J. Prediction of the mechanism of Dachengqi Decoction treating colorectal cancer based on the analysis method of “ into serum components -action target-key pathway”. J. Ethnopharmacol. 2022, 293, 115286. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Li, B.; Lam, S.M.; Shui, G. Integration of lipidomics and metabolomics for in-depth understanding of cellular mechanism and disease progression. J. Genet. Genom. 2020, 47, 69–83. [Google Scholar] [CrossRef] [PubMed]
- Kyle, J.E. Extracting Biological Insight from Untargeted Lipidomics Data. Methods Mol. Biol. 2020, 2104, 121–137. [Google Scholar] [CrossRef] [PubMed]
- Zaytseva, Y. Lipid Metabolism as a Targetable Metabolic Vulnerability in Colorectal Cancer. Cancers 2021, 13, 301. [Google Scholar] [CrossRef] [PubMed]
- Pakiet, A.; Kobiela, J.; Stepnowski, P.; Sledzinski, T.; Mika, A. Changes in lipids composition and metabolism in colorectal cancer: A review. Lipids Health Dis. 2019, 18, 29. [Google Scholar] [CrossRef]
- Kondo, Y.; Nishiumi, S.; Shinohara, M.; Hatano, N.; Ikeda, A.; Yoshie, T.; Kobayashi, T.; Shiomi, Y.; Irino, Y.; Takenawa, T.; et al. Serum fatty acid profiling of colorectal cancer by gas chromatography/mass spectrometry. Biomark Med. 2011, 5, 451–460. [Google Scholar] [CrossRef]
- Liu, T.; Peng, F.; Yu, J.; Tan, Z.; Rao, T.; Chen, Y.; Wang, Y.; Liu, Z.; Zhou, H.; Peng, J. LC-MS-based lipid profile in colorectal cancer patients: TAGs are the main disturbed lipid markers of colorectal cancer progression. Anal. Bioanal. Chem. 2019, 411, 5079–5088. [Google Scholar] [CrossRef]
- Tevini, J.; Eder, S.K.; Huber-Schönauer, U.; Niederseer, D.; Strebinger, G.; Gostner, J.M.; Aigner, E.; Datz, C.; Felder, T.K. Changing Metabolic Patterns along the Colorectal Adenoma-Carcinoma Sequence. J. Clin. Med. 2022, 11, 721. [Google Scholar] [CrossRef] [PubMed]
- Ecker, J.; Benedetti, E.; Kindt, A.S.D.; Höring, M.; Perl, M.; Machmüller, A.C.; Sichler, A.; Plagge, J.; Wang, Y.; Zeissig, S.; et al. The Colorectal Cancer Lipidome: Identification of a Robust Tumor-Specific Lipid Species Signature. Gastroenterology 2021, 161, 910–923.e919. [Google Scholar] [CrossRef]
- Dehairs, J.; Derua, R.; Rueda-Rincon, N.; Swinnen, J.V. Lipidomics in drug development. Drug Discov. Today Technol. 2015, 13, 33–38. [Google Scholar] [CrossRef]
- Moreno, L.O.; Sánchez, P.N.; Abalo, R. Lipidomics as Tools for Finding Biomarkers of Intestinal Pathology: From Irritable Bowel Syndrome to Colorectal Cancer. Curr. Drug Targets 2022, 23, 636–655. [Google Scholar] [CrossRef] [PubMed]
- Lv, J.; Zhang, L.; Yan, F.; Wang, X. Clinical lipidomics: A new way to diagnose human diseases. Clin. Transl. Med. 2018, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, I.; Verma, S.; Kumar, S.; Jere, A.; Anamika, K. Multi-omics Data Integration, Interpretation, and Its Application. Bioinform. Biol. Insights 2020, 14, 1177932219899051. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Risacher, S.L.; Shen, L.; Saykin, A.J. Network approaches to systems biology analysis of complex disease: Integrative methods for multi-omics data. Brief Bioinform. 2018, 19, 1370–1381. [Google Scholar] [CrossRef]
- O’Connor, J.P.; Aboagye, E.O.; Adams, J.E.; Aerts, H.J.; Barrington, S.F.; Beer, A.J.; Boellaard, R.; Bohndiek, S.E.; Brady, M.; Brown, G.; et al. Imaging biomarker roadmap for cancer studies. Nat. Rev. Clin. Oncol. 2017, 14, 169–186. [Google Scholar] [CrossRef]
- Koyande, N.; Gangopadhyay, M.; Thatikonda, S.; Rengan, A.K. The role of gut microbiota in the development of colorectal cancer: A review. Int. J. Colorectal. Dis. 2022, 37, 1509–1523. [Google Scholar] [CrossRef]
- Rebersek, M. Gut microbiome and its role in colorectal cancer. BMC Cancer 2021, 21, 1325. [Google Scholar] [CrossRef]
- Pączek, S.; Łukaszewicz-Zając, M.; Mroczko, B. Granzymes-Their Role in Colorectal Cancer. Int. J. Mol. Sci. 2022, 23, 5277. [Google Scholar] [CrossRef] [PubMed]
- Eylem, C.C.; Yilmaz, M.; Derkus, B.; Nemutlu, E.; Camci, C.B.; Yilmaz, E.; Turkoglu, M.A.; Aytac, B.; Ozyurt, N.; Emregul, E. Untargeted multi-omic analysis of colorectal cancer-specific exosomes reveals joint pathways of colorectal cancer in both clinical samples and cell culture. Cancer Lett. 2020, 469, 186–194. [Google Scholar] [CrossRef]
- Nam, A.S.; Chaligne, R.; Landau, D.A. Integrating genetic and non-genetic determinants of cancer evolution by single-cell multi-omics. Nat. Rev. Genet. 2021, 22, 3–18. [Google Scholar] [CrossRef] [PubMed]
- Vasaikar, S.V.; Straub, P.; Wang, J.; Zhang, B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018, 46, D956–D963. [Google Scholar] [CrossRef] [PubMed]
- Correa-Aguila, R.; Alonso-Pupo, N.; Hernández-Rodríguez, E.W. Multi-omics data integration approaches for precision oncology. Mol. Omics 2022, 18, 469–479. [Google Scholar] [CrossRef] [PubMed]
| Biomarker | Sample Type | Change | Application | References |
|---|---|---|---|---|
| CBX8, CD96 | datasets | downregulated | diagnostic | [54] |
| MTUS1 | tissue | downregulated | diagnostic and prognostic | [55] |
| SDC2, NDRG4 | stool | upregulated | Screening | [56] |
| SOX21 | stool | upregulated | diagnostic | [57] |
| BDNF, PTGS2, GSK3B and CTNNB1 | tissue | upregulated | prognostic and diagnostic | [33] |
| HPGD | tissue | downregulated | prognostic and diagnostic | [33] |
| YWHAB, MCM4, and FBXO46 | datasets | overexpress | prognostic | [58] |
| DPP72 | datasets | lower expression | prognostic | [58] |
| SDC2, TFPI2 | stool | hypermethylated | screening | [59] |
| SNORD15B, SNORA5C | tissue | upregulated | diagnostic and prognostic | [60] |
| GALR1 | tissue | hypermethylation | screening | [61] |
| LRRC19 | datasets | downregulated | prognosis | [62] |
| KRAS, BRAF, PIK3CA | tissue | mutation | detection | [63] |
| Biomarker | Sample Type | Change | Application | References |
|---|---|---|---|---|
| miR-92a, miR-21 | serum | upregulated | diagnostic and prognostic | [91] |
| hsa_circ_0000567 | CRC tissue and cell lines | downregulated | diagnostic | [92] |
| hsa-circ-0006282 | plasma | upregulated | Diagnostic | [93] |
| hsa_circ_000592, hsa_circ_0001900 and hsa_circ_0001178 | plasma | upregulated | diagnostic | [94] |
| miR-129-1-3p mmiR-566 | urine | upregulated | detection | [95] |
| GPR55 | CRC tissue and cell lines | downregulated | prognostic | [96] |
| miR-1290 | plasma | upregulated | prognostic | [97] |
| miR-320d | plasma | downregulated | diagnostic | [98] |
| miR-103a-3p, miR-127-3p, miR-17-5p, miR151a5p, miR-181a-5p, miR-18a-5p and miR-18b-5p | plasma | upregulated | diagnostic | [99] |
| CCAT2, CCAT1, H19, MALAT1, MEG3, HULC, HOTAIR, PCAT1, PTENP1 and TUSC7 | stool | upregulated | detection | [100] |
| miR-214, miR-199a-3p, miR-196a, miR-106a, miR-183, miR-134, miR-92a, miR-96, miR-20a, miR-21, miR-17, miR-7. | stool | upregulated | screening | [101] |
| miR-138, miR-143, miR-29b, miR-9, miR-146a, miR-127-5p, miR-938, miR-222. | stool | downregulated | screening | [101] |
| Biomarker | Sample Type | Change | Application | References |
|---|---|---|---|---|
| CHD 9 | tissue | upregulated | prognostic | [117] |
| ACTBL2 | tissue | upregulated | diagnostic | [102] |
| CDK3, CDK5, and CDK8 | tissue | upregulated | diagnostic | [118] |
| STK4 or MST1 | serum | downregulated | detection | [119] |
| MRC1 and S100A90 | serum | upregulated | diagnostic | [114] |
| CEACAM-7 | tissue | downregulated | predictive | [120] |
| CEA | plasma | upregulated | predictive and prognostic | [121] |
| SPG20 and STK31 | blood | upregulated | diagnostic | [122] |
| TPM3 | tissue/plasma | upregulated | detection | [123] |
| FJX1 | serum | upregulated | prognostic and diagnostic | [124] |
| NOP14 | datasets | upregulated | Prognosis | [125] |
| SPARCL1 | datasets | Downregulated | diagnosis | [126] |
| Biomarker | Sample | Change | Application | References |
|---|---|---|---|---|
| F.nucleatum, P. anaerobius and P. Micra | stool | increase | detection | [155] |
| P. micra, Streptococcus anginosus | stool | increase | diagnosis | [156] |
| P. Micra F. nucleatum | stool | increase | diagnosis | [157] |
| norvaline and myristic acid | stool | upregulated | diagnosis | [158] |
| menaquinone-10 | stool | upregulated | diagnosis | [159] |
| F. nucleatum | stool | upregulated | detection | [160] |
| Oleic acid | stool | Upregulated | screening | [161] |
| Succinate, Butyrate, Lactate, Glutamate, and Alanine. | tumour tissue/feces | Upregulated (excluding Butyrate downregulated) | detection | [152] |
| Cholesteryl esters, Sphingomyelins | stool | Upregulated | diagnosis | [134] |
| Fusobacterium, Parvimonas and Staphylococcus | stool | increase | diagnosis | [134] |
| Pyruvic acid, lysine, glycolic acid, fumaric acid, ornithine | blood | upregulated | detection | [162] |
| tryptophan, Palmitoleic acid, lysine, 3hydroxyisovaleric acid | blood | decrease | detection | [162] |
| octadecanoic acid, citric acid, hexadecanoic acid, and propanoic acid-2-methyl-1-(1,1-dimethylethyl)-2-methyl-1,3-propanediyl este | urine | downregulated | screening | [163] |
| Hydroxyproline dipeptide, tyrosine, tryptophan, pseudouridine, glucuronic acid, glycine, histidine, glucose, 5-oxoproline, threonic acid, and isocitric acid | urine | upregulated | screening | [163] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ullah, I.; Yang, L.; Yin, F.-T.; Sun, Y.; Li, X.-H.; Li, J.; Wang, X.-J. Multi-Omics Approaches in Colorectal Cancer Screening and Diagnosis, Recent Updates and Future Perspectives. Cancers 2022, 14, 5545. https://doi.org/10.3390/cancers14225545
Ullah I, Yang L, Yin F-T, Sun Y, Li X-H, Li J, Wang X-J. Multi-Omics Approaches in Colorectal Cancer Screening and Diagnosis, Recent Updates and Future Perspectives. Cancers. 2022; 14(22):5545. https://doi.org/10.3390/cancers14225545
Chicago/Turabian StyleUllah, Ihsan, Le Yang, Feng-Ting Yin, Ye Sun, Xing-Hua Li, Jing Li, and Xi-Jun Wang. 2022. "Multi-Omics Approaches in Colorectal Cancer Screening and Diagnosis, Recent Updates and Future Perspectives" Cancers 14, no. 22: 5545. https://doi.org/10.3390/cancers14225545
APA StyleUllah, I., Yang, L., Yin, F.-T., Sun, Y., Li, X.-H., Li, J., & Wang, X.-J. (2022). Multi-Omics Approaches in Colorectal Cancer Screening and Diagnosis, Recent Updates and Future Perspectives. Cancers, 14(22), 5545. https://doi.org/10.3390/cancers14225545






