A Deep Learning Model System for Diagnosis and Management of Adnexal Masses
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Participants and Datasets
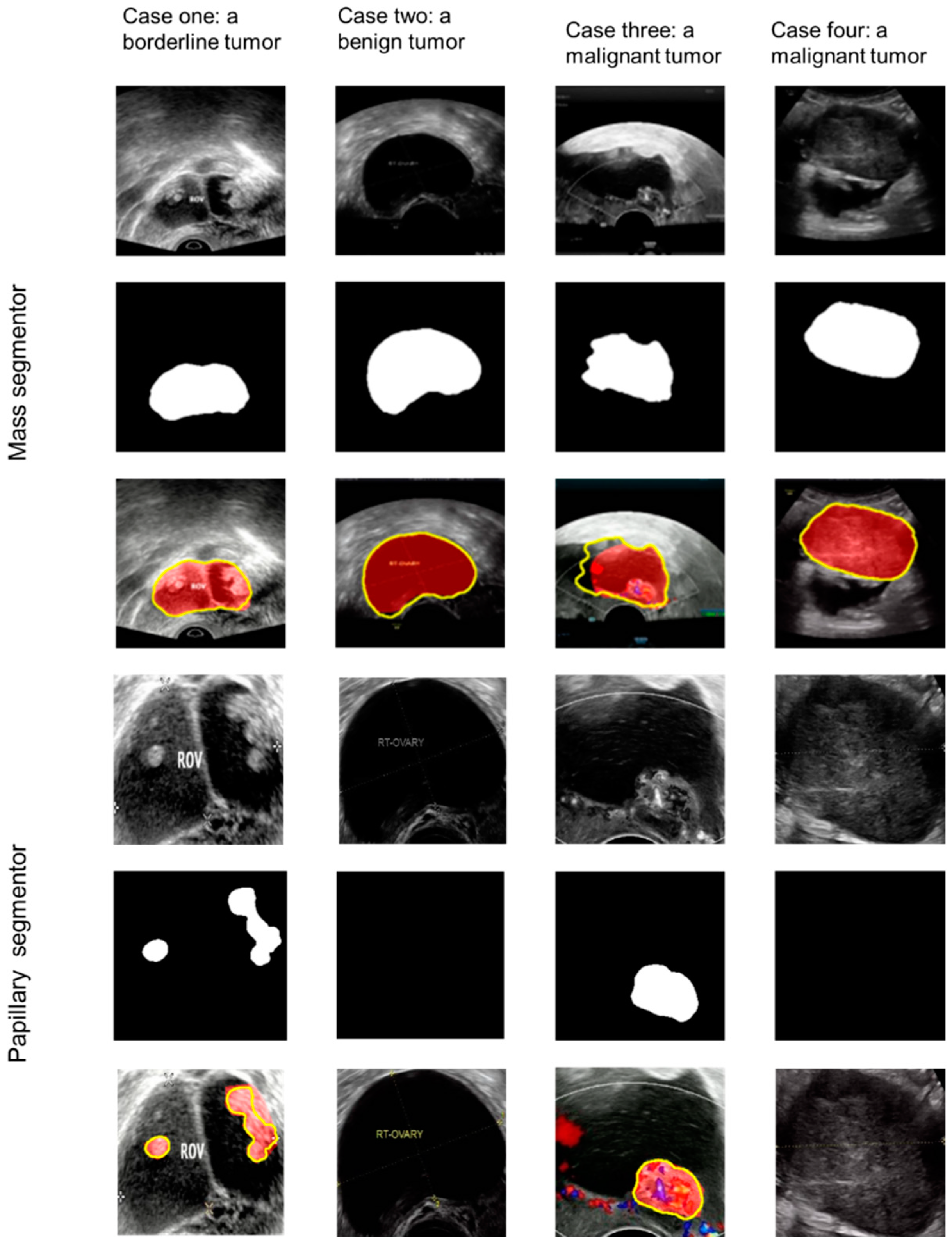
2.3. Annotation and Framework
2.4. Model Architecture
2.5. Evaluation and Comparison with Sonographers
2.6. Statistical Analysis
3. Results
3.1. Data and Patients
3.2. Papillary Projections
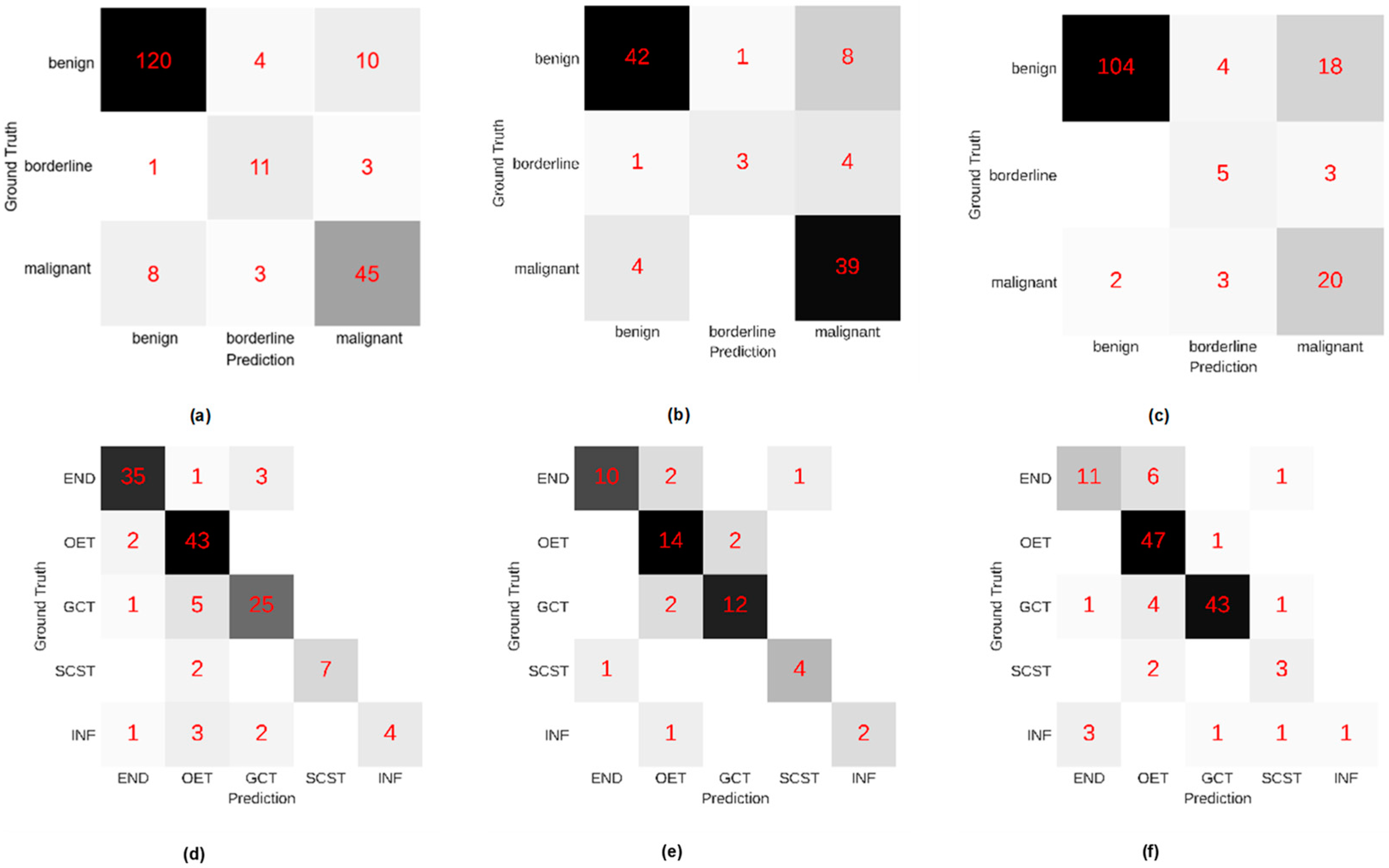
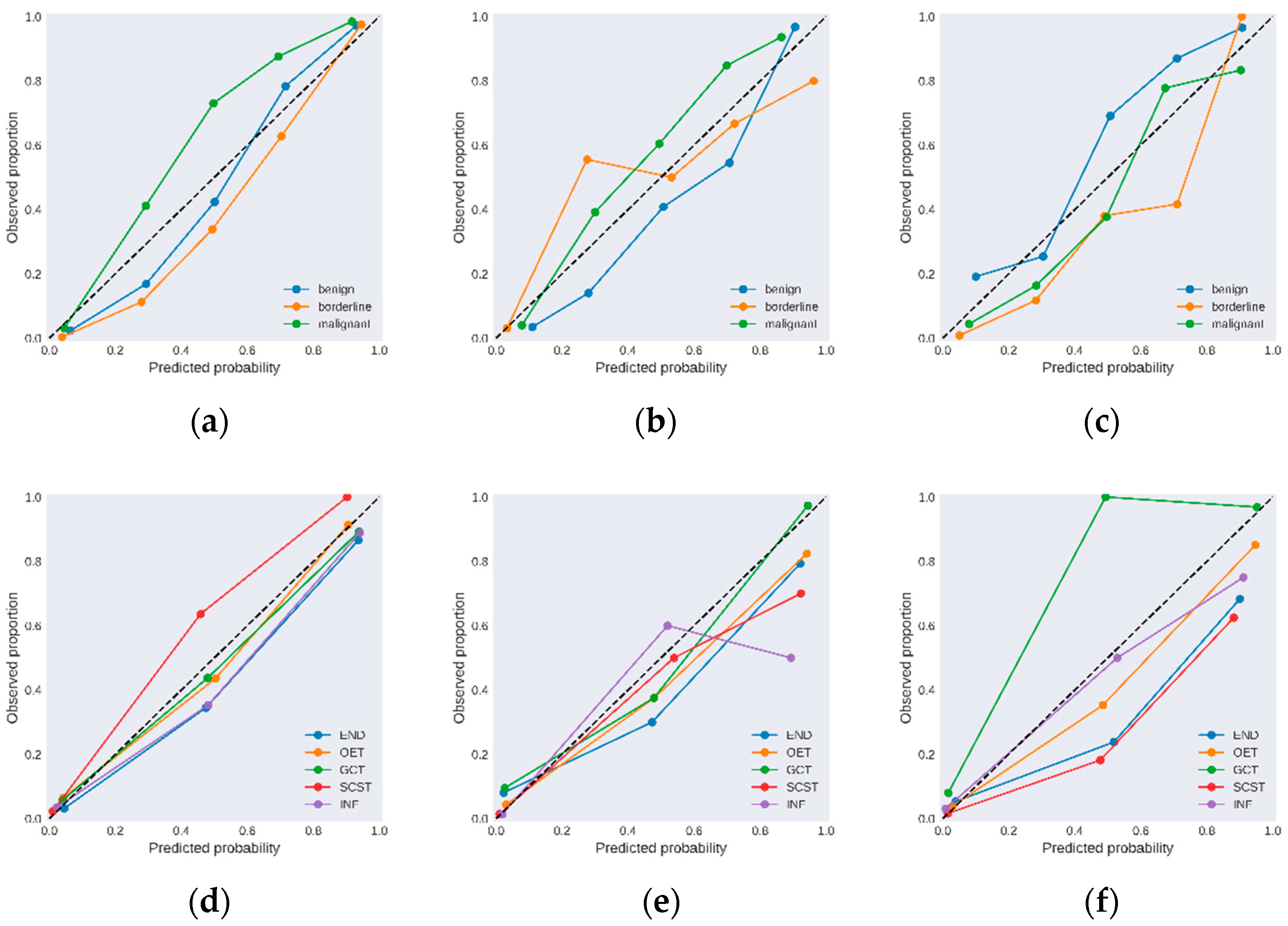
3.3. Diagnostic Performance for the DL Model System
3.4. Comparison with Sonographers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Froyman, W.; Landolfo, C.; De Cock, B.; Wynants, L.; Sladkevicius, P.; Testa, A.; Van Holsbeke, C.; Domali, E.; Fruscio, R.; Epstein, E.; et al. Risk of complications in patients with conservatively managed ovarian tumours (IOTA5): A 2-year interim analysis of a multicentre, prospective, cohort study. Lancet. Oncol. 2019, 20, 448–458. [Google Scholar] [CrossRef]
- American College of Obstetricians and Gynecologists’ Committee on Practice Bulletins—Gynecology. Practice Bulletin No. 174: Evaluation and Management of Adnexal Masses. Obstet. Gynecol. 2016, 128, e210–e226. [Google Scholar] [CrossRef] [PubMed]
- Andreotti, R.; Timmerman, D.; Strachowski, L.; Froyman, W.; Benacerraf, B.; Bennett, G.; Bourne, T.; Brown, D.; Coleman, B.; Frates, M.; et al. O-RADS US Risk Stratification and Management System: A Consensus Guideline from the ACR Ovarian-Adnexal Reporting and Data System Committee. Radiology 2020, 294, 168–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heintz, A.; Odicino, F.; Maisonneuve, P.; Quinn, M.; Benedet, J.; Creasman, W.; Ngan, H.; Pecorelli, S.; Beller, U. Carcinoma of the ovary. FIGO 26th Annual Report on the Results of Treatment in Gynecological Cancer. Int. J. Gynaecol. Obstet. Off. Organ Int. Fed. Gynaecol. Obstet. 2006, 95, S161–S192. [Google Scholar] [CrossRef]
- Armstrong, D.; Alvarez, R.; Bakkum-Gamez, J.; Barroilhet, L.; Behbakht, K.; Berchuck, A.; Chen, L.; Cristea, M.; DeRosa, M.; Eisenhauer, E.; et al. Ovarian Cancer, Version 2.2020, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. JNCCN 2021, 19, 191–226. [Google Scholar] [CrossRef]
- Timmerman, D.; Testa, A.; Bourne, T.; Ameye, L.; Jurkovic, D.; Van Holsbeke, C.; Paladini, D.; Van Calster, B.; Vergote, I.; Van Huffel, S.; et al. Simple ultrasound-based rules for the diagnosis of ovarian cancer. Ultrasound Obstet. Gynecol. Off. J. Int. Soc. Ultrasound Obstet. Gynecol. 2008, 31, 681–690. [Google Scholar] [CrossRef]
- Chapron, C.; Marcellin, L.; Borghese, B.; Santulli, P. Rethinking mechanisms, diagnosis and management of endometriosis. Nat. Rev. Endocrinol. 2019, 15, 666–682. [Google Scholar] [CrossRef]
- Anfelter, P.; Testa, A.; Chiappa, V.; Froyman, W.; Fruscio, R.; Guerriero, S.; Alcazar, J.; Mascillini, F.; Pascual, M.; Sibal, M.; et al. Imaging in gynecological disease (17): Ultrasound features of malignant ovarian yolk sac tumors (endodermal sinus tumors). Ultrasound Obstet. Gynecol. Off. J. Int. Soc. Ultrasound Obstet. Gynecol. 2020, 56, 276–284. [Google Scholar] [CrossRef] [Green Version]
- Schultz, K.; Harris, A.; Schneider, D.; Young, R.; Brown, J.; Gershenson, D.; Dehner, L.; Hill, D.; Messinger, Y.; Frazier, A. Ovarian Sex Cord-Stromal Tumors. J. Oncol. Pract. 2016, 12, 940–946. [Google Scholar] [CrossRef] [Green Version]
- Bristow, R.; Smith, A.; Zhang, Z.; Chan, D.; Crutcher, G.; Fung, E.; Munroe, D. Ovarian malignancy risk stratification of the adnexal mass using a multivariate index assay. Gynecol. Oncol. 2013, 128, 252–259. [Google Scholar] [CrossRef]
- Moore, R.; Brown, A.; Miller, M.; Skates, S.; Allard, W.; Verch, T.; Steinhoff, M.; Messerlian, G.; DiSilvestro, P.; Granai, C.; et al. The use of multiple novel tumor biomarkers for the detection of ovarian carcinoma in patients with a pelvic mass. Gynecol. Oncol. 2008, 108, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Prat, J. Staging classification for cancer of the ovary, fallopian tube, and peritoneum. Int. J. Gynaecol. Obstet. Off. Organ Int. Fed. Gynaecol. Obstet. 2014, 124, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Alcázar, J.; Pascual, M.; Graupera, B.; Aubá, M.; Errasti, T.; Olartecoechea, B.; Ruiz-Zambrana, A.; Hereter, L.; Ajossa, S.; Guerriero, S. External validation of IOTA simple descriptors and simple rules for classifying adnexal masses. Ultrasound Obstet. Gynecol. Off. J. Int. Soc. Ultrasound Obstet. Gynecol. 2016, 48, 397–402. [Google Scholar] [CrossRef] [PubMed]
- Amor, F.; Vaccaro, H.; Alcázar, J.; León, M.; Craig, J.; Martinez, J. Gynecologic imaging reporting and data system: A new proposal for classifying adnexal masses on the basis of sonographic findings. J. Ultrasound Med. Off. J. Am. Inst. Ultrasound Med. 2009, 28, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Basha, M.; Refaat, R.; Ibrahim, S.; Madkour, N.; Awad, A.; Mohamed, E.; El Sammak, A.; Zaitoun, M.; Dawoud, H.; Khamis, M.; et al. Gynecology Imaging Reporting and Data System (GI-RADS): Diagnostic performance and inter-reviewer agreement. Eur. Radiol. 2019, 29, 5981–5990. [Google Scholar] [CrossRef] [PubMed]
- Timmerman, D.; Testa, A.; Bourne, T.; Ferrazzi, E.; Ameye, L.; Konstantinovic, M.; Van Calster, B.; Collins, W.; Vergote, I.; Van Huffel, S.; et al. Logistic regression model to distinguish between the benign and malignant adnexal mass before surgery: A multicenter study by the International Ovarian Tumor Analysis Group. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2005, 23, 8794–8801. [Google Scholar] [CrossRef]
- Van Calster, B.; Van Hoorde, K.; Valentin, L.; Testa, A.; Fischerova, D.; Van Holsbeke, C.; Savelli, L.; Franchi, D.; Epstein, E.; Kaijser, J.; et al. Evaluating the risk of ovarian cancer before surgery using the ADNEX model to differentiate between benign, borderline, early and advanced stage invasive, and secondary metastatic tumours: Prospective multicentre diagnostic study. BMJ Clin. Res. Ed. 2014, 349, g5920. [Google Scholar] [CrossRef] [Green Version]
- Kaijser, J.; Sayasneh, A.; Van Hoorde, K.; Ghaem-Maghami, S.; Bourne, T.; Timmerman, D.; Van Calster, B. Presurgical diagnosis of adnexal tumours using mathematical models and scoring systems: A systematic review and meta-analysis. Hum. Reprod. Update 2014, 20, 449–462. [Google Scholar] [CrossRef] [Green Version]
- Sayasneh, A.; Ferrara, L.; De Cock, B.; Saso, S.; Al-Memar, M.; Johnson, S.; Kaijser, J.; Carvalho, J.; Husicka, R.; Smith, A.; et al. Evaluating the risk of ovarian cancer before surgery using the ADNEX model: A multicentre external validation study. Br. J. Cancer 2016, 115, 542–548. [Google Scholar] [CrossRef] [Green Version]
- Van Calster, B.; Valentin, L.; Froyman, W.; Landolfo, C.; Ceusters, J.; Testa, A.; Wynants, L.; Sladkevicius, P.; Van Holsbeke, C.; Domali, E.; et al. Validation of models to diagnose ovarian cancer in patients managed surgically or conservatively: Multicentre cohort study. BMJ Clin. Res. Ed. 2020, 370, m2614. [Google Scholar] [CrossRef]
- Arnaout, R.; Curran, L.; Zhao, Y.; Levine, J.; Chinn, E.; Moon-Grady, A. An ensemble of neural networks provides expert-level prenatal detection of complex congenital heart disease. Nat. Med. 2021, 27, 882–891. [Google Scholar] [CrossRef] [PubMed]
- Gulshan, V.; Peng, L.; Coram, M.; Stumpe, M.; Wu, D.; Narayanaswamy, A.; Venugopalan, S.; Widner, K.; Madams, T.; Cuadros, J.; et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA 2016, 316, 2402–2410. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.; Sun, S.; Mun, J.; Kim, C.; Kim, S.; Cho, S.; Youn, S.; Kim, H.; Chung, J. Dermatologist-level classification of malignant lip diseases using a deep convolutional neural network. Br. J. Dermatol. 2020, 182, 1388–1394. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, S.; Zhang, Q.; Wei, X.; Pan, Y.; Zhao, J.; Xin, X.; Qin, C.; Wang, X.; Li, J.; et al. Diagnosis of thyroid cancer using deep convolutional neural network models applied to sonographic images: A retrospective, multicohort, diagnostic study. Lancet. Oncol. 2019, 20, 193–201. [Google Scholar] [CrossRef]
- Gao, Y.; Zeng, S.; Xu, X.; Li, H.; Yao, S.; Song, K.; Li, X.; Chen, L.; Tang, J.; Xing, H.; et al. Deep learning-enabled pelvic ultrasound images for accurate diagnosis of ovarian cancer in China: A retrospective, multicentre, diagnostic study. Lancet Digit. Health 2022, 4, e179–e187. [Google Scholar] [CrossRef]
- Chen, H.; Yang, B.W.; Qian, L.; Meng, Y.S.; Bai, X.H.; Hong, X.W.; He, X.; Jiang, M.J.; Yuan, F.; Du, Q.W.; et al. Deep Learning Prediction of Ovarian Malignancy at US Compared with O-RADS and Expert Assessment. Radiology 2022, 304, 106–113. [Google Scholar] [CrossRef]
- Christiansen, F.; Epstein, E.; Smedberg, E.; Åkerlund, M.; Smith, K.; Epstein, E. Ultrasound image analysis using deep neural networks for discriminating between benign and malignant ovarian tumors: Comparison with expert subjective assessment. Ultrasound Obstet. Gynecol. Off. J. Int. Soc. Ultrasound Obstet. Gynecol. 2021, 57, 155–163. [Google Scholar] [CrossRef]
- Delle Marchette, M.; Ceppi, L.; Andreano, A.; Bonazzi, C.; Buda, A.; Grassi, T.; Giuliani, D.; Sina, F.; Lamanna, M.; Bianchi, T.; et al. Oncologic and fertility impact of surgical approach for borderline ovarian tumours treated with fertility sparing surgery. Eur. J. Cancer 2019, 111, 61–68. [Google Scholar] [CrossRef]
- Timmerman, D.; Valentin, L.; Bourne, T.H.; Collins, W.P.; Verrelst, H.; Vergote, I. Terms, definitions and measurements to describe the sonographic features of adnexal tumors: A consensus opinion from the International Ovarian Tumor Analysis (IOTA) Group. Ultrasound Obs. Gynecol. 2000, 16, 500–505. [Google Scholar] [CrossRef] [Green Version]
- Timor-Tritsch, I.E.; Foley, C.E.; Brandon, C.; Yoon, E.; Ciaffarrano, J.; Monteagudo, A.; Mittal, K.; Boyd, L. New sonographic marker of borderline ovarian tumor: Microcystic pattern of papillae and solid components. Ultrasound Obs. Gynecol. 2019, 54, 395–402. [Google Scholar] [CrossRef]
- Gupta, A.; Jha, P.; Baran, T.M.; Maturen, K.E.; Patel-Lippmann, K.; Zafar, H.M.; Kamaya, A.; Antil, N.; Barroilhet, L.; Sadowski, E. Ovarian Cancer Detection in Average-Risk Women: Classic- versus Nonclassic-appearing Adnexal Lesions at US. Radiology 2022, 303, 603–610. [Google Scholar] [CrossRef] [PubMed]
- Barnett, A.J.; Schwartz, F.R.; Tao, C.; Chen, C.; Ren, Y.; Lo, J.Y.; Rudin, C. A case-based interpretable deep learning model for classification of mass lesions in digital mammography. Nat. Mach. Intell. 2021, 3, 1061–1070. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar] [CrossRef] [Green Version]
- Ding, X.; Zhang, X.; Zhou, Y.; Han, J.; Ding, G.; Sun, J. Scaling Up Your Kernels to 31 × 31: Revisiting Large Kernel Design in CNNs. In Proceedings of the 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), New Orleans, LA, USA, 18–24 June 2022. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Cham, Switzerland, 2015; Volume 9351, pp. 234–241. [Google Scholar] [CrossRef] [Green Version]
- Bokhovkin, A.; Burnaev, E. Boundary Loss for Remote Sensing Imagery Semantic Segmentation. In International Symposium on Neural Networks; Springer: Cham, Switzerland, 2019; Volume 11555, pp. 388–401. [Google Scholar] [CrossRef] [Green Version]
- Loshchilov, I.; Hutter, F. Fixing Weight Decay Regularization in Adam. In Proceedings of the Sixth International Conference on Learning Representations, Vancouver, BC, Canada, 30 April–3 May 2018. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. In Proceedings of the 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–23 June 2018. [Google Scholar]
- Grandini, M.; Bagli, E.; Visani, G. Metrics for Multi-Class Classification: An Overview. arXiv 2020, arXiv:2008.05756. [Google Scholar]
- Jia, S.; Xiang, Y.; Yang, J.; Shi, J.; Jia, C.; Leng, J. Oncofertility outcomes after fertility-sparing treatment of bilateral serous borderline ovarian tumors: Results of a large retrospective study. Hum. Reprod. 2020, 35, 328–339. [Google Scholar] [CrossRef] [PubMed]
- Alcázar, J.; Olartecoechea, B.; Guerriero, S.; Jurado, M. Expectant management of adnexal masses in selected premenopausal women: A prospective observational study. Ultrasound Obstet. Gynecol. Off. J. Int. Soc. Ultrasound Obstet. Gynecol. 2013, 41, 582–588. [Google Scholar] [CrossRef] [Green Version]
- May, T.; Oza, A. Conservative management of adnexal masses. Lancet Oncol. 2019, 20, 326–327. [Google Scholar] [CrossRef]
- Chen, L.-C.; Papandreou, G.; Schroff, F.; Adam, H. Rethinking atrous convolution for semantic image segmentation. arXiv 2017, arXiv:1706.05587. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. arXiv 2021, arXiv:2103.14030. [Google Scholar]



| Patients | Type Classification | Pathological Subtype Classification | Training Dataset | Internal Validation Dataset | External Test Dataset 1 | External Test Dataset 2 |
|---|---|---|---|---|---|---|
| Mean age | 43.43 (11, 81) | 44.50 (11, 81) | 48.87 (15, 77) | 43.26 (19, 50) | ||
| Healthy cases * (images) | 508 (1100) | 255 (564) | 96 (178) | 105 (233) | ||
| Cases (images) with adnexal masses † | 591 (3397) | 205 (653) | 102 (312) | 159 (528) | ||
| Benign | 374 (1117) | 134 (377) | 51 (151) | 126 (401) | ||
| END † | 117 (332) | 39 (102) | 13 (35) | 18 (51) | ||
| OET | 103 (342) | 45 (127) | 16 (49) | 48 (157) | ||
| GCT | 102 (272) | 31 (82) | 14 (50) | 49 (156) | ||
| SCST | 33 (107) | 9 (32) | 5 (11) | 5 (18) | ||
| INF | 19 (64) | 10 (34) | 3 (6) | 6 (19) | ||
| Borderline | 50 (917) | 15 (57) | 8 (27) | 8 (30) | ||
| Malignant | 167 (1363) | 56 (219) | 43 (134) | 25 (97) | ||
| Total | 1099 (4497) | 460 (1217) | 198 (490) | 264 (761) |
| Variable | Internal Validation Dataset | External Test Dataset 1 | External Test Dataset 2 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Benign | Borderline | Malignant | Benign | Borderline | Malignant | Benign | Borderline | Malignant | |
| Accuracy | 0.888 (0.875–0.902) | 0.946 (0.940–0.957) | 0.883 (0.875–0.897) | 0.863 (0.846–0.879) | 0.941 (0.934–0.956) | 0.843 (0.824–0.868) | 0.849 (0.832–0.867) | 0.937 (0.930–0.951) | 0.836 (0.825–0.853) |
| Sensitivity | 0.896 (0.882–0.913) | 0.733 (0.667–0.786) | 0.804 (0.780–0.837) | 0.824 (0.795–0.864) | 0.375 (0.286–0.500) | 0.907 (0.892–0.944) | 0.825 (0.809–0.847) | 0.625 (0.571–0.714) | 0.800 (0.762–0.857) |
| Specificity | 0.873 (0.855–0.902) | 0.963 (0.959–0.971) | 0.913 (0.902–0.926) | 0.902 (0.886–0.933) | 0.989 (0.988–1.000) | 0.797 (0.769–0.830) | 0.939 (0.926–0.967) | 0.954 (0.948–0.964) | 0.843 (0.828–0.860) |
| Positive predictive value | 0.930 (0.921–0.947) | 0.611 (0.563–0.688) | 0.776 (0.750–0.811) | 0.894 (0.875–0.927) | 0.750 (0.667–1.000) | 0.765 (0.733–0.800) | 0.981 (0.978–0.990) | 0.417 (0.364–0.500) | 0.488 (0.444–0.528) |
| Negative predictive value | 0.816 (0.794–0.843) | 0.979 (0.976–0.983) | 0.925 (0.916–0.939) | 0.836 (0.813–0.872) | 0.949 (0.943–0.966) | 0.922 (0.909–0.953) | 0.584 (0.551–0.625) | 0.980 (0.977–0.985) | 0.958 (0.952–0.972) |
| Variable | Internal Validation Dataset | External Test Dataset 1 | External Test Dataset 2 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| END | OET | GCT | SCST | INF | END | OET | GCT | SCST | INF | END | OET | GCT | SCST | INF | |
| Accuracy | 0.940 (0.933–0.950) | 0.903 (0.892–0.917) | 0.918 (0.908–0.933) | 0.985 (0.983–0.992) | 0.955 (0.950–0.967) | 0.922 (0.911–0.956) | 0.863 (0.844–0.889) | 0.922 (0.911–0.956) | 0.961 (0.956–0.978) | 0.980 (0.978–1.000) | 0.913 (0.903–0.929) | 0.897 (0.885–0.912) | 0.937 (0.929–0.947) | 0.960 (0.956–0.973) | 0.960 (0.956–0.973) |
| Sensitivity | 0.897 (0.879–0.939) | 0.956 (0.947–0.976) | 0.806 (0.778–0.852) | 0.778 (0.714–0.875) | 0.400 (0.333–0.500) | 0.769 (0.700–0.833) | 0.875 (0.846–0.933) | 0.857 (0.818–0.923) | 0.800 (0.667–1.000) | 0.667 (0.500–1.000) | 0.611 (0.538–0.688) | 0.979 (0.976–1.000) | 0.878 (0.857–0.907) | 0.600 (0.500–0.750) | 0.167 (0.000–0.250) |
| Specificity | 0.958 (0.952–0.976) | 0.876 (0.859–0.899) | 0.951 (0.944–0.967) | 1.000 (1.000–1.000) | 1.000 (1.000–1.000) | 0.974 (0.969–1.000) | 0.857 (0.833–0.900) | 0.946 (0.935–0.970) | 0.978 (0.975–1.000) | 1.000 (1.000–1.000) | 0.962 (0.958–0.979) | 0.846 (0.826–0.871) | 0.974 (0.970–0.986) | 0.975 (0.972–0.982) | 1.000 (1.000–1.000) |
| Positive predictive value | 0.897 (0.879–0.938) | 0.796 (0.771–0.833) | 0.833 (0.808–0.885) | 1.000 (1.000–1.000) | 1.000 (1.000–1.000) | 0.909 (0.875–1.000) | 0.737 (0.688–0.813) | 0.857 (0.818–0.923) | 0.800 (0.667–1.000) | 1.000 (1.000–1.000) | 0.733 (0.667–0.800) | 0.797 (0.769–0.830) | 0.956 (0.947–0.976) | 0.500 (0.400–0.600) | 1.000 (1.000–1.000) |
| Negative predictive value | 0.958 (0.952–0.976) | 0.975 (0.971–0.986) | 0.942 (0.934–0.957) | 0.984 (0.982–0.991) | 0.954 (0.948–0.966) | 0.925 (0.912–0.946) | 0.938 (0.926–0.966) | 0.946 (0.935–0.970) | 0.978 (0.975–1.000) | 0.980 (0.977–1.000) | 0.937 (0.929–0.950) | 0.985 (0.982–1.000) | 0.926 (0.915–0945) | 0.983 (0.981–0.991) | 0.960 (0.955–0.973) |
| Variables | Benign | Borderline | Malignant | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| External Test Dataset 1 | External Test Dataset 2 | External Test Dataset 1 | External Test Dataset 2 | External Test Dataset 1 | External Test Dataset 2 | |||||||||||||
| Reviewer A | Reviewer B | Reviewer C | Reviewer A | Reviewer B | Reviewer C | Reviewer A | Reviewer B | Reviewer C | Reviewer A | Reviewer B | Reviewer C | Reviewer A | Reviewer B | Reviewer C | Reviewer A | Reviewer B | Reviewer C | |
| Accuracy | 0.814 | 0.843 | 0.578 | 0.855 | 0.818 | 0.780 | 0.892 | 0.912 | 0.902 | 0.943 | 0.893 | 0.918 | 0.882 | 0.873 | 0.559 | 0.899 | 0.824 | 0.811 |
| Sensitivity | 0.824 | 0.784 | 0.451 | 0.921 | 0.825 | 0.802 | 0.500 | 0.375 | 0.000 | 0.250 | 0.125 | 0.000 | 0.814 | 0.930 | 0.698 | 0.680 | 0.680 | 0.760 |
| Specificity | 0.804 | 0.902 | 0.706 | 0.606 | 0.788 | 0.697 | 0.926 | 0.957 | 0.978 | 0.980 | 0.934 | 0.967 | 0.932 | 0.831 | 0.458 | 0.940 | 0.851 | 0.821 |
| Positive predictive value | 0.808 | 0.889 | 0.605 | 0.899 | 0.937 | 0.910 | 0.364 | 0.429 | 0.000 | 0.400 | 0.091 | 0.000 | 0.897 | 0.800 | 0.484 | 0.680 | 0.459 | 0.442 |
| Negative predictive value | 0.820 | 0.807 | 0.563 | 0.667 | 0.542 | 0.479 | 0.956 | 0.947 | 0.920 | 0.961 | 0.953 | 0.948 | 0.873 | 0.942 | 0.675 | 0.940 | 0.934 | 0.948 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Chen, Y.; Zhang, M.; Zhang, P.; He, K.; Yan, F.; Li, J.; Xu, H.; Burkhoff, D.; Luo, Y.; et al. A Deep Learning Model System for Diagnosis and Management of Adnexal Masses. Cancers 2022, 14, 5291. https://doi.org/10.3390/cancers14215291
Li J, Chen Y, Zhang M, Zhang P, He K, Yan F, Li J, Xu H, Burkhoff D, Luo Y, et al. A Deep Learning Model System for Diagnosis and Management of Adnexal Masses. Cancers. 2022; 14(21):5291. https://doi.org/10.3390/cancers14215291
Chicago/Turabian StyleLi, Jianan, Yixin Chen, Minyu Zhang, Peifang Zhang, Kunlun He, Fengqin Yan, Jingbo Li, Hong Xu, Daniel Burkhoff, Yukun Luo, and et al. 2022. "A Deep Learning Model System for Diagnosis and Management of Adnexal Masses" Cancers 14, no. 21: 5291. https://doi.org/10.3390/cancers14215291
APA StyleLi, J., Chen, Y., Zhang, M., Zhang, P., He, K., Yan, F., Li, J., Xu, H., Burkhoff, D., Luo, Y., Wang, L., & Li, Q. (2022). A Deep Learning Model System for Diagnosis and Management of Adnexal Masses. Cancers, 14(21), 5291. https://doi.org/10.3390/cancers14215291







