Genome-Wide DNA Methylation and Gene Expression Profiling Characterizes Molecular Subtypes of Esophagus Squamous Cell Carcinoma for Predicting Patient Survival and Immunotherapy Efficacy
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
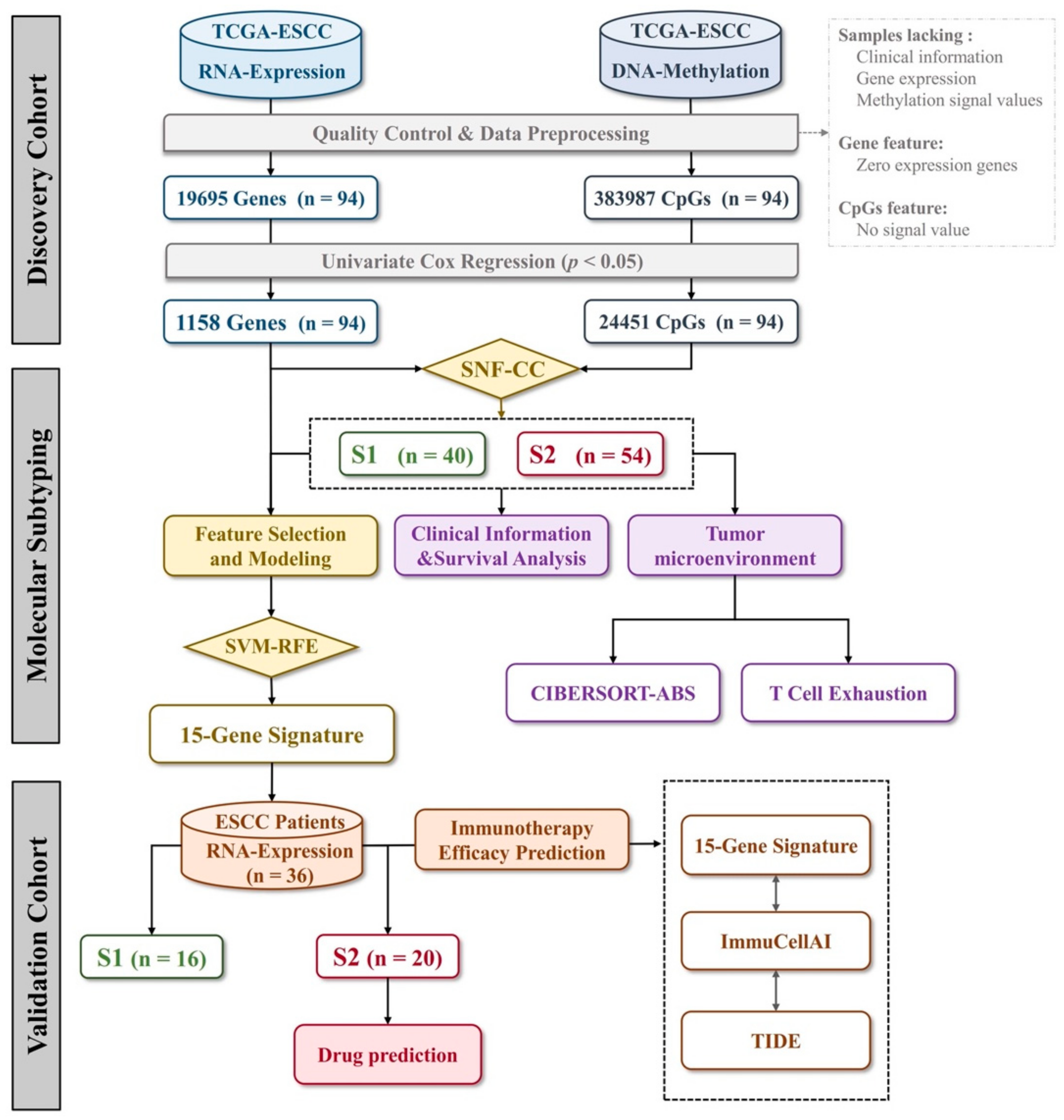
2.1. Acquisition of TCGA Cohort and Multiomics Data Processing
2.2. Prognostic Features Selection and Survival Analysis
2.3. Similarity Network Fusion and Consensus Clustering Analysis
2.4. Assessment of Tumor-Infiltrating Immune Cells
2.5. Identification of Differentially Expressed Genes between Molecular Subtypes and Functional Enrichment Analysis
2.6. Signature Gene Identification and Classification Model Construction
2.7. Acquisition of Chinese ESCC Patient Samples
2.8. Nucleic Acid Extraction and Gene Expression Profiling
2.9. Prediction of Immune Checkpoint Blockade Therapy Response and Drug Repurposing
3. Results
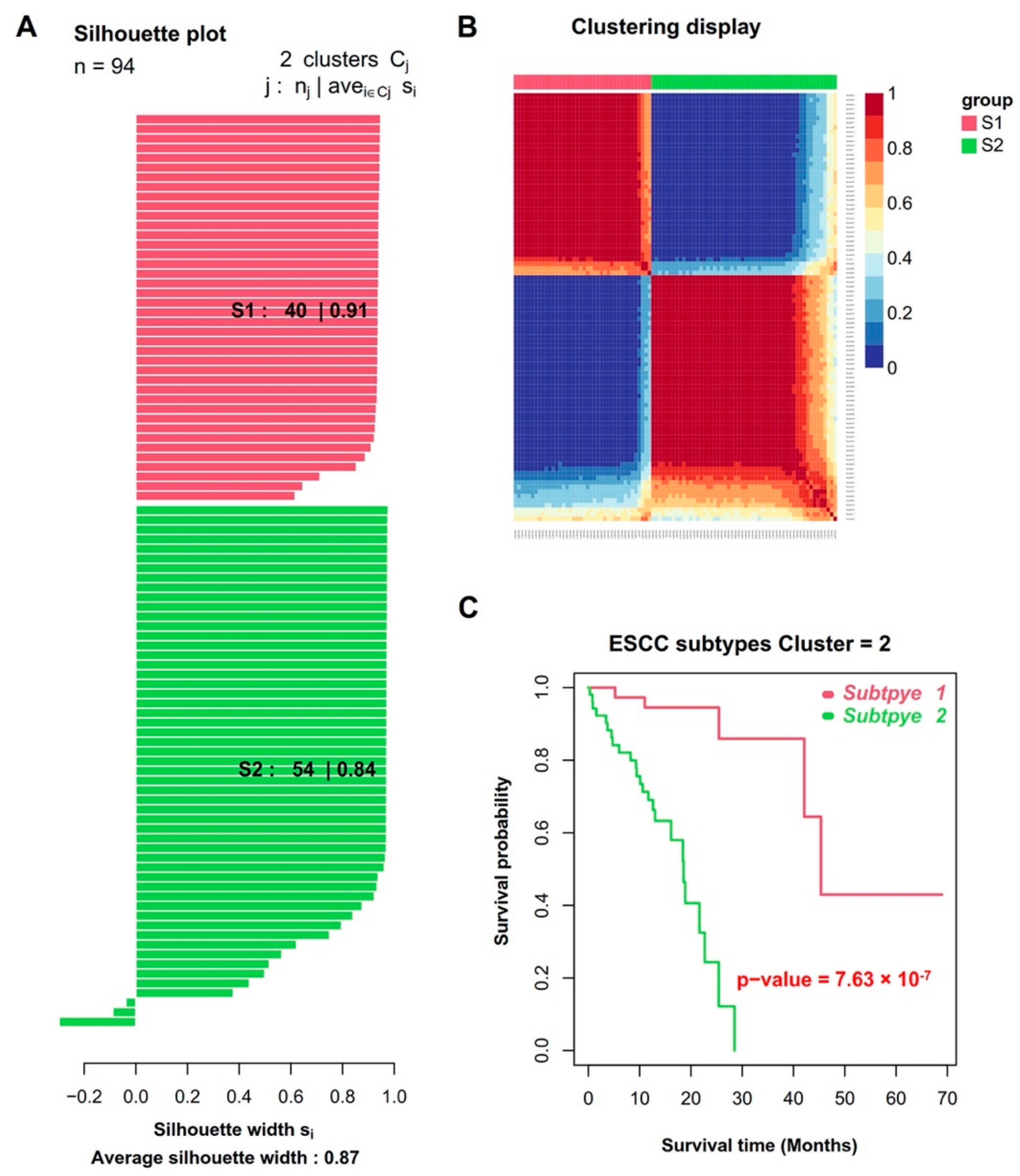
3.1. Integrative Analysis of DNA Methylation and Gene Expression Profiles Reveals Two Molecular Subtypes of ESCC
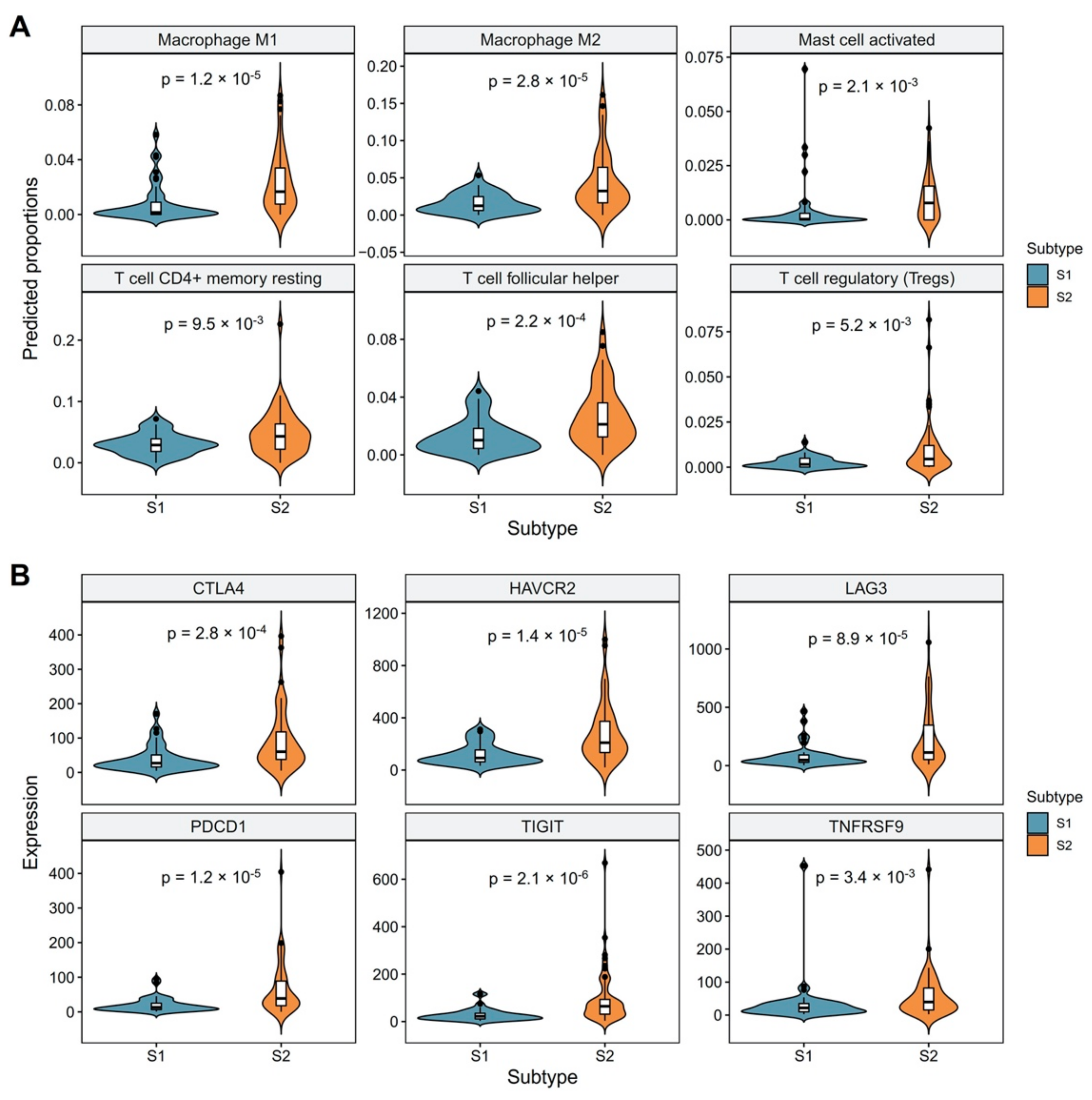
3.2. Revealing the Relationship between Molecular Subtypes and Tumor Microenvironment
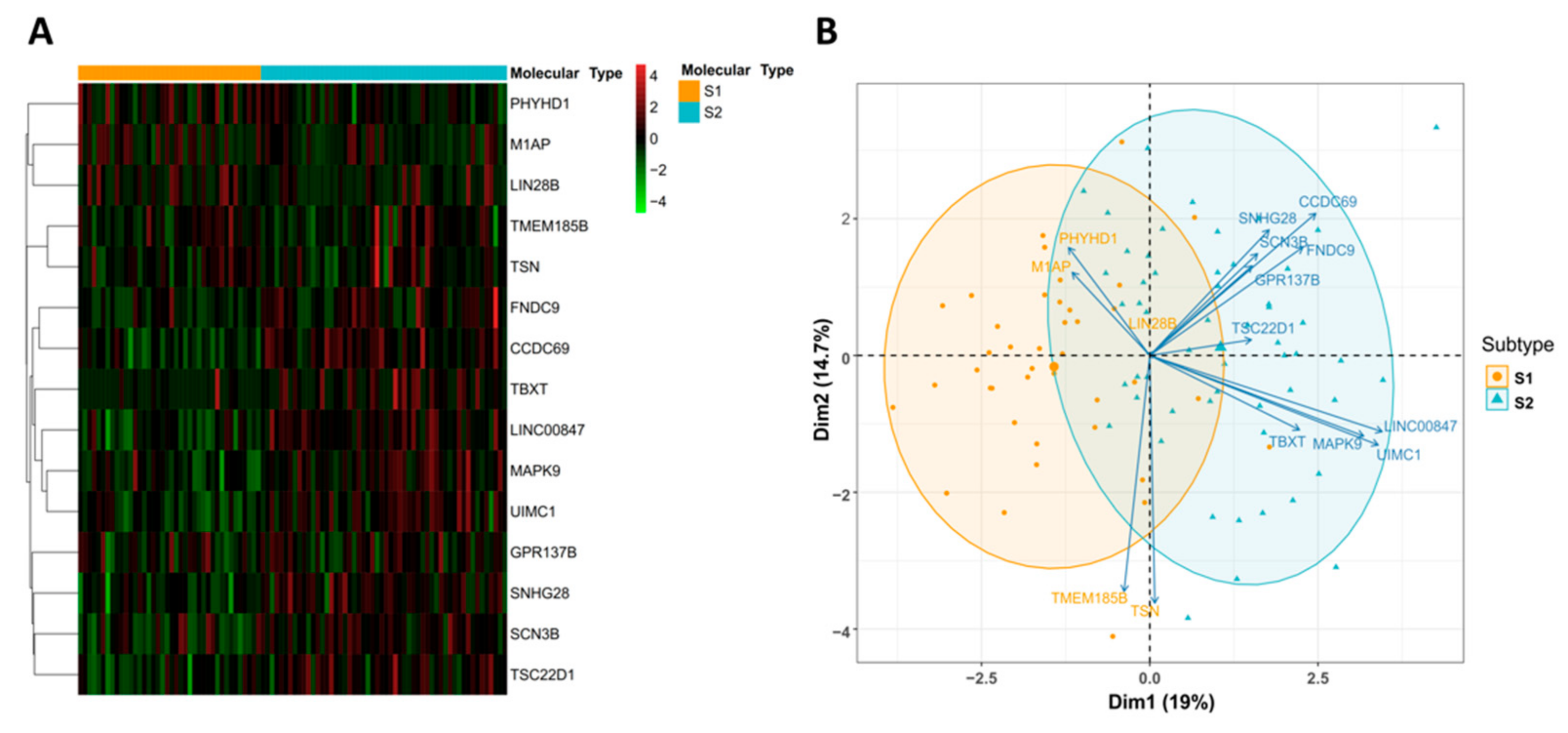
3.3. Identification and Evaluation of a 15-Gene Signature for Subtype Classification
3.4. Independent Validation of the Predictive Power of Molecular Subtypes for Immunotherapy Efficacy
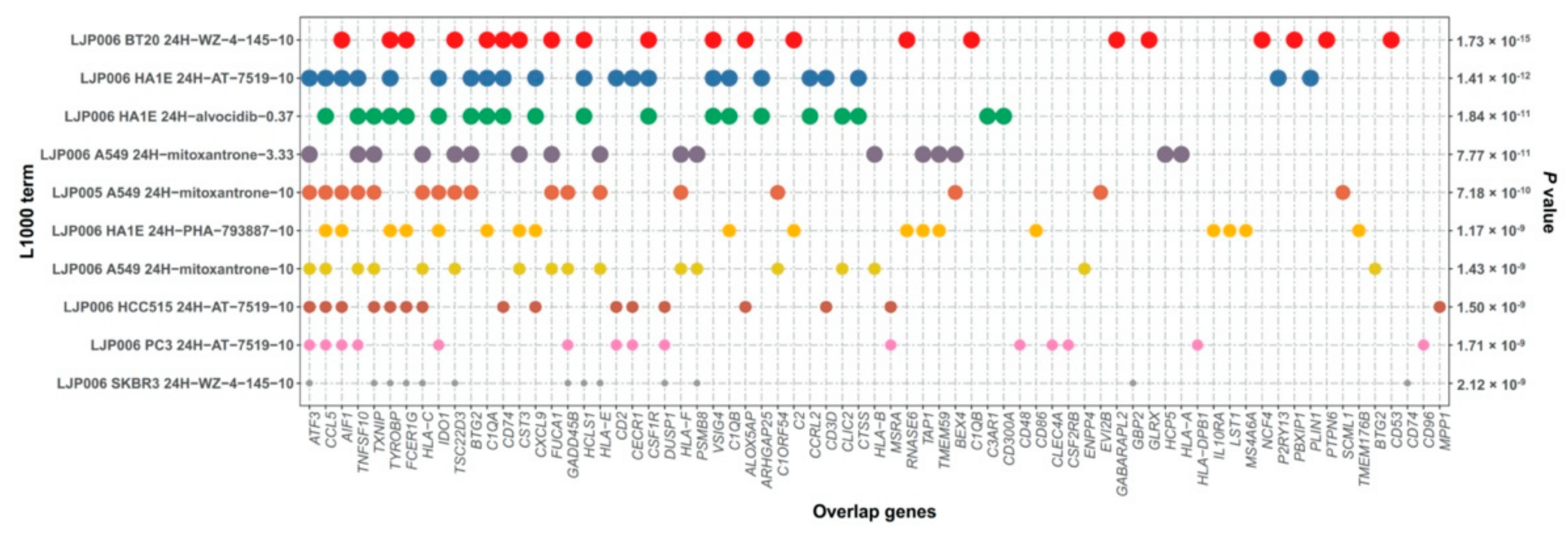
3.5. Drug Prediction for S2 Subtype ESCC Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Liang, D.; Du, L.; Guo, T.; Liu, Y.; Sun, X.; Wang, N.; Zhang, M.; Wei, K.; Shan, B.; et al. Clinical characteristics and survival of 5283 esophageal cancer patients: A multicenter study from eighteen hospitals across six regions in China. Cancer Commun. 2020, 40, 531–544. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Lin, S.; Li, N.; Deng, Y.; Wang, M.; Xiang, D.; Xiang, G.; Wang, S.; Ye, X.; Zheng, Y.; et al. Burden, trends, and risk factors of esophageal cancer in China from 1990 to 2017: An up-to-date overview and comparison with those in Japan and South Korea. J. Hematol. Oncol. 2020, 13, 146. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Tao, H.; Song, D.; Chen, C. Recurrence risk model for esophageal cancer after radical surgery. Chin. J. Cancer Res. 2013, 25, 549–555. [Google Scholar]
- Guo, X.-F.; Mao, T.; Gu, Z.-T.; Ji, C.-Y.; Fang, W.-T.; Chen, W.-H. Clinical study on postoperative recurrence in patients with pN0 esophageal squamous cell carcinoma. J. Cardiothorac. Surg. 2014, 9, 150–157. [Google Scholar] [CrossRef]
- Liu, Y.; Ren, Z.; Yuan, L.; Xu, S.; Yao, Z.; Qiao, L.; Li, K. Paclitaxel plus cisplatin vs. 5-fluorouracil plus cisplatin as first-line treatment for patients with advanced squamous cell esophageal cancer. Am. J. Cancer Res. 2016, 6, 2345–2350. [Google Scholar]
- Lee, S.J.; Kim, S.; Kim, M.; Lee, J.; Park, Y.H.; Im, Y.-H.; Park, S.H. Capecitabine in combination with either cisplatin or weekly paclitaxel as a first-line treatment for metastatic esophageal squamous cell carcinoma: A randomized phase II study. BMC Cancer 2015, 15, 693–697. [Google Scholar] [CrossRef]
- Moehler, M.; Maderer, A.; Thuss-Patience, P.C.; Brenner, B.; Meiler, J.; Ettrich, T.J.; Hofheinz, R.-D.; Al-Batran, S.E.; Vogel, A.; Mueller, L.; et al. Cisplatin and 5-fluorouracil with or without epidermal growth factor receptor inhibition panitumumab for patients with non-resectable, advanced or metastatic oesophageal squamous cell cancer: A prospective, open-label, randomised phase III AIO/EORTC trial (POWER). Ann. Oncol. 2020, 31, 228–235. [Google Scholar]
- Sun, J.-M.; Shen, L.; Shah, M.A.; Enzinger, P.; Adenis, A.; Doi, T.; Kojima, T.; Metges, J.-P.; Li, Z.; Kim, S.-B.; et al. KEYNOTE-590 Investigators Pembrolizumab plus chemotherapy versus chemotherapy alone for first-line treatment of advanced oesophageal cancer (KEYNOTE-590): A randomised, placebo-controlled, phase 3 study. Lancet 2021, 398, 759–771. [Google Scholar] [CrossRef]
- Luo, H.; Lu, J.; Bai, Y.; Mao, T.; Wang, J.; Fan, Q.; Zhang, Y.; Zhao, K.; Chen, Z.; Gao, S.; et al. ESCORT—1st Investigators Effect of Camrelizumab vs. Placebo Added to Chemotherapy on Survival and Progression-Free Survival in Patients with Advanced or Metastatic Esophageal Squamous Cell Carcinoma: The ESCORT-1st Randomized Clinical Trial. JAMA 2021, 326, 916–925. [Google Scholar] [CrossRef]
- Wang, Z.-X.; Cui, C.; Yao, J.; Zhang, Y.; Li, M.; Feng, J.; Yang, S.; Fan, Y.; Shi, J.; Zhang, X.; et al. Toripalimab plus chemotherapy in treatment-naïve, advanced esophageal squamous cell carcinoma (JUPITER-06): A multi-center phase 3 trial. Cancer Cell 2022, 40, 277–288.e3. [Google Scholar] [CrossRef]
- Doki, Y.; Ajani, J.A.; Kato, K.; Xu, J.; Wyrwicz, L.; Motoyama, S.; Ogata, T.; Kawakami, H.; Hsu, C.-H.; Adenis, A.; et al. CheckMate 648 Trial Investigators Nivolumab Combination Therapy in Advanced Esophageal Squamous-Cell Carcinoma. N. Engl. J. Med. 2022, 386, 449–462. [Google Scholar] [CrossRef]
- Paver, E.C.; Cooper, W.A.; Colebatch, A.J.; Ferguson, P.M.; Hill, S.K.; Lum, T.; Shin, J.-S.; O’Toole, S.; Anderson, L.; Scolyer, R.A.; et al. Programmed death ligand-1 (PD-L1) as a predictive marker for immunotherapy in solid tumours: A guide to immunohistochemistry implementation and interpretation. Pathology 2021, 53, 141–156. [Google Scholar] [CrossRef]
- Rong, L.; Liu, Y.; Hui, Z.; Zhao, Z.; Zhang, Y.; Wang, B.; Yuan, Y.; Li, W.; Guo, L.; Ying, J.; et al. PD-L1 expression and its clinicopathological correlation in advanced esophageal squamous cell carcinoma in a Chinese population. Diagn. Pathol. 2019, 14, 6–10. [Google Scholar] [CrossRef]
- Yang, H.; Wang, K.; Wang, T.; Li, M.; Li, B.; Li, S.; Yuan, L. The Combination Options and Predictive Biomarkers of PD-1/PD-L1 Inhibitors in Esophageal Cancer. Front. Oncol. 2020, 10, 300. [Google Scholar] [CrossRef]
- Cavalli, F.M.G.; Remke, M.; Rampasek, L.; Peacock, J.; Shih, D.J.H.; Luu, B.; Garzia, L.; Torchia, J.; Nor, C.; Morrissy, A.S.; et al. Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell 2017, 31, 737–754.e6. [Google Scholar] [CrossRef]
- Wang, B.; Mezlini, A.M.; Demir, F.; Fiume, M.; Tu, Z.; Brudno, M.; Haibe-Kains, B.; Goldenberg, A. Similarity network fusion for aggregating data types on a genomic scale. Nat. Methods 2014, 11, 333–337. [Google Scholar] [CrossRef]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Chen, B.; Khodadoust, M.S.; Liu, C.L.; Newman, A.M.; Alizadeh, A.A. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol. Biol. 2018, 1711, 243–259. [Google Scholar]
- Simon, R.; Lam, A.; Li, M.-C.; Ngan, M.; Menenzes, S.; Zhao, Y. Analysis of Gene Expression Data Using BRB-Array Tools. Cancer Inform. 2007, 3, 11–17. [Google Scholar] [CrossRef]
- Sulakhe, D.; Balasubramanian, S.; Xie, B.; Feng, B.; Taylor, A.; Wang, S.; Berrocal, E.; Dave, U.; Xu, J.; Börnigen, D.; et al. Lynx: A database and knowledge extraction engine for integrative medicine. Nucleic Acids Res. 2014, 42, D1007–D1012. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M. Building Predictive Models in R Using the caret Package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Guyon, I.; Weston, J.; Barnhill, S.; Vapnik, V. Gene selection for cancer classification using support vector machines. Mach. Learn. 2002, 46, 389–422. [Google Scholar] [CrossRef]
- Eisenhauer, E.A.; Therasse, P.; Bogaerts, J.; Schwartz, L.H.; Sargent, D.; Ford, R.; Dancey, J.; Arbuck, S.; Gwyther, S.; Mooney, M.; et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur. J. Cancer 2009, 45, 228–247. [Google Scholar] [CrossRef]
- Fu, J.; Li, K.; Zhang, W.; Wan, C.; Zhang, J.; Jiang, P.; Liu, X.S. Large-scale public data reuse to model immunotherapy response and resistance. Genome Med. 2020, 12, 21–28. [Google Scholar] [CrossRef]
- Miao, Y.-R.; Zhang, Q.; Lei, Q.; Luo, M.; Xie, G.-Y.; Wang, H.; Guo, A.-Y. ImmuCellAI: A Unique Method for Comprehensive T-Cell Subsets Abundance Prediction and its Application in Cancer Immunotherapy. Adv. Sci. 2020, 7, 1902880. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef]
- Yue, Y.; Lian, J.; Wang, T.; Luo, C.; Yuan, Y.; Qin, G.; Zhang, B.; Zhang, Y. Interleukin-33-nuclear factor-κB-CCL2 signaling pathway promotes progression of esophageal squamous cell carcinoma by directing regulatory T cells. Cancer Sci. 2020, 111, 795–806. [Google Scholar] [CrossRef]
- Nabeki, B.; Ishigami, S.; Uchikado, Y.; Sasaki, K.; Kita, Y.; Okumura, H.; Arigami, T.; Kijima, Y.; Kurahara, H.; Maemura, K.; et al. Interleukin-32 expression and Treg infiltration in esophageal squamous cell carcinoma. Anticancer Res. 2015, 35, 2941–2947. [Google Scholar]
- Yao, J.; Duan, L.; Huang, X.; Liu, J.; Fan, X.; Xiao, Z.; Yan, R.; Liu, H.; An, G.; Hu, B.; et al. Development and Validation of a Prognostic Gene Signature Correlated with M2 Macrophage Infiltration in Esophageal Squamous Cell Carcinoma. Front. Oncol. 2021, 11, 769727. [Google Scholar] [CrossRef]
- Fakhrjou, A.; Niroumand-Oscoei, S.M.; Somi, M.H.; Ghojazadeh, M.; Naghashi, S.; Samankan, S. Prognostic value of tumor-infiltrating mast cells in outcome of patients with esophagus squamous cell carcinoma. J. Gastrointest. Cancer 2014, 45, 48–53. [Google Scholar] [CrossRef]
- Zheng, L.; Qin, S.; Si, W.; Wang, A.; Xing, B.; Gao, R.; Ren, X.; Wang, L.; Wu, X.; Zhang, J.; et al. Pan-cancer single-cell landscape of tumor-infiltrating T cells. Science 2021, 374, 6474. [Google Scholar] [CrossRef]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. Lymphoma/Leukemia Molecular Profiling Project Stromal gene signatures in large-B-cell lymphomas. N. Engl. J. Med. 2008, 359, 2313–2323. [Google Scholar]
- Sparano, J.A.; Gray, R.J.; Makower, D.F.; Pritchard, K.I.; Albain, K.S.; Hayes, D.F.; Geyer, C.E.; Dees, E.C.; Goetz, M.P.; Olson, J.A.; et al. Adjuvant Chemotherapy Guided by a 21-Gene Expression Assay in Breast Cancer. N. Engl. J. Med. 2018, 379, 111–121. [Google Scholar] [CrossRef]
- Zheng, Y.; Chen, Z.; Han, Y.; Han, L.; Zou, X.; Zhou, B.; Hu, R.; Hao, J.; Bai, S.; Xiao, H.; et al. Immune suppressive landscape in the human esophageal squamous cell carcinoma microenvironment. Nat. Commun. 2020, 11, 6268. [Google Scholar] [CrossRef]
- Evison, B.J.; Sleebs, B.E.; Watson, K.G.; Phillips, D.R.; Cutts, S.M. Mitoxantrone, More Than Just Another Topoisomerase II Poison. Med. Res. Rev. 2016, 36, 248–299. [Google Scholar] [CrossRef]
- Allegra, J.C.; Woodcock, T.; Woolf, S.; Henderson, I.C.; Bryan, S.; Reisman, A.; Dukart, G. A randomized trial comparing mitoxantrone with doxorubicin in patients with stage IV breast cancer. Investig. New Drugs 1985, 3, 153–161. [Google Scholar] [CrossRef]
- Henderson, I.C.; Allegra, J.C.; Woodcock, T.; Wolff, S.; Bryan, S.; Cartwright, K.; Dukart, G.; Henry, D. Randomized clinical trial comparing mitoxantrone with doxorubicin in previously treated patients with metastatic breast cancer. Am. Soc. Clin. Oncol. Educ. Book 1989, 7, 560–571. [Google Scholar] [CrossRef]
- Faulds, D.; Balfour, J.A.; Chrisp, P.; Langtry, H.D. Mitoxantrone. A review of its pharmacodynamic and pharmacokinetic properties, and therapeutic potential in the chemotherapy of cancer. Drugs 1991, 41, 400–449. [Google Scholar] [CrossRef]
- Dunn, C.J.; Goa, K.L. Mitoxantrone: A review of its pharmacological properties and use in acute nonlymphoblastic leukaemia. Drugs Aging 1996, 9, 122–147. [Google Scholar] [CrossRef]
- Weiss, M.A. Treatment of adult patients with relapsed or refractory acute lymphoblastic leukemia (ALL). Leukemia 1997, 11 (Suppl. S4), S28–S30. [Google Scholar]
- Tallman, M.S.; Gilliland, D.G.; Rowe, J.M. Drug therapy for acute myeloid leukemia. Blood 2005, 106, 1154–1163. [Google Scholar] [CrossRef]
- Kantoff, P.W.; Halabi, S.; Conaway, M.; Picus, J.; Kirshner, J.; Hars, V.; Trump, D.; Winer, E.P.; Vogelzang, N.J. Hydrocortisone with or without mitoxantrone in men with hormone-refractory prostate cancer: Results of the cancer and leukemia group B 9182 study. Am. Soc. Clin. Oncol. Educ. Book 1999, 17, 2506–2513. [Google Scholar] [CrossRef]
- Tannock, I.F.; Osoba, D.; Stockler, M.R.; Ernst, D.S.; Neville, A.J.; Moore, M.J.; Armitage, G.R.; Wilson, J.J.; Venner, P.M.; Coppin, C.M.; et al. Chemotherapy with mitoxantrone plus prednisone or prednisone alone for symptomatic hormone-resistant prostate cancer: A Canadian randomized trial with palliative end points. Am. Soc. Clin. Oncol. Educ. Book 1996, 14, 1756–1764. [Google Scholar] [CrossRef]
- Hoffmanns, H.W.; Altmeier, G. Local application of mitoxantrone in inoperable, stenosing esophageal carcinoma. Preliminary report. Onkologie 1986, 9, 27–29. [Google Scholar]
- Wang, L.; Wei, Q.; Zhang, M.; Chen, L.; Li, Z.; Zhou, C.; He, M.; Wei, M.; Zhao, L. Identification of the prognostic value of immune gene signature and infiltrating immune cells for esophageal cancer patients. Int. Immunopharmacol. 2020, 87, 106795. [Google Scholar] [CrossRef]
- Mao, Y.; Fu, Z.; Zhang, Y.; Dong, L.; Zhang, Y.; Zhang, Q.; Li, X.; Liu, J. A seven-lncRNA signature predicts overall survival in esophageal squamous cell carcinoma. Sci. Rep. 2018, 8, 8823. [Google Scholar] [CrossRef]
- Lu, T.; Chen, D.; Wang, Y.; Sun, X.; Li, S.; Miao, S.; Wo, Y.; Dong, Y.; Leng, X.; Du, W.; et al. Identification of DNA methylation-driven genes in esophageal squamous cell carcinoma: A study based on The Cancer Genome Atlas. Cancer Cell Int. 2019, 19, 52. [Google Scholar] [CrossRef]
- Zhang, C.; Luo, Y.; Zhang, Z.; Zhang, Z.; Zhang, G.; Wang, F.; Che, Y.; Fang, L.; Zhang, Y.; Sun, N.; et al. Identification of a Prognostic Immune Signature for Esophageal Squamous Cell Carcinoma to Predict Survival and Inflammatory Landscapes. Front. Cell Dev. Biol. 2020, 8, 580005. [Google Scholar] [CrossRef]
- Gao, J.; Tang, T.; Zhang, B.; Li, G. A Prognostic Signature Based on Immunogenomic Profiling Offers Guidance for Esophageal Squamous Cell Cancer Treatment. Front. Oncol. 2021, 11, 603634. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Xie, L.; He, Y.-H.; Wu, Z.-Y.; Liu, L.-X.; Bai, X.-F.; Deng, D.-X.; Xu, X.-E.; Liao, L.-D.; Lin, W.; et al. Large-scale and high-resolution mass spectrometry-based proteomics profiling defines molecular subtypes of esophageal cancer for therapeutic targeting. Nat. Commun. 2021, 12, 4961. [Google Scholar] [CrossRef] [PubMed]

| Characteristics | Total Cohort | S1 (%) | S2 (%) | χ2 | p-Value |
|---|---|---|---|---|---|
| Number of patients | 94 | 40 | 54 | ||
| Status | |||||
| Alive | 75 | 36 (90.0) | 39 (72.2) | 3.468 | 0.062 |
| Dead | 19 | 4 (10.0) | 15 (27.8) | ||
| Gender | |||||
| Male | 81 | 33 (82.5) | 48 (88.9) | 0.342 | 0.559 |
| Female | 13 | 7 (17.5) | 6 (11.1) | ||
| Age at diagnosis | |||||
| Mean | 58 | 58 | 59 | ||
| Range | 36–90 | 36–84 | 36–90 | ||
| Race | |||||
| Asian | 45 | 21 (52.5) | 24 (44.4) | 3.365 | 0.339 |
| White | 41 | 16 (40.0) | 25 (46.3) | ||
| Black (black or African American) | 5 | 3 (7.5) | 2 (3.7) | ||
| NA | 3 | 0 (0.0) | 3 (5.6) | ||
| Tumor central location | |||||
| Mid | 44 | 18 (45.0) | 26 (48.1) | 1.543 | 0.672 |
| Distal | 43 | 18 (45.0) | 25 (46.3) | ||
| Proximal | 6 | 3 (7.5) | 3 (5.6) | ||
| Not specified | 1 | 1 (2.5) | 0 (0.0) | ||
| Stage | |||||
| I | 6 | 3 (7.5) | 3 (5.6) | 5.045 | 0.283 |
| II | 55 | 27 (67.5) | 28 (51.9) | ||
| III | 27 | 9 (22.5) | 18 (33.3) | ||
| IV | 4 | 0 (0.0) | 4 (7.4) | ||
| NA | 2 | 1 (2.5) | 1 (1.8) | ||
| Grade | |||||
| Grade 1 | 16 | 11 (27.5) | 5 (9.3) | 5.806 | 0.122 |
| Grade 2 | 48 | 17 (42.5) | 31 (57.4) | ||
| Grade 3 | 21 | 9 (22.5) | 12 (22.2) | ||
| Grade X | 9 | 3 (7.5) | 6 (11.1) | ||
| Alcohol | |||||
| Yes | 68 | 31 (77.5) | 37 (68.5) | 1.136 | 0.567 |
| Never | 24 | 8 (20.0) | 16 (29.6) | ||
| NA | 2 | 1 (2.5) | 1 (1.9) | ||
| Smoking | |||||
| Never | 32 | 15 (37.5) | 17 (31.5) | 0.896 | 0.826 |
| Current | 28 | 10 (25.0) | 18 (33.3) | ||
| Reformed ≤ 15 years | 21 | 8 (20.0) | 13 (24.1) | ||
| Reformed > 15 years | 9 | 4 (10.0) | 5 (9.3) | ||
| NA | 4 | 3 (7.5) | 1 (1.8) | ||
| Radiation treatment | |||||
| Yes | 30 | 14 (35.0) | 16 (29.6) | 4.140 | 0.126 |
| No | 40 | 20 (50.0) | 20 (37.1) | ||
| NA | 24 | 6 (15.0) | 18 (33.3) | ||
| Pharmaceutical treatment | |||||
| Yes | 8 | 5 (12.5) | 3 (5.5) | 1.676 | 0.433 |
| No | 69 | 29 (72.5) | 40 (74.1) | ||
| NA | 17 | 6 (15.0) | 11 (20.4) | ||
| Characteristics | Univariate Analysis | Multivariate Analysis | ||||
|---|---|---|---|---|---|---|
| Hazard Ratio | 95% CI | p-Value | Hazard Ratio | 95% CI | p-Value | |
| Gender | ||||||
| Male | Reference | |||||
| Female | 0.14 | 0.02–1.00 | 5.01 × 10−2 | 0.35 | 0.03–4.00 | 4.02 × 10−1 |
| Age at diagnosis | 1.03 | 0.99–1.08 | 1.26 × 10−1 | 1.03 | 0.97–1.11 | 3.07 × 10−1 |
| Race | ||||||
| Asian | Reference | |||||
| White | 1.53 | 0.65–3.63 | 3.31 × 10−1 | 1.37 | 0.45–4.16 | 5.80 × 10−1 |
| Black (black or African American) | 3.05 | 0.81–11.44 | 9.89 × 10−2 | 0.18 | 0.01–3.20 | 2.40 × 10−1 |
| Tumor central location | ||||||
| Mid | Reference | |||||
| Distal | 0.81 | 0.38–1.74 | 5.92 × 10−1 | 1.51 | 0.53–4.28 | 4.36 × 10−1 |
| Proximal | 0 | 0–Inf | 9.98 × 10−1 | 0 | 0-Inf | 9.98 × 10−1 |
| Stage | ||||||
| I–II | Reference | |||||
| III–IV | 2.59 | 1.23–5.46 | 1.26 × 10−2 | 1.25 | 0.46–3.39 | 6.59 × 10−1 |
| Histologic grade | ||||||
| Grade 1 | Reference | |||||
| Grade 2 | 1.84 | 0.62–5.46 | 2.71 × 10−1 | 0.61 | 0.15–2.47 | 4.89 × 10−1 |
| Grade 3 | 0.93 | 0.23–3.71 | 9.13 × 10−1 | 0.24 | 0.05–1.20 | 8.24 × 10−2 |
| Alcohol consumption | ||||||
| Never | Reference | |||||
| Yes | 2.02 | 0.7–5.85 | 1.93 × 10−1 | 1.57 | 0.39–6.27 | 5.2 × 10−1 |
| Tobacco smoking history | ||||||
| Never | Reference | |||||
| Yes | 1.51 | 0.64–3.55 | 3.46 × 10−1 | 0.74 | 0.21–2.62 | 6.4 × 10−1 |
| Molecular types | ||||||
| Subtype 1 | Reference | |||||
| Subtype 2 | 13.53 | 3.85–47.57 | 4.91 × 10−5 | 51.60 | 3.99–667.48 | 2.5 × 10−3 |
| Characteristics | Total | S1 (%) | S2 (%) | χ2 | p-Value |
|---|---|---|---|---|---|
| Number of patients | 36 | 16 | 20 | ||
| Gender | |||||
| Male | 31 | 14 (87.5) | 17 (85.0) | 6.390 | 1.000 |
| Female | 5 | 2 (12.5) | 3 (15.0) | ||
| Age at diagnosis | |||||
| Median | 64.5 | 63.5 | 65 | ||
| Range | 47–79 | 52–79 | 47–79 | ||
| Stage | |||||
| II | 8 | 4 (25.0) | 4 (20.0) | 4.950 | 0.084 |
| III | 12 | 8 (50.0) | 4 (20.0) | ||
| IV | 16 | 4 (25.0) | 12 (60.0) | ||
| Differentiation 1 | |||||
| Well | 5 | 3 (30.0) | 2 (16.7) | 0.054 | 0.816 |
| Moderately/poorly | 17 | 7 (70.0) | 10 (83.3) | ||
| Alcohol 2 | |||||
| Yes | 16 | 9 (56.2) | 7 (38.9) | 0.446 | 0.504 |
| Never | 18 | 7 (43.8) | 11 (61.1) | ||
| Smoking 3 | |||||
| Never | 18 | 6 (37.5) | 12 (66.7) | 4.183 | 0.124 |
| Current | 14 | 8 (50.0) | 6 (33.3) | ||
| Reformed ≤ 15 years | 0 | 0 (0.0) | 0 (0.0) | ||
| Reformed > 15 years | 2 | 2(12.5) | 0(0.0) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, Y.; Gao, Q.; Su, X.; Xiao, C.; Yu, B.; Huang, S.; Sun, Y.; Wu, S.; Wo, Y.; Xu, Q.; et al. Genome-Wide DNA Methylation and Gene Expression Profiling Characterizes Molecular Subtypes of Esophagus Squamous Cell Carcinoma for Predicting Patient Survival and Immunotherapy Efficacy. Cancers 2022, 14, 4970. https://doi.org/10.3390/cancers14204970
Zheng Y, Gao Q, Su X, Xiao C, Yu B, Huang S, Sun Y, Wu S, Wo Y, Xu Q, et al. Genome-Wide DNA Methylation and Gene Expression Profiling Characterizes Molecular Subtypes of Esophagus Squamous Cell Carcinoma for Predicting Patient Survival and Immunotherapy Efficacy. Cancers. 2022; 14(20):4970. https://doi.org/10.3390/cancers14204970
Chicago/Turabian StyleZheng, Yulong, Qiqi Gao, Xingyun Su, Cheng Xiao, Bo Yu, Shenglin Huang, Yifeng Sun, Sheng Wu, Yixin Wo, Qinghua Xu, and et al. 2022. "Genome-Wide DNA Methylation and Gene Expression Profiling Characterizes Molecular Subtypes of Esophagus Squamous Cell Carcinoma for Predicting Patient Survival and Immunotherapy Efficacy" Cancers 14, no. 20: 4970. https://doi.org/10.3390/cancers14204970
APA StyleZheng, Y., Gao, Q., Su, X., Xiao, C., Yu, B., Huang, S., Sun, Y., Wu, S., Wo, Y., Xu, Q., Xu, N., & Yu, H. (2022). Genome-Wide DNA Methylation and Gene Expression Profiling Characterizes Molecular Subtypes of Esophagus Squamous Cell Carcinoma for Predicting Patient Survival and Immunotherapy Efficacy. Cancers, 14(20), 4970. https://doi.org/10.3390/cancers14204970






