TNTdetect.AI: A Deep Learning Model for Automated Detection and Counting of Tunneling Nanotubes in Microscopy Images
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Microscopy Imaging
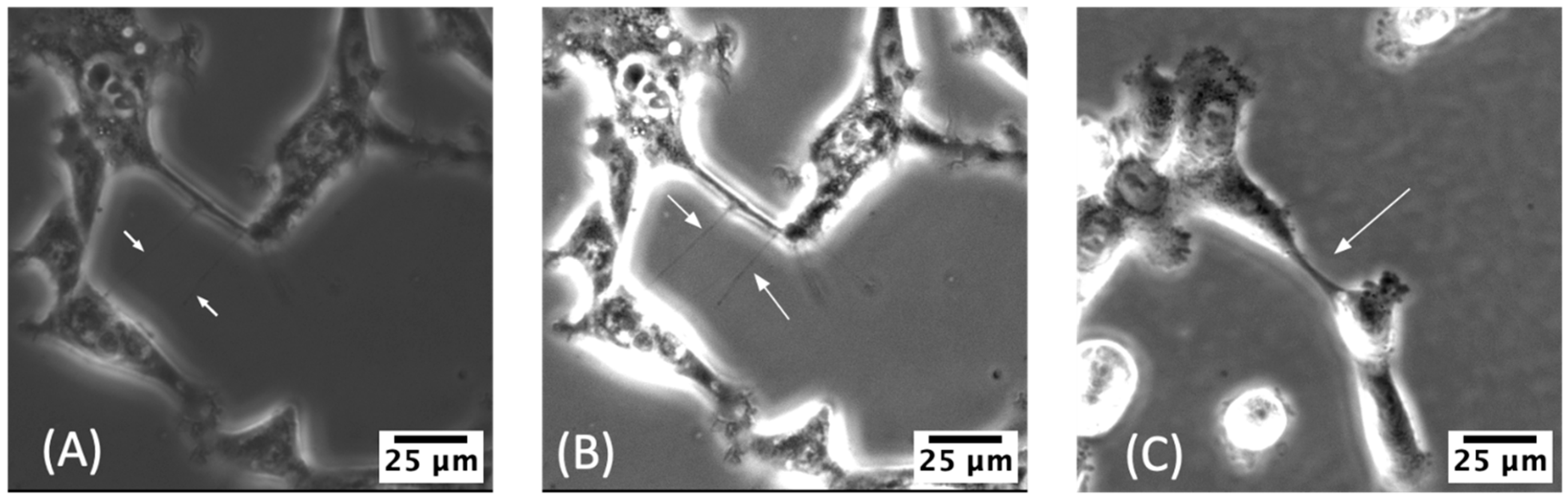
2.3. Manual Identification of TNTs
2.4. Initial Verification of TNTs Using Current Standard Methodology: Visual Identification
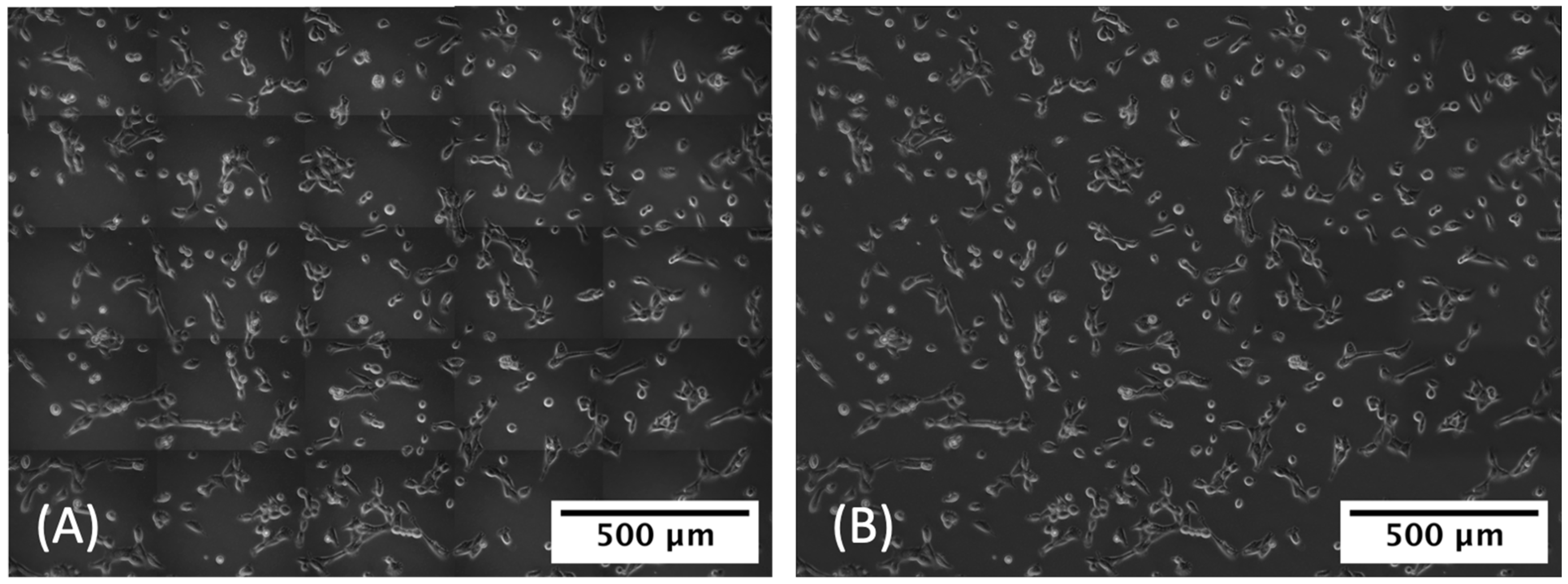
2.5. Human Expert Review of Stitched MSTO Images and Identification of TNTs
3. Results
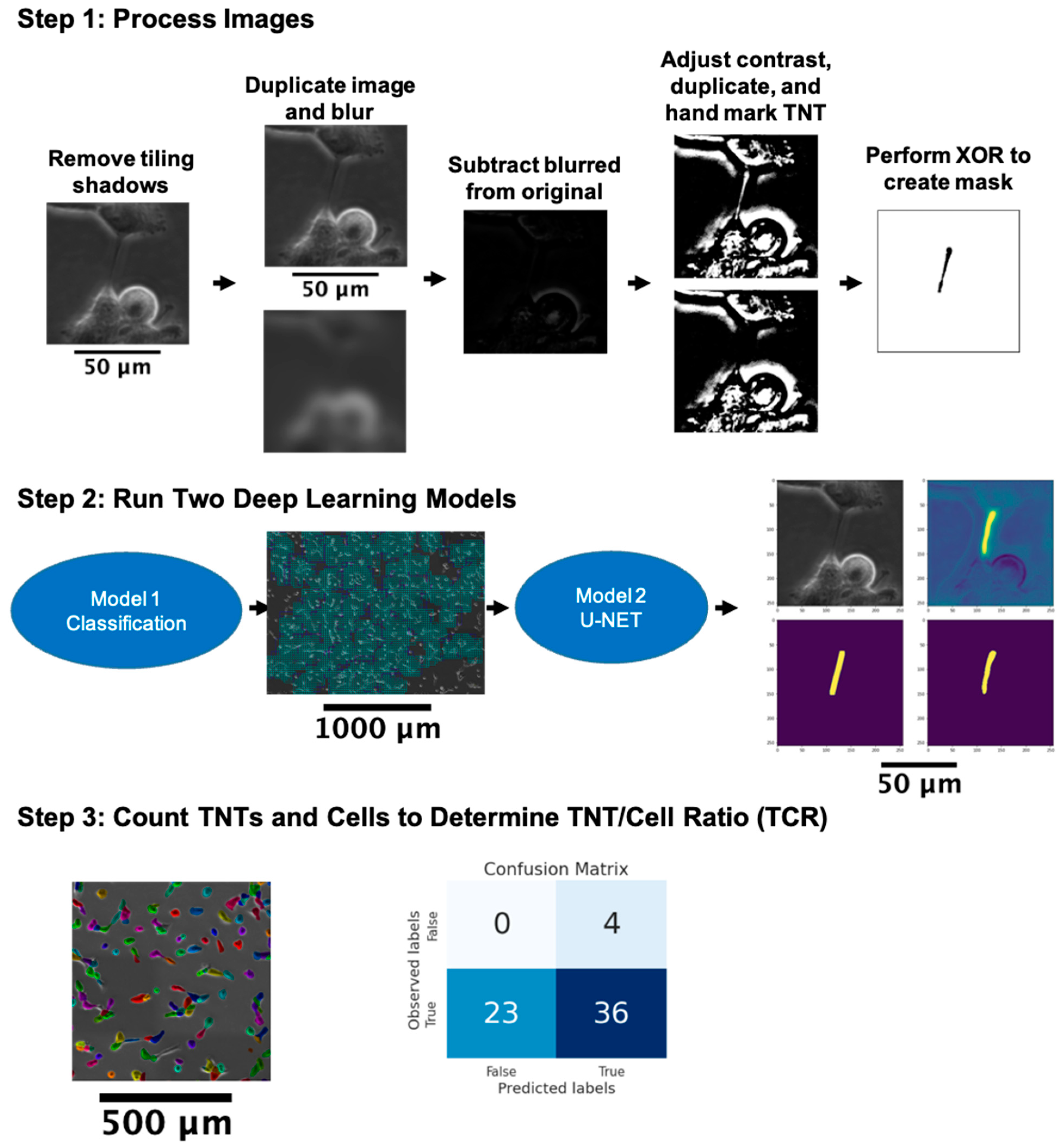
3.1. General Approach to the Automated Detection of TNTs
3.2. Pre-Processing
3.2.1. Removal of Tile Shadows
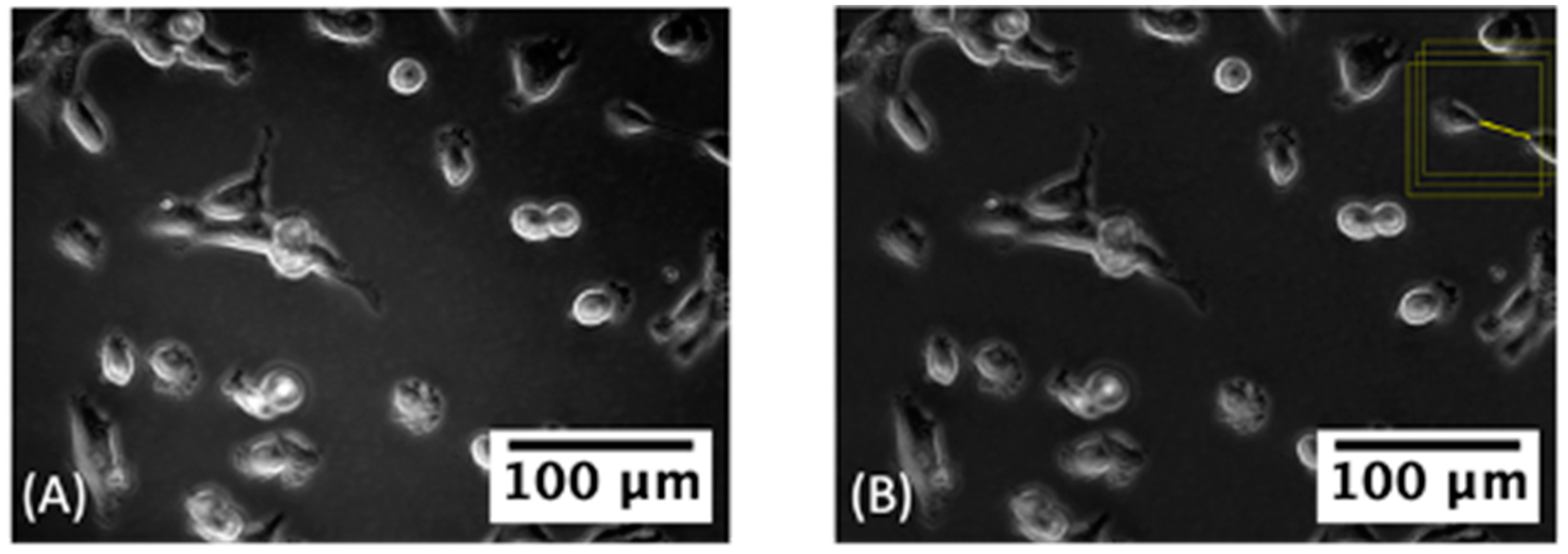
3.2.2. Label Correction
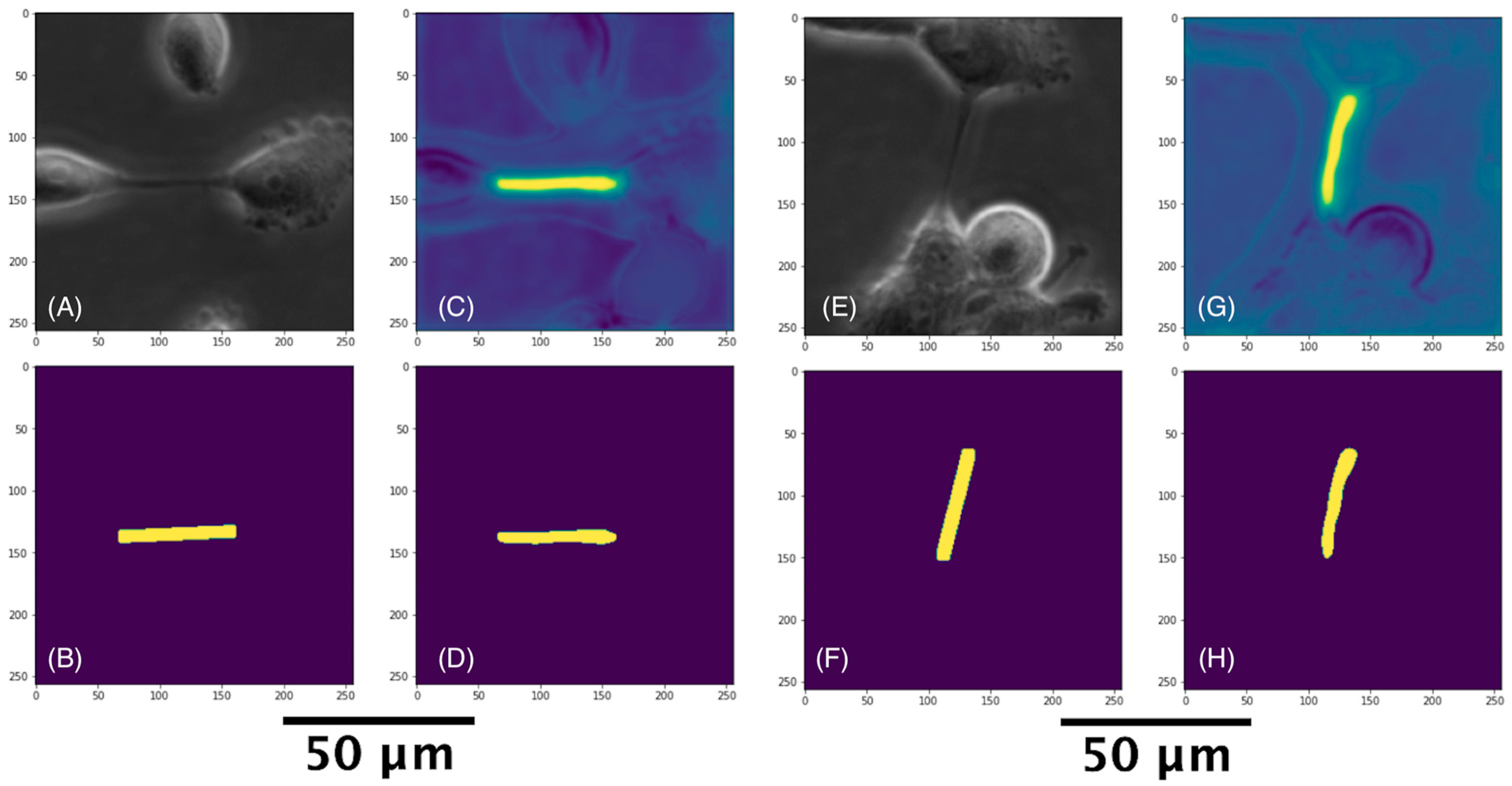
3.3. Detecting TNT Regions
3.3.1. Classifying TNT-Inclusive Regions
3.3.2. U-Net with Attention Architecture for Segmentation
3.4. Cell and TNT Counting
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AC | active contour |
| AI | artificial intelligence |
| AURA-net | modified U-Net architecture designed to answer the problem of segmentation for small datasets of phase contrast microscopy images |
| BCE | binary cross-entropy |
| CNN | convolutional neural network |
| FP | false positive |
| ML | machine learning |
| OpenCV | open-source computer vision library |
| PPV | positive predictive value |
| ReLU | rectified linear unit |
| ResNET | residual neural network |
| TCR | TNT-to-cell ratio |
| TNT | tunnelling nanotube |
| U-Net | a convolutional network architecture for fast and precise image segmentation |
| VGGNet | a convolutional neural network architecture to increase ML model performance |
References
- Lau, R.P.; Kim, T.H.; Rao, J. Advances in Imaging Modalities, Artificial Intelligence, and Single Cell Biomarker Analysis, and Their Applications in Cytopathology. Front. Med. 2021, 8, 689954. [Google Scholar] [CrossRef] [PubMed]
- Nagaki, K.; Furuta, T.; Yamaji, N.; Kuniyoshi, D.; Ishihara, M.; Kishima, Y.; Murata, M.; Hoshino, A.; Takatsuka, H. Effectiveness of Create ML in microscopy image classifications: A simple and inexpensive deep learning pipeline for non-data scientists. Chromosome Res. 2021, 29, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Durkee, M.S.; Abraham, R.; Clark, M.R.; Giger, M.L. Artificial Intelligence and Cellular Segmentation in Tissue Microscopy Images. Am. J. Pathol. 2021, 191, 1693–1701. [Google Scholar] [CrossRef] [PubMed]
- von Chamier, L.; Laine, R.F.; Jukkala, J.; Spahn, C.; Krentzel, D.; Nehme, E.; Lerche, M.; Hernandez-Perez, S.; Mattila, P.K.; Karinou, E.; et al. Democratising deep learning for microscopy with ZeroCostDL4Mic. Nat. Commun. 2021, 12, 2276. [Google Scholar] [CrossRef] [PubMed]
- von Chamier, L.; Laine, R.F.; Henriques, R. Artificial intelligence for microscopy: What you should know. Biochem. Soc. Trans. 2019, 47, 1029–1040. [Google Scholar] [CrossRef] [PubMed]
- Falk, T.; Mai, D.; Bensch, R.; Cicek, O.; Abdulkadir, A.; Marrakchi, Y.; Bohm, A.; Deubner, J.; Jackel, Z.; Seiwald, K.; et al. U-Net: Deep learning for cell counting, detection, and morphometry. Nat. Methods 2019, 16, 67–70. [Google Scholar] [CrossRef]
- Zinchuk, V.; Grossenbacher-Zinchuk, O. Machine Learning for Analysis of Microscopy Images: A Practical Guide. Curr. Protoc. Cell Biol. 2020, 86, e101. [Google Scholar] [CrossRef] [PubMed]
- Xing, F.; Yang, L. Chapter 4—Machine learning and its application in microscopic image analysis. In Machine Learning and Medical Imaging; Wu, G., Shen, D., Sabuncu, M.R., Eds.; Academic Press: Cambridge, MA, USA, 2016; pp. 97–127. [Google Scholar]
- Li, Y.; Nowak, C.M.; Pham, U.; Nguyen, K.; Bleris, L. Cell morphology-based machine learning models for human cell state classification. NPJ Syst. Biol. Appl. 2021, 7, 23. [Google Scholar] [CrossRef]
- Chen, D.; Sarkar, S.; Candia, J.; Florczyk, S.J.; Bodhak, S.; Driscoll, M.K.; Simon, C.G., Jr.; Dunkers, J.P.; Losert, W. Machine learning based methodology to identify cell shape phenotypes associated with microenvironmental cues. Biomaterials 2016, 104, 104–118. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Min, Y.; Oh, J.M.; Cho, Y.K. AI-powered transmitted light microscopy for functional analysis of live cells. Sci. Rep. 2019, 9, 18428. [Google Scholar] [CrossRef]
- Li, Y.; Di, J.; Wang, K.; Wang, S.; Zhao, J. Classification of cell morphology with quantitative phase microscopy and machine learning. Opt. Express 2020, 28, 23916–23927. [Google Scholar] [CrossRef] [PubMed]
- Aida, S.; Okugawa, J.; Fujisaka, S.; Kasai, T.; Kameda, H.; Sugiyama, T. Deep Learning of Cancer Stem Cell Morphology Using Conditional Generative Adversarial Networks. Biomolecules 2020, 10, 931. [Google Scholar] [CrossRef] [PubMed]
- Moen, E.; Bannon, D.; Kudo, T.; Graf, W.; Covert, M.; Van Valen, D. Deep learning for cellular image analysis. Nat. Methods 2019, 16, 1233–1246. [Google Scholar] [CrossRef] [PubMed]
- Scheeder, C.; Heigwer, F.; Boutros, M. Machine learning and image-based profiling in drug discovery. Curr. Opin. Syst. Biol. 2018, 10, 43–52. [Google Scholar] [CrossRef]
- Chandrasekaran, S.N.; Ceulemans, H.; Boyd, J.D.; Carpenter, A.E. Image-based profiling for drug discovery: Due for a machine-learning upgrade? Nat. Rev. Drug Discov. 2021, 20, 145–159. [Google Scholar] [CrossRef] [PubMed]
- Way, G.P.; Kost-Alimova, M.; Shibue, T.; Harrington, W.F.; Gill, S.; Piccioni, F.; Becker, T.; Shafqat-Abbasi, H.; Hahn, W.C.; Carpenter, A.E.; et al. Predicting cell health phenotypes using image-based morphology profiling. Mol. Biol. Cell. 2021, 32, 995–1005. [Google Scholar] [CrossRef]
- Helgadottir, S.; Midtvedt, B.; Pineda, J.; Sabirsh, A.; Adiels, C.B.; Romeo, S.; Midtvedt, D.; Volpe, G. Extracting quantitative biological information from bright-field cell images using deep learning. Biophys. Rev. 2021, 2, 031401. [Google Scholar] [CrossRef]
- Belting, M.; Wittrup, A. Nanotubes, exosomes, and nucleic acid-binding peptides provide novel mechanisms of intercellular communication in eukaryotic cells: Implications in health and disease. J. Cell Biol. 2008, 183, 1187–1191. [Google Scholar] [CrossRef]
- Bobrie, A.; Colombo, M.; Raposo, G.; Thery, C. Exosome secretion: Molecular mechanisms and roles in immune responses. Traffic 2011, 12, 1659–1668. [Google Scholar] [CrossRef]
- Zhang, H.G.; Grizzle, W.E. Exosomes and cancer: A newly described pathway of immune suppression. Clin. Cancer Res. 2011, 17, 959–964. [Google Scholar] [CrossRef]
- Guescini, M. Microvesicle and tunneling nanotube mediated intercellular transfer of g-protein coupled receptors in cell cultures. Exp. Cell Res. 2012, 318, 603–613. [Google Scholar] [CrossRef] [PubMed]
- Demory Beckler, M.; Higginbotham, J.N.; Franklin, J.L.; Ham, A.J.; Halvey, P.J.; Imasuen, I.E.; Whitwell, C.; Li, M.; Liebler, D.C.; Coffey, R.J. Proteomic analysis of exosomes from mutant KRAS colon cancer cells identifies intercellular transfer of mutant KRAS. Mol. Cell. Proteom. 2013, 12, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Fedele, C.; Singh, A.; Zerlanko, B.J.; Iozzo, R.V.; Languino, L.R. The alphavbeta6 integrin is transferred intercellularly via exosomes. J. Biol. Chem. 2015, 290, 4545–4551. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R. The biology and function of exosomes in cancer. J. Clin. Investig. 2016, 126, 1208–1215. [Google Scholar] [CrossRef] [PubMed]
- Eugenin, E.A.; Gaskill, P.J.; Berman, J.W. Tunneling nanotubes (TNT) are induced by HIV-infection of macrophages: A potential mechanism for intercellular HIV trafficking. Cell. Immunol. 2009, 254, 142–148. [Google Scholar] [CrossRef]
- Wang, X.; Veruki, M.L.; Bukoreshtliev, N.V.; Hartveit, E.; Gerdes, H.H. Animal cells connected by nanotubes can be electrically coupled through interposed gap-junction channels. Proc. Natl. Acad. Sci. USA 2010, 107, 17194–17199. [Google Scholar] [CrossRef]
- Wang, Y.; Cui, J.; Sun, X.; Zhang, Y. Tunneling-nanotube development in astrocytes depends on p53 activation. Cell Death Differ. 2011, 18, 732–742. [Google Scholar] [CrossRef]
- Ady, J.W.; Desir, S.; Thayanithy, V.; Vogel, R.I.; Moreira, A.L.; Downey, R.J.; Fong, Y.; Manova-Todorova, K.; Moore, M.A.; Lou, E. Intercellular communication in malignant pleural mesothelioma: Properties of tunneling nanotubes. Front. Physiol. 2014, 5, 400. [Google Scholar] [CrossRef]
- Osswald, M.; Jung, E.; Sahm, F.; Solecki, G.; Venkataramani, V.; Blaes, J.; Weil, S.; Horstmann, H.; Wiestler, B.; Syed, M.; et al. Brain tumour cells interconnect to a functional and resistant network. Nature 2015, 528, 93–98. [Google Scholar] [CrossRef]
- Buszczak, M.; Inaba, M.; Yamashita, Y.M. Signaling by Cellular Protrusions: Keeping the Conversation Private. Trends Cell Biol. 2016, 26, 526–534. [Google Scholar] [CrossRef]
- Lou, E. Intercellular conduits in tumours: The new social network. Trends Cancer 2016, 2, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Malik, S.; Eugenin, E.A. Mechanisms of HIV Neuropathogenesis: Role of Cellular Communication Systems. Curr. HIV Res. 2016, 14, 400–411. [Google Scholar] [CrossRef] [PubMed]
- Osswald, M.; Solecki, G.; Wick, W.; Winkler, F. A malignant cellular network in gliomas: Potential clinical implications. Neuro-Oncology 2016, 18, 479–485. [Google Scholar] [CrossRef] [PubMed]
- Ariazi, J.; Benowitz, A.; De Biasi, V.; Den Boer, M.L.; Cherqui, S.; Cui, H.; Douillet, N.; Eugenin, E.A.; Favre, D.; Goodman, S.; et al. Tunneling Nanotubes and Gap Junctions-Their Role in Long-Range Intercellular Communication during Development, Health, and Disease Conditions. Front. Mol. Neurosci. 2017, 10, 333. [Google Scholar] [CrossRef]
- Jung, E.; Osswald, M.; Blaes, J.; Wiestler, B.; Sahm, F.; Schmenger, T.; Solecki, G.; Deumelandt, K.; Kurz, F.T.; Xie, R.; et al. Tweety-Homolog 1 Drives Brain Colonization of Gliomas. J. Neurosci. 2017, 37, 6837–6850. [Google Scholar] [CrossRef]
- Lou, E.; Gholami, S.; Romin, Y.; Thayanithy, V.; Fujisawa, S.; Desir, S.; Steer, C.J.; Subramanian, S.; Fong, Y.; Manova-Todorova, K.; et al. Imaging Tunneling Membrane Tubes Elucidates Cell Communication in Tumors. Trends Cancer 2017, 3, 678–685. [Google Scholar] [CrossRef]
- Thayanithy, V.; O’Hare, P.; Wong, P.; Zhao, X.; Steer, C.J.; Subramanian, S.; Lou, E. A transwell assay that excludes exosomes for assessment of tunneling nanotube-mediated intercellular communication. Cell Commun. Signal. 2017, 15, 46. [Google Scholar] [CrossRef]
- Weil, S.; Osswald, M.; Solecki, G.; Grosch, J.; Jung, E.; Lemke, D.; Ratliff, M.; Hanggi, D.; Wick, W.; Winkler, F. Tumor microtubes convey resistance to surgical lesions and chemotherapy in gliomas. Neuro-Oncology 2017, 19, 1316–1326. [Google Scholar] [CrossRef]
- Lou, E.; Zhai, E.; Sarkari, A.; Desir, S.; Wong, P.; Iizuka, Y.; Yang, J.; Subramanian, S.; McCarthy, J.; Bazzaro, M.; et al. Cellular and Molecular Networking within the Ecosystem of Cancer Cell Communication via Tunneling Nanotubes. Front. Cell Dev. Biol. 2018, 6, 95. [Google Scholar] [CrossRef]
- Valdebenito, S.; Lou, E.; Baldoni, J.; Okafo, G.; Eugenin, E. The Novel Roles of Connexin Channels and Tunneling Nanotubes in Cancer Pathogenesis. Int. J. Mol. Sci. 2018, 19, 1270. [Google Scholar] [CrossRef]
- Lou, E. A Ticket to Ride: The Implications of Direct Intercellular Communication via Tunneling Nanotubes in Peritoneal and Other Invasive Malignancies. Front. Oncol. 2020, 10, 559548. [Google Scholar] [CrossRef] [PubMed]
- Venkataramani, V.; Schneider, M.; Giordano, F.A.; Kuner, T.; Wick, W.; Herrlinger, U.; Winkler, F. Disconnecting multicellular networks in brain tumours. Nat. Rev. Cancer 2022, 22, 481–491. [Google Scholar] [CrossRef]
- Rustom, A.; Saffrich, R.; Markovic, I.; Walther, P.; Gerdes, H.H. Nanotubular highways for intercellular organelle transport. Science 2004, 303, 1007–1010. [Google Scholar] [CrossRef] [PubMed]
- Desir, S.; Wong, P.; Turbyville, T.; Chen, D.; Shetty, M.; Clark, C.; Zhai, E.; Romin, Y.; Manova-Todorova, K.; Starr, T.K.; et al. Intercellular Transfer of Oncogenic KRAS via Tunneling Nanotubes Introduces Intracellular Mutational Heterogeneity in Colon Cancer Cells. Cancers 2019, 11, 892. [Google Scholar] [CrossRef]
- Sartori-Rupp, A.; Cordero Cervantes, D.; Pepe, A.; Gousset, K.; Delage, E.; Corroyer-Dulmont, S.; Schmitt, C.; Krijnse-Locker, J.; Zurzolo, C. Correlative cryo-electron microscopy reveals the structure of TNTs in neuronal cells. Nat. Commun. 2019, 10, 342. [Google Scholar] [CrossRef] [PubMed]
- Antanaviciute, I.; Rysevaite, K.; Liutkevicius, V.; Marandykina, A.; Rimkute, L.; Sveikatiene, R.; Uloza, V.; Skeberdis, V.A. Long-Distance Communication between Laryngeal Carcinoma Cells. PLoS ONE 2014, 9, e99196. [Google Scholar] [CrossRef]
- Onfelt, B.; Nedvetzki, S.; Yanagi, K.; Davis, D.M. Cutting edge: Membrane nanotubes connect immune cells. J. Immunol. 2004, 173, 1511–1513. [Google Scholar] [CrossRef]
- Rudnicka, D.; Feldmann, J.; Porrot, F.; Wietgrefe, S.; Guadagnini, S.; Prevost, M.C.; Estaquier, J.; Haase, A.T.; Sol-Foulon, N.; Schwartz, O. Simultaneous cell-to-cell transmission of human immunodeficiency virus to multiple targets through polysynapses. J. Virol. 2009, 83, 6234–6246. [Google Scholar] [CrossRef] [PubMed]
- Naphade, S. Brief reports: Lysosomal cross-correction by hematopoietic stem cell-derived macrophages via tunneling nanotubes. Stem Cells 2015, 33, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Desir, S.; Dickson, E.L.; Vogel, R.I.; Thayanithy, V.; Wong, P.; Teoh, D.; Geller, M.A.; Steer, C.J.; Subramanian, S.; Lou, E. Tunneling nanotube formation is stimulated by hypoxia in ovarian cancer cells. Oncotarget 2016, 7, 43150–43161. [Google Scholar] [CrossRef] [PubMed]
- Omsland, M.; Bruserud, O.; Gjertsen, B.T.; Andresen, V. Tunneling nanotube (TNT) formation is downregulated by cytarabine and NF-kappaB inhibition in acute myeloid leukemia (AML). Oncotarget 2017, 8, 7946–7963. [Google Scholar] [CrossRef] [PubMed]
- Desir, S.; O’Hare, P.; Vogel, R.I.; Sperduto, W.; Sarkari, A.; Dickson, E.L.; Wong, P.; Nelson, A.C.; Fong, Y.; Steer, C.J.; et al. Chemotherapy-Induced Tunneling Nanotubes Mediate Intercellular Drug Efflux in Pancreatic Cancer. Sci. Rep. 2018, 8, 9484. [Google Scholar] [CrossRef]
- Lou, E.; Fujisawa, S.; Morozov, A.; Barlas, A.; Romin, Y.; Dogan, Y.; Gholami, S.; Moreira, A.L.; Manova-Todorova, K.; Moore, M.A. Tunneling nanotubes provide a unique conduit for intercellular transfer of cellular contents in human malignant pleural mesothelioma. PLoS ONE 2012, 7, e33093. [Google Scholar] [CrossRef] [PubMed]
- Azorin, D.D.; Winkler, F. Two routes of direct intercellular communication in brain cancer. Biochem. J. 2021, 478, 1283–1286. [Google Scholar] [CrossRef]
- Chinnery, H.R.; Pearlman, E.; McMenamin, P.G. Cutting edge: Membrane nanotubes in vivo: A feature of MHC class II+ cells in the mouse cornea. J. Immunol. 2008, 180, 5779–5783. [Google Scholar] [CrossRef]
- Hase, K.; Kimura, S.; Takatsu, H.; Ohmae, M.; Kawano, S.; Kitamura, H.; Ito, M.; Watarai, H.; Hazelett, C.C.; Yeaman, C.; et al. M-Sec promotes membrane nanotube formation by interacting with Ral and the exocyst complex. Nat. Cell Biol. 2009, 11, 1427–1432. [Google Scholar] [CrossRef]
- Islam, M.N.; Das, S.R.; Emin, M.T.; Wei, M.; Sun, L.; Westphalen, K.; Rowlands, D.J.; Quadri, S.K.; Bhattacharya, S.; Bhattacharya, J. Mitochondrial transfer from bone-marrow-derived stromal cells to pulmonary alveoli protects against acute lung injury. Nat. Med. 2012, 18, 759–765. [Google Scholar] [CrossRef]
- Pasquier, J.; Galas, L.; Boulange-Lecomte, C.; Rioult, D.; Bultelle, F.; Magal, P.; Webb, G.; Le Foll, F. Different modalities of intercellular membrane exchanges mediate cell-to-cell p-glycoprotein transfers in MCF-7 breast cancer cells. J. Biol. Chem. 2012, 287, 7374–7387. [Google Scholar] [CrossRef]
- Pasquier, J.; Guerrouahen, B.S.; Al Thawadi, H.; Ghiabi, P.; Maleki, M.; Abu-Kaoud, N.; Jacob, A.; Mirshahi, M.; Galas, L.; Rafii, S.; et al. Preferential transfer of mitochondria from endothelial to cancer cells through tunneling nanotubes modulates chemoresistance. J. Transl. Med. 2013, 11, 94. [Google Scholar] [CrossRef]
- Guo, L.; Zhang, Y.; Yang, Z.; Peng, H.; Wei, R.; Wang, C.; Feng, M. Tunneling Nanotubular Expressways for Ultrafast and Accurate M1 Macrophage Delivery of Anticancer Drugs to Metastatic Ovarian Carcinoma. ACS Nano 2019, 13, 1078–1096. [Google Scholar] [CrossRef]
- Hurtig, J.; Chiu, D.T.; Onfelt, B. Intercellular nanotubes: Insights from imaging studies and beyond. WIREs Nanomed. Nanobiotechnol. 2010, 2, 260–276. [Google Scholar] [CrossRef] [PubMed]
- Ady, J.; Thayanithy, V.; Mojica, K.; Wong, P.; Carson, J.; Rao, P.; Fong, Y.; Lou, E. Tunneling nanotubes: An alternate route for propagation of the bystander effect following oncolytic viral infection. Mol. Ther. Oncolytics. 2016, 3, 16029. [Google Scholar] [CrossRef]
- Lou, E.; Subramanian, S. Tunneling Nanotubes: Intercellular Conduits for Direct Cell-to-Cell Communication in Cancer; Springer: Berlin/Heidelberg, Germany; New York, NY, USA, 2016. [Google Scholar] [CrossRef]
- Dilsizoglu Senol, A.; Pepe, A.; Grudina, C.; Sassoon, N.; Reiko, U.; Bousset, L.; Melki, R.; Piel, J.; Gugger, M.; Zurzolo, C. Effect of tolytoxin on tunneling nanotube formation and function. Sci. Rep. 2019, 9, 5741. [Google Scholar] [CrossRef]
- Kolba, M.D.; Dudka, W.; Zareba-Koziol, M.; Kominek, A.; Ronchi, P.; Turos, L.; Chroscicki, P.; Wlodarczyk, J.; Schwab, Y.; Klejman, A.; et al. Tunneling nanotube-mediated intercellular vesicle and protein transfer in the stroma-provided imatinib resistance in chronic myeloid leukemia cells. Cell Death Dis. 2019, 10, 817. [Google Scholar] [CrossRef]
- Jana, A.; Ladner, K.; Lou, E.; Nain, A.S. Tunneling Nanotubes between Cells Migrating in ECM Mimicking Fibrous Environments. Cancers 2022, 14, 1989. [Google Scholar] [CrossRef]
- Vignjevic, D.; Yarar, D.; Welch, M.D.; Peloquin, J.; Svitkina, T.; Borisy, G.G. Formation of filopodia-like bundles in vitro from a dendritic network. J. Cell Biol. 2003, 160, 951–962. [Google Scholar] [CrossRef] [PubMed]
- Vignjevic, D.; Kojima, S.; Aratyn, Y.; Danciu, O.; Svitkina, T.; Borisy, G.G. Role of fascin in filopodial protrusion. J. Cell Biol. 2006, 174, 863–875. [Google Scholar] [CrossRef] [PubMed]
- Thayanithy, V.; Babatunde, V.; Dickson, E.L.; Wong, P.; Oh, S.; Ke, X.; Barlas, A.; Fujisawa, S.; Romin, Y.; Moreira, A.L.; et al. Tumor exosomes induce tunneling nanotubes in lipid raft-enriched regions of human mesothelioma cells. Exp. Cell Res. 2014, 323, 178–188. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Veranic, P.; Lokar, M.; Schutz, G.J.; Weghuber, J.; Wieser, S.; Hagerstrand, H.; Kralj-Iglic, V.; Iglic, A. Different types of cell-to-cell connections mediated by nanotubular structures. Biophys. J. 2008, 95, 4416–4425. [Google Scholar] [CrossRef]
- Peng, T.; Thorn, K.; Schroeder, T.; Wang, L.; Theis, F.J.; Marr, C.; Navab, N. A BaSiC tool for background and shading correction of optical microscopy images. Nat. Commun. 2017, 8, 14836. [Google Scholar] [CrossRef] [PubMed]
- Ingaramo, M.; York, A.G.; Hoogendoorn, E.; Postma, M.; Shroff, H.; Patterson, G.H. Richardson-Lucy deconvolution as a general tool for combining images with complementary strengths. ChemPhysChem 2014, 15, 794–800. [Google Scholar] [CrossRef] [PubMed]
- Laasmaa, M.; Vendelin, M.; Peterson, P. Application of regularized Richardson-Lucy algorithm for deconvolution of confocal microscopy images. J. Microsc. 2011, 243, 124–140. [Google Scholar] [CrossRef] [PubMed]
- Hodneland, E.; Lundervold, A.; Gurke, S.; Tai, X.C.; Rustom, A.; Gerdes, H.H. Automated detection of tunneling nanotubes in 3D images. Cytom. A 2006, 69, 961–972. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Kai, L.; Li, F.-F. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Lee, C.-Y.; Xie, S.; Gallagher, P.; Zhang, Z.; Tu, Z. Deeply-Supervised Nets. In Proceedings of the Eighteenth International Conference on Artificial Intelligence and Statistics, San Diego, CA, USA, 10–12 May 2015; pp. 562–570. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Cohen, E.; Uhlmann, V. aura-net: Robust segmentation of phase-contrast microscopy images with few annotations. In Proceedings of the 2021 IEEE 18th International Symposium on Biomedical Imaging (ISBI), Nice, France, 13–16 April 2021; pp. 640–644. [Google Scholar]
- Rectified Linear Units. Available online: https://paperswithcode.com/method/relu (accessed on 20 August 2022).
- Max Pooling. Available online: https://paperswithcode.com/method/max-pooling (accessed on 20 August 2022).
- Yosinski, J.; Clune, J.; Bengio, Y.; Lipson, H. How transferable are features in deep neural networks? In Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2014; Volume 27. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2017; Volume 30. [Google Scholar]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.-A. V-net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 fourth international conference on 3D vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Chen, X.; Williams, B.M.; Vallabhaneni, S.R.; Czanner, G.; Williams, R.; Zheng, Y. Learning Active Contour Models for Medical Image Segmentation. In Proceedings of the 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019; pp. 11624–11632. [Google Scholar]
- Stringer, C.; Pachitariu, M. Cellpose 2.0: How to train your own model. bioRxiv 2022. [Google Scholar] [CrossRef]
- Contours: Getting Started. Available online: https://docs.opencv.org/4.x/d4/d73/tutorial_py_contours_begin.html (accessed on 20 August 2022).
- Meijering, E. Cell Segmentation: 50 Years Down the Road [Life Sciences]. IEEE Signal Process. Mag. 2012, 29, 140–145. [Google Scholar] [CrossRef]
- Vembadi, A.; Menachery, A.; Qasaimeh, M.A. Cell Cytometry: Review and Perspective on Biotechnological Advances. Front. Bioeng. Biotechnol. 2019, 7, 147. [Google Scholar] [CrossRef]
- Zhao, X.; May, A.; Lou, E.; Subramanian, S. Genotypic and phenotypic signatures to predict immune checkpoint blockade therapy response in patients with colorectal cancer. Transl. Res. 2018, 196, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Vignjevic, D.; Peloquin, J.; Borisy, G.G. In vitro assembly of filopodia-like bundles. Methods Enzymol. 2006, 406, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Bailey, K.M.; Airik, M.; Krook, M.A.; Pedersen, E.A.; Lawlor, E.R. Micro-Environmental Stress Induces Src-Dependent Activation of Invadopodia and Cell Migration in Ewing Sarcoma. Neoplasia 2016, 18, 480–488. [Google Scholar] [CrossRef] [PubMed]
- Schoumacher, M.; Goldman, R.D.; Louvard, D.; Vignjevic, D.M. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J. Cell Biol. 2010, 189, 541–556. [Google Scholar] [CrossRef] [PubMed]
- Perestrelo, T.; Chen, W.; Correia, M.; Le, C.; Pereira, S.; Rodrigues, A.S.; Sousa, M.I.; Ramalho-Santos, J.; Wirtz, D. Pluri-IQ: Quantification of Embryonic Stem Cell Pluripotency through an Image-Based Analysis Software. Stem Cell Rep. 2017, 9, 697–709. [Google Scholar] [CrossRef]
- Nilufar, S.; Morrow, A.A.; Lee, J.M.; Perkins, T.J. FiloDetect: Automatic detection of filopodia from fluorescence microscopy images. BMC Syst. Biol. 2013, 7, 66. [Google Scholar] [CrossRef] [PubMed]
- Jacquemet, G.; Paatero, I.; Carisey, A.F.; Padzik, A.; Orange, J.S.; Hamidi, H.; Ivaska, J. FiloQuant reveals increased filopodia density during breast cancer progression. J. Cell Biol. 2017, 216, 3387–3403. [Google Scholar] [CrossRef] [PubMed]
- Tsygankov, D.; Bilancia, C.G.; Vitriol, E.A.; Hahn, K.M.; Peifer, M.; Elston, T.C. CellGeo: A computational platform for the analysis of shape changes in cells with complex geometries. J. Cell Biol. 2014, 204, 443–460. [Google Scholar] [CrossRef] [PubMed]
- Barry, D.J.; Durkin, C.H.; Abella, J.V.; Way, M. Open source software for quantification of cell migration, protrusions, and fluorescence intensities. J. Cell Biol. 2015, 209, 163–180. [Google Scholar] [CrossRef]
- Seo, J.H.; Yang, S.; Kang, M.S.; Her, N.G.; Nam, D.H.; Choi, J.H.; Kim, M.H. Automated stitching of microscope images of fluorescence in cells with minimal overlap. Micron 2019, 126, 102718. [Google Scholar] [CrossRef]
| Image Set | No. of TNTs (True *) | PPV (Precision) | Sensitivity (Recall) | No. of FPs | No. of Human Expert-Corrected FPs | f-1 Score |
|---|---|---|---|---|---|---|
| Training 1 (stitched image MSTO2) | 43 | 0.67 | 0.70 | 14 | 0 | 0.68 |
| Training 2 (stitched image MSTO3) | 18 | 0.38 | 0.61 | 17 | 1 | 0.47 |
| Training 3 (stitched image MSTO4) | 33 | 0.52 | 0.42 | 13 | 1 | 0.47 |
| Test 1 (stitched image MSTO5) | 42 | 0.41 | 0.26 | 16 | 2 | 0.32 |
| Image Set | No. of TNTs (True *) | No. of TNTs (Predicted **) | No. of Cells (from Cellpose) | TCR × 100 (True *) | TCR × 100 (Predicted **) |
|---|---|---|---|---|---|
| Training 1 (stitched image MSTO2) | 43 | 45 | 897 | 4.79 | 5.02 |
| Training 2 (stitched image MSTO3) | 18 | 29 | 777 | 2.32 | 3.73 |
| Training 3 (stitched image MSTO4) | 33 | 27 | 754 | 4.38 | 3.58 |
| Test 1 (stitched image MSTO5) | 42 | 27 | 897 | 4.68 | 3.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ceran, Y.; Ergüder, H.; Ladner, K.; Korenfeld, S.; Deniz, K.; Padmanabhan, S.; Wong, P.; Baday, M.; Pengo, T.; Lou, E.; et al. TNTdetect.AI: A Deep Learning Model for Automated Detection and Counting of Tunneling Nanotubes in Microscopy Images. Cancers 2022, 14, 4958. https://doi.org/10.3390/cancers14194958
Ceran Y, Ergüder H, Ladner K, Korenfeld S, Deniz K, Padmanabhan S, Wong P, Baday M, Pengo T, Lou E, et al. TNTdetect.AI: A Deep Learning Model for Automated Detection and Counting of Tunneling Nanotubes in Microscopy Images. Cancers. 2022; 14(19):4958. https://doi.org/10.3390/cancers14194958
Chicago/Turabian StyleCeran, Yasin, Hamza Ergüder, Katherine Ladner, Sophie Korenfeld, Karina Deniz, Sanyukta Padmanabhan, Phillip Wong, Murat Baday, Thomas Pengo, Emil Lou, and et al. 2022. "TNTdetect.AI: A Deep Learning Model for Automated Detection and Counting of Tunneling Nanotubes in Microscopy Images" Cancers 14, no. 19: 4958. https://doi.org/10.3390/cancers14194958
APA StyleCeran, Y., Ergüder, H., Ladner, K., Korenfeld, S., Deniz, K., Padmanabhan, S., Wong, P., Baday, M., Pengo, T., Lou, E., & Patel, C. B. (2022). TNTdetect.AI: A Deep Learning Model for Automated Detection and Counting of Tunneling Nanotubes in Microscopy Images. Cancers, 14(19), 4958. https://doi.org/10.3390/cancers14194958






