CircKIF4A Is a Prognostic Factor and Modulator of Natural Killer/T-Cell Lymphoma Progression
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Cell Cultures and Transfection
2.2. Real-Time Quantitative PCR Analysis (qPCR)
2.3. RNase R Resistant Assay
2.4. Actinomycin D Digestion
2.5. Luciferase Reporter Assay
2.6. RNA Immunoprecipitation (RIP)
2.7. Glucose Uptake and Lactate Production
2.8. Immunohistochemistry (IHC)
2.9. Patients Samples and Ethical Standards
2.10. Statistical Analysis
3. Results
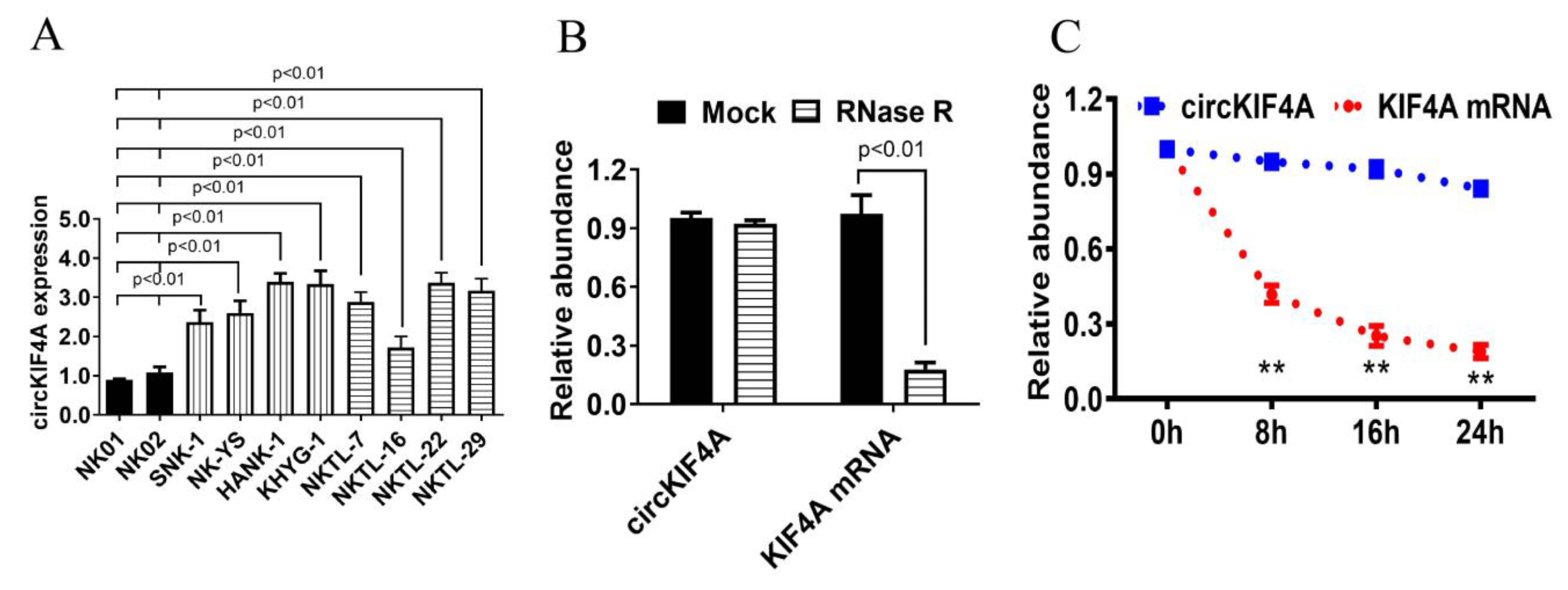
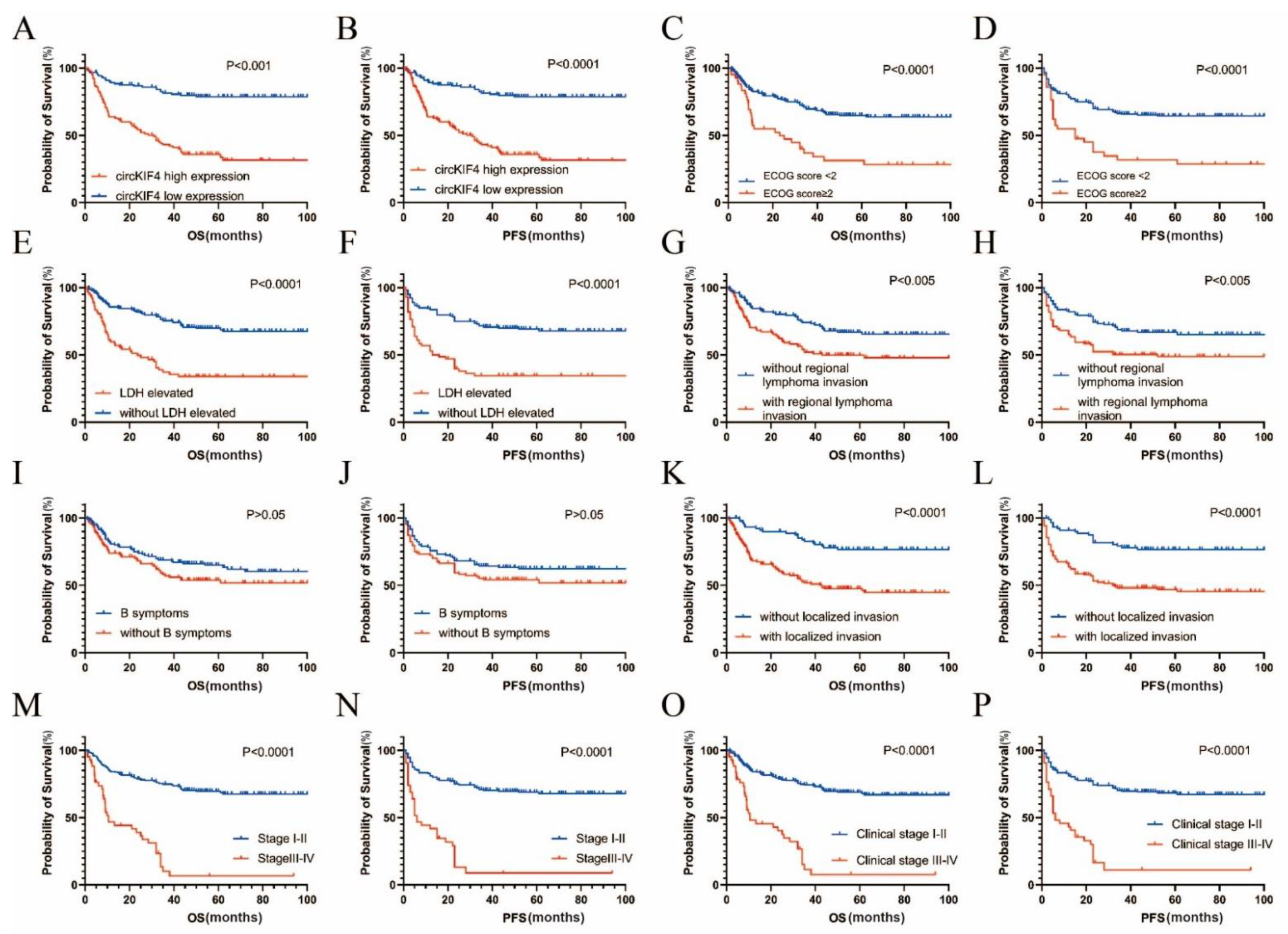
3.1. CircKIF4A Was Upregulated in NKTL and Correlated with Poor Survival
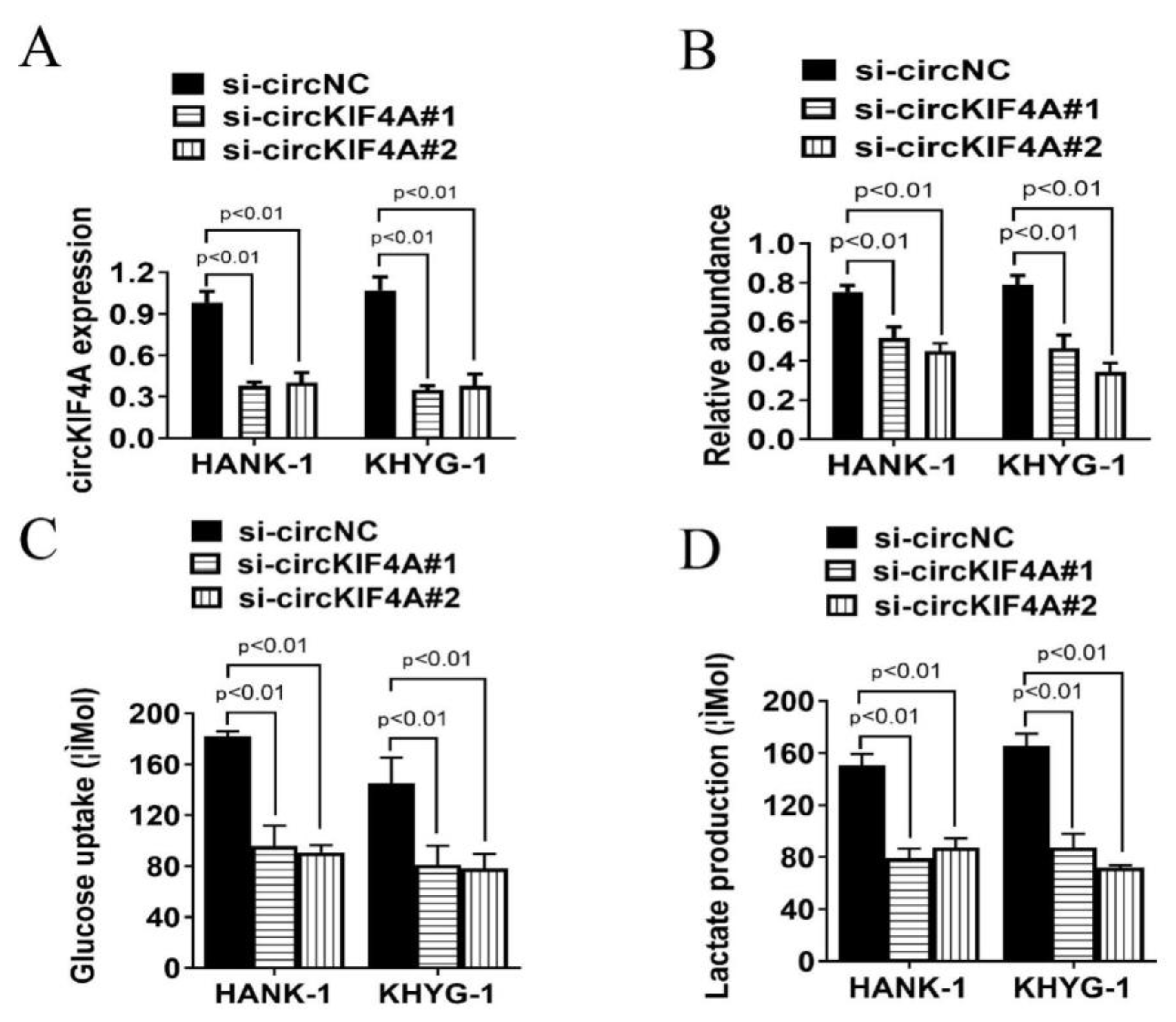
3.2. CircKIF4A Silencing Inhibits Glucose Uptake and Lactate Production of NKTL Cells
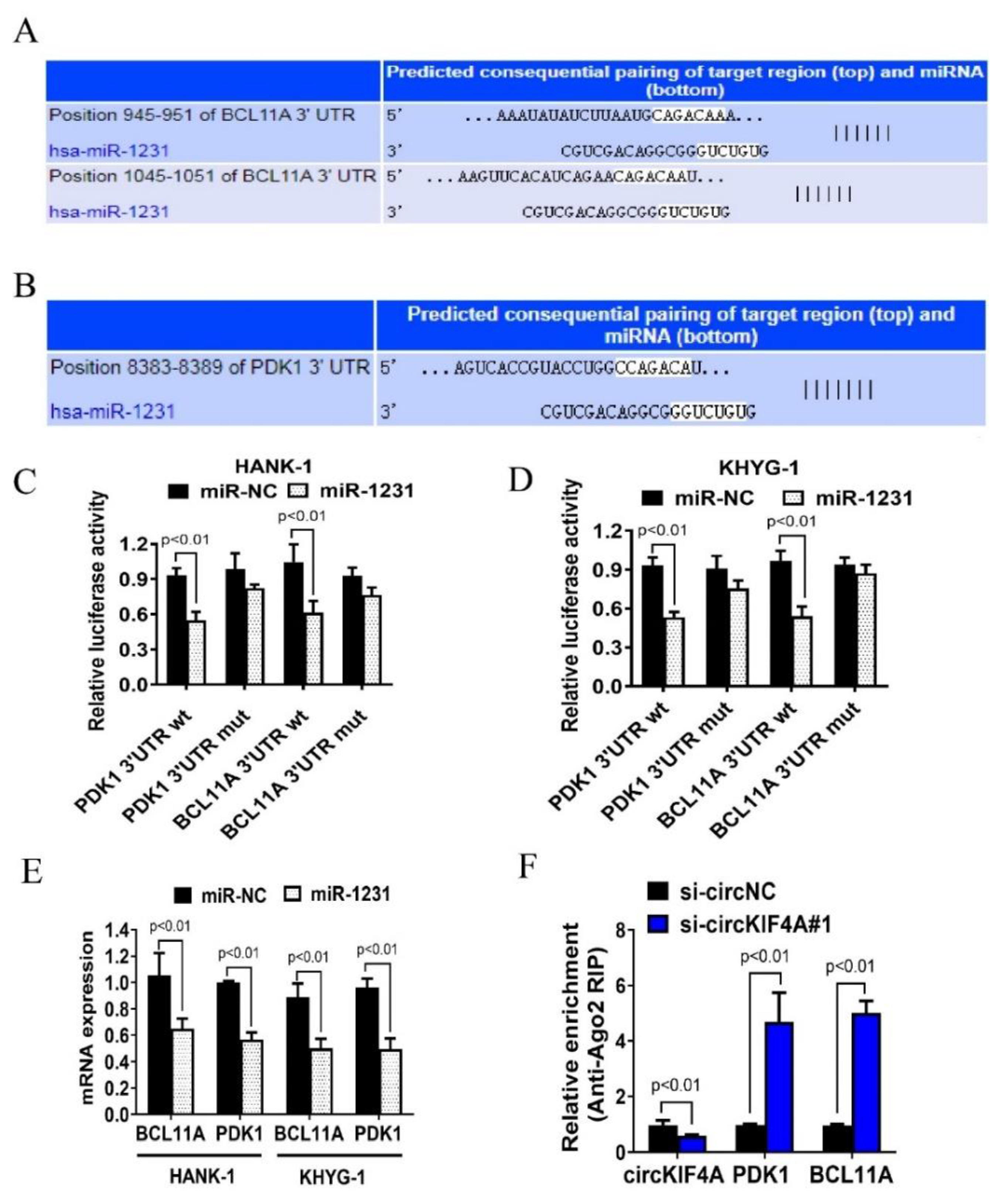
3.3. CircKIF4A Functions as a Sponge for miR-1231 in NKTL
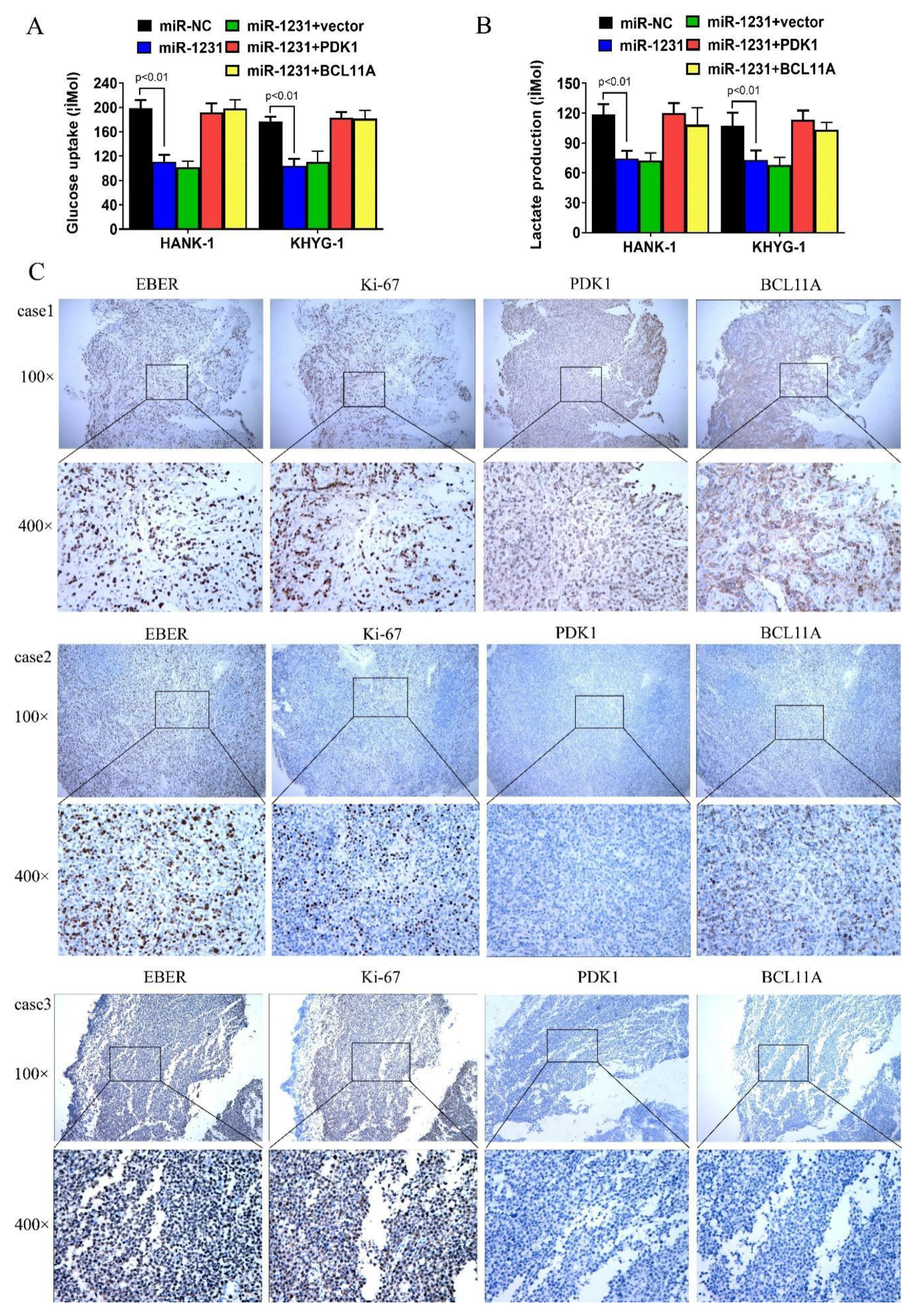
3.4. CircKIF4A Regulates NKTL through BCL11A and PDK1
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Xue, W.; Zhang, M. Updating targets for natural killer/T-cell lymphoma immunotherapy. Cancer Biol. Med. 2021, 18, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Fu, B.B.; Gale, R.P.; Liang, Y. NK-/T-cell lymphomas. Leukemia 2021, 35, 2460–2468. [Google Scholar] [CrossRef] [PubMed]
- Tse, E.; Kwong, Y.L. NK/T-cell lymphomas. Best Pract. Res. Clin. Haematol. 2019, 32, 253–261. [Google Scholar] [CrossRef]
- Chan, J.Y.; Lim, J.Q.; Ong, C.K. Towards Next Generation Biomarkers in Natural Killer/T-Cell Lymphoma. Life 2021, 11, 838. [Google Scholar] [CrossRef]
- Zhao, X.; Cai, Y.; Xu, J. Circular RNAs: Biogenesis, Mechanism, and Function in Human Cancers. Int. J. Mol. Sci. 2019, 20, 3926. [Google Scholar] [CrossRef]
- Zang, J.; Lu, D.; Xu, A. The interaction of circRNAs and RNA binding proteins: An important part of circRNA maintenance and function. J. Neurosci. Res. 2018, 98, 87–97. [Google Scholar] [CrossRef]
- Cen, Y.; Zhu, T.; Zhang, Y.; Zhao, L.; Zhu, J.; Wang, L.; Xu, J.; Ding, T.; Xie, X.; Wang, X.; et al. hsa_circ_0005358 suppresses cervical cancer metastasis by interacting with PTBP1 protein to destabilize CDCP1 mRNA. Mol. Ther. Nucleic Acids 2021, 27, 227–240. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zheng, Z.; Xie, Y.; Zhong, Q.; Shangguan, W.; Zhang, Y.; Zhu, D.; Xie, W. Circular RNA circPPP6R3 upregulates CD44 to promote the progression of clear cell renal cell carcinoma via sponging miR-1238-3p. Cell Death Dis. 2021, 13, 22. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, L.; Wu, P.; Li, X.; Tang, Y.; Ou, X.; Zhang, Y.; Xiao, X.; Wang, J.; Tang, H. The circROBO1/KLF5/FUS feedback loop regulates the liver metastasis of breast cancer by inhibiting the selective autophagy of afadin. Mol. Cancer 2022, 21, 29. [Google Scholar] [CrossRef]
- Hu, G.; Yan, Z.; Zhang, C.; Cheng, M.; Yan, Y.; Wang, Y.; Deng, L.; Lu, Q.; Luo, S. FOXM1 promotes hepatocellular carcinoma progression by regulating KIF4A expression. J. Exp. Clin. Cancer Res. 2019, 38, 188. [Google Scholar] [CrossRef]
- Cao, Q.; Song, Z.; Ruan, H.; Wang, C.; Yang, X.; Bao, L.; Wang, K.; Cheng, G.; Xu, T.; Xiao, W.; et al. Targeting the KIF4A/AR Axis to Reverse Endocrine Therapy Resistance in Castration-resistant Prostate Cancer. Clin. Cancer Res. 2020, 26, 1516–1528. [Google Scholar] [CrossRef] [PubMed]
- Mazumdar, M.; Sundareshan, S.; Misteli, T. Human chromokinesin KIF4A functions in chromosome condensation and segregation. J. Cell Biol. 2004, 166, 613–620. [Google Scholar] [CrossRef] [PubMed]
- Sheng, S.; Hu, Y.; Yu, F.; Tong, W.; Wang, S.; Cai, Y.; Zhu, J. circKIF4A sponges miR-127 to promote ovarian cancer progression. Aging 2020, 12, 17921–17929. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Huang, X.; Wang, J.; Yang, L.; Kong, Y.; Gao, G.; Zhang, L.; Chen, Z.-S.; Xie, X. circKIF4A acts as a prognostic factor and mediator to regulate the progression of triple-negative breast cancer. Mol. Cancer 2019, 18, 23. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, Y.; Liu, X.; He, X.; Zhang, L.-L.; Wu, G.; Qu, B.-L.; Qian, L.-T.; Hou, X.-R.; Zhang, F.-Q.; et al. Progression-free survival at 24 months and subsequent survival of patients with extranodal NK/T-cell lymphoma: A China Lymphoma Collaborative Group (CLCG) study. Leukemia 2020, 35, 1671–1682. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.Z.; Wang, Y. The factors associated with the early diagnosis of nasal NK/T-cell lymphoma with prominent ocu-lar symptoms and general nasal NKTL. Am. J. Otolaryngol. 2019, 40, 353–357. [Google Scholar] [CrossRef]
- Huang, H.; Zhu, J.; Yao, M.; Kim, T.M.; Yoon, D.H.; Cho, S.-G.; Eom, H.S.; Lim, S.T.; Yeh, S.-P.; Song, Y.; et al. Daratumumab monotherapy for patients with relapsed or refractory natural killer/T-cell lymphoma, nasal type: An open-label, single-arm, multicenter, phase 2 study. J. Hematol. Oncol. 2021, 14, 25. [Google Scholar] [CrossRef]
- Haverkos, B.M.; Pan, Z.; Gru, A.A.; Freud, A.G.; Rabinovitch, R.; Xu-Welliver, M.; Otto, B.; Barrionuevo, C.; Baiocchi, R.A.; Rochford, R.; et al. Extranodal NK/T Cell Lymphoma, Nasal Type (ENKTL-NT): An Update on Epidemiology, Clinical Presentation, and Natural History in North American and European Cases. Curr. Hematol. Malign-Rep. 2016, 11, 514–527. [Google Scholar] [CrossRef]
- He, R.; Liu, P.; Xie, X.; Zhou, Y.; Liao, Q.; Xiong, W.; Li, X.; Li, G.; Zeng, Z.; Tang, H. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J. Exp. Clin. Cancer Res. 2017, 36, 145. [Google Scholar] [CrossRef]
- Bazhabayi, M.; Qiu, X.; Li, X.; Yang, A.; Wen, W.; Zhang, X.; Xiao, X.; He, R.; Liu, P. CircGFRA1 facilitates the malignant progression of HER-2-positive breast cancer via acting as a sponge of miR-1228 and enhancing AIFM2 expression. J. Cell. Mol. Med. 2021, 25, 10248–10256. [Google Scholar] [CrossRef]
- Huo, L.-W.; Wang, Y.-F.; Bai, X.-B.; Zheng, H.-L.; Wang, M.-D. circKIF4A promotes tumorogenesis of glioma by targeting miR-139-3p to activate Wnt5a signaling. Mol. Med. 2020, 26, 29. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Fu, J.; Chen, Y.; Li, Y.; Ning, L.; Huang, D.; Yan, S.; Zhang, Q. Circular RNA circKIF4A facilitates the malignant progression and suppresses ferroptosis by sponging miR-1231 and upregulating GPX4 in papillary thyroid cancer. Aging 2021, 13, 16500–16512. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Q.; Guo, B.; Feng, J.; Zhao, D. miR-1231 Is Downregulated in Prostate Cancer with Prognostic and Functional Implications. Oncol. Res. Treat. 2019, 43, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Xie, J.R.; Jiang, Y.Y.; Xu, W.; Tao, J.Z. MiR-1231 correlates tumor prognosis and inhibits cell growth in ovarian cancer. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 8308–8313. [Google Scholar]
- Gagliardi, P.A.; di Blasio, L.; Primo, L. PDK1: A signaling hub for cell migration and tumor invasion. Biochim. Biophys. Acta 2015, 1856, 178–188. [Google Scholar] [CrossRef]
- Peng, F.; Wang, J.-H.; Fan, W.-J.; Meng, Y.-T.; Li, M.-M.; Li, T.-T.; Cui, B.; Wang, H.-F.; Zhao, Y.; An, F.; et al. Glycolysis gatekeeper PDK1 reprograms breast cancer stem cells under hypoxia. Oncogene 2017, 37, 1062–1074. [Google Scholar] [CrossRef]
- Wang, J.; Liu, F.; Ao, P.; Li, X.; Zheng, H.; Wu, D.; Zhang, B.; She, J.; Yuan, J.; Wu, X. Correlation of PDK1 expression with clinicopathologic features and prognosis of hepatocellular carcino-ma. Onco. Targets Ther. 2016, 9, 5597–5602. [Google Scholar]
- McFate, T.; Mohyeldin, A.; Lu, H.; Thakar, J.; Henriques, J.; Halim, N.D.; Wu, H.; Schell, M.J.; Tsang, T.M.; Teahan, O.; et al. Pyruvate Dehydrogenase Complex Activity Controls Metabolic and Malignant Phenotype in Cancer Cells. J. Biol. Chem. 2008, 283, 22700–22708. [Google Scholar] [CrossRef]
- Kato, M.; Li, J.; Chuang, J.L.; Chuang, D.T. Distinct Structural Mechanisms for Inhibition of Pyruvate Dehydrogenase Kinase Isoforms by AZD7545, Dichloroacetate, and Radicicol. Structure 2007, 15, 992–1004. [Google Scholar] [CrossRef]
- Hitosugi, T.; Fan, J.; Chung, T.-W.; Lythgoe, K.; Wang, X.; Xie, J.; Ge, Q.; Gu, T.-L.; Polakiewicz, R.D.; Roesel, J.L.; et al. Tyrosine Phosphorylation of Mitochondrial Pyruvate Dehydrogenase Kinase 1 Is Important for Cancer Metabolism. Mol. Cell 2011, 44, 864–877. [Google Scholar] [CrossRef]
- Zhou, J.; Zhou, L.; Zhang, D.; Tang, W.-J.; Tang, D.; Shi, X.-L.; Yang, Y.; Zhou, L.; Liu, F.; Yu, Y.; et al. BCL11A Promotes the Progression of Laryngeal Squamous Cell Carcinoma. Front. Oncol. 2020, 10, 375. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Xie, X.; Ye, Y.; Wang, L.; Che, F. BCL11A: A potential diagnostic biomarker and therapeutic target in human diseases. Biosci. Rep. 2019, 39, BSR20190604. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Li, C.; Feng, W.; Yue, J.; Song, J.; Peng, A.; Wang, H. BCL11A Is Oncogenic and Predicts Poor Outcomes in Natural Killer/T-Cell Lymphoma. Front. Pharmacol. 2020, 11, 820. [Google Scholar] [CrossRef] [PubMed]

| Variables | Hazard Ratio | Std. Err. | p | [95% Conf. Interval] | |
|---|---|---|---|---|---|
| Gender (male vs. female) | 0.752 | 0.231 | 0.219 | 0.478 | 1.184 |
| Age (<60 vs. ≥60) | 1.333 | 0.262 | 0.274 | 0.797 | 2.229 |
| clinical stage (I–II vs. III–IV) | 5.575 | 0.223 | <0.001 * | 3.599 | 8.637 |
| ECOG score (<2 vs. ≥2) | 3.960 | 0.211 | <0.001 * | 2.618 | 5.991 |
| circKIF4 (high expression vs. low expression) | 0.606 | 0.208 | 0.016 * | 0.403 | 0.912 |
| LDH (<60U/L vs ≥60U/L) | 2.962 | 0.210 | <0.001 * | 1.964 | 4.466 |
| B symptoms (positive vs. negative) | 1.379 | 0.210 | 0.122 | 0.918 | 2.073 |
| regional lymph nodes invasion (negative vs. positive) | 1.962 | 0.245 | 0.001 * | 1.299 | 2.964 |
| Multivariate analysis | |||||
| clinical stage (I–II vs. III–IV) | 1.747 | 0.290 | 0.055 | 0.989 | 3.084 |
| ECOG score (<2 vs. ≥2) | 1.166 | 0.296 | 0.603 | 0.654 | 2.081 |
| circKIF4 (high expression vs. low expression) | 0.500 | 0.223 | 0.002 * | 0.323 | 0.774 |
| LDH (<60U/L vs ≥60U/L) | 0.741 | 0.232 | 0.196 | 0.471 | 1.167 |
| regional lymph nodes invasion (negative vs. positive) | 0.000 | 52.764 | 0.803 | 0.001 | 1.577 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, R.; Wen, W.; Fu, B.; Zhu, R.; Chen, G.; Bai, S.; Cao, X.; Wang, H. CircKIF4A Is a Prognostic Factor and Modulator of Natural Killer/T-Cell Lymphoma Progression. Cancers 2022, 14, 4950. https://doi.org/10.3390/cancers14194950
He R, Wen W, Fu B, Zhu R, Chen G, Bai S, Cao X, Wang H. CircKIF4A Is a Prognostic Factor and Modulator of Natural Killer/T-Cell Lymphoma Progression. Cancers. 2022; 14(19):4950. https://doi.org/10.3390/cancers14194950
Chicago/Turabian StyleHe, Rongfang, Wei Wen, Bibo Fu, Renjie Zhu, Guanjun Chen, Shenrui Bai, Xi Cao, and Hua Wang. 2022. "CircKIF4A Is a Prognostic Factor and Modulator of Natural Killer/T-Cell Lymphoma Progression" Cancers 14, no. 19: 4950. https://doi.org/10.3390/cancers14194950
APA StyleHe, R., Wen, W., Fu, B., Zhu, R., Chen, G., Bai, S., Cao, X., & Wang, H. (2022). CircKIF4A Is a Prognostic Factor and Modulator of Natural Killer/T-Cell Lymphoma Progression. Cancers, 14(19), 4950. https://doi.org/10.3390/cancers14194950







