SF3B1 Mutations in Hematological Malignancies
Simple Summary
Abstract
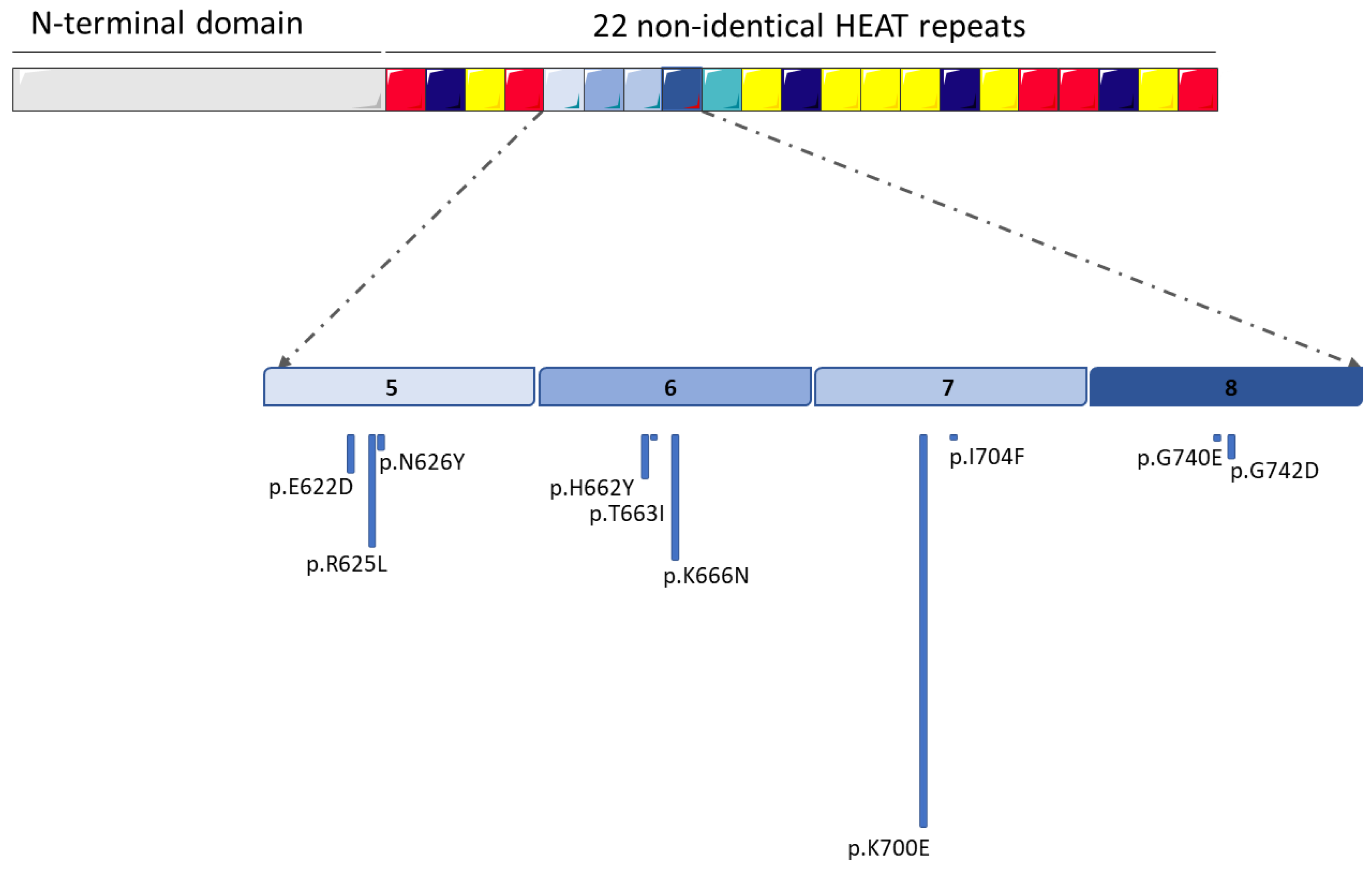
1. Introduction
2. SF3B1 in Myelodysplastic Syndromes
3. SF3B1 in Myeloproliferative Neoplasms
4. SF3B1 in Myelodysplastic/Myeloproliferative Neoplasms
5. SF3B1 in Chronic Myelomonocytic Leukemia
6. SF3B1 in Acute Myeloid Leukemia
7. SF3B1 in Chronic Lymphocytic Leukemia
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Wang, Z.; Burge, C.B. Splicing regulation: From a parts list of regulatory elements to an integrated splicing code. RNA 2008, 14, 802–813. [Google Scholar] [CrossRef] [PubMed]
- Yoshimi, A.; Abdel-Wahab, O. Molecular Pathways: Understanding and Targeting Mutant Spliceosomal Proteins. Clin. Cancer Res. 2017, 23, 336–341. [Google Scholar] [CrossRef]
- Visconte, V.; Makishima, H.; Maciejewski, J.P.; Tiu, R.V. Emerging roles of the spliceosomal machinery in myelodysplastic syndromes and other hematological disorders. Leukemia 2012, 26, 2447–2454. [Google Scholar] [CrossRef]
- Fujita, K.I.; Ishizuka, T.; Mitsukawa, M.; Kurata, M.; Masuda, S. Regulating Divergent Transcriptomes through mRNA Splicing and Its Modulation Using Various Small Compounds. Int. J. Mol. Sci. 2020, 21, 2026. [Google Scholar] [CrossRef]
- Yoshimoto, R.; Kaida, D.; Furuno, M.; Burroughs, A.M.; Noma, S.; Suzuki, H.; Kawamura, Y.; Hayashizaki, Y.; Mayeda, A.; Yoshida, M. Global analysis of pre-mRNA subcellular localization following splicing inhibition by spliceostatin A. RNA 2017, 23, 47–57. [Google Scholar] [CrossRef]
- Alsafadi, S.; Houy, A.; Battistella, A.; Popova, T.; Wassef, M.; Henry, E.; Tirode, F.; Constantinou, A.; Piperno-Neumann, S.; Roman-Roman, S.; et al. Cancer-associated SF3B1 mutations affect alternative splicing by promoting alternative branchpoint usage. Nat. Commun. 2016, 7, 10615. [Google Scholar] [CrossRef] [PubMed]
- Malcovati, L.; Papaemmanuil, E.; Bowen, D.T.; Boultwood, J.; Della Porta, M.G.; Pascutto, C.; Travaglino, E.; Groves, M.J.; Godfrey, A.L.; Ambaglio, I.; et al. Clinical significance of SF3B1 mutations in myelodysplastic syndromes and myelodysplastic/myeloproliferative neoplasms. Blood 2011, 118, 6239–6246. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Gong, Q.; Wang, Y.; Li, M.; Wang, L.; Ding, H.; Li, P. The biological function and clinical significance of SF3B1 mutations in cancer. Biomark. Res. 2020, 8, 38. [Google Scholar] [CrossRef]
- Nickless, A.; Bailis, J.M.; You, Z. Control of gene expression through the nonsense-mediated RNA decay pathway. Cell Biosci. 2017, 7, 26. [Google Scholar] [CrossRef]
- Bergot, T.; Lippert, E.; Douet-Guilbert, N.; Commet, S.; Corcos, L.; Bernard, D.G. Human Cancer-Associated Mutations of SF3B1 Lead to a Splicing Modification of Its Own RNA. Cancers 2020, 12, 652. [Google Scholar] [CrossRef]
- Obeng, E.A.; Chappell, R.J.; Seiler, M.; Chen, M.C.; Campagna, D.R.; Schmidt, P.J.; Schneider, R.K.; Lord, A.M.; Wang, L.; Gambe, R.G.; et al. Physiologic Expression of Sf3b1(K700E) Causes Impaired Erythropoiesis, Aberrant Splicing, and Sensitivity to Therapeutic Spliceosome Modulation. Cancer Cell 2016, 30, 404–417. [Google Scholar] [CrossRef] [PubMed]
- Mupo, A.; Seiler, M.; Sathiaseelan, V.; Pance, A.; Yang, Y.; Agrawal, A.A.; Iorio, F.; Bautista, R.; Pacharne, S.; Tzelepis, K.; et al. Hemopoietic-specific Sf3b1-K700E knock-in mice display the splicing defect seen in human MDS but develop anemia without ring sideroblasts. Leukemia 2017, 31, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Mizuta, S.; Yamane, N.; Komai, T.; Koba, Y.; Ukyo, N.; Tamekane, A.; Watanabe, M. Evaluation of SF3B1 Mutation Screening by High-Resolution Melting Analysis and its Clinical Utility for Myelodysplastic Syndrome with Ring Sideroblasts at the Point of Diagnosis. Lab. Med. 2019, 50, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Petiti, J.; Itri, F.; Signorino, E.; Frolli, A.; Fava, C.; Armenio, M.; Marini, S.; Giugliano, E.; Lo Iacono, M.; Saglio, G.; et al. Detection of SF3B1 p.Lys700Glu Mutation by PNA-PCR Clamping in Myelodysplastic Syndromes and Myeloproliferative Neoplasms. J. Clin. Med. 2022, 11, 1267. [Google Scholar] [CrossRef] [PubMed]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef] [PubMed]
- Malcovati, L.; Stevenson, K.; Papaemmanuil, E.; Neuberg, D.; Bejar, R.; Boultwood, J.; Bowen, D.T.; Campbell, P.J.; Ebert, B.L.; Fenaux, P.; et al. SF3B1-mutant MDS as a distinct disease subtype: A proposal from the International Working Group for the Prognosis of MDS. Blood 2020, 136, 157–170. [Google Scholar] [CrossRef]
- Khoury, J.D.; Solary, E.; Abla, O.; Akkari, Y.; Alaggio, R.; Apperley, J.F.; Bejar, R.; Berti, E.; Busque, L.; Chan, J.K.C.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia 2022, 36, 1703–1719. [Google Scholar] [CrossRef] [PubMed]
- Clough, C.A.; Pangallo, J.; Sarchi, M.; Ilagan, J.O.; North, K.; Bergantinos, R.; Stolla, M.C.; Naru, J.; Nugent, P.; Kim, E.; et al. Coordinated missplicing of TMEM14C and ABCB7 causes ring sideroblast formation in SF3B1-mutant myelodysplastic syndrome. Blood 2022, 139, 2038–2049. [Google Scholar] [CrossRef] [PubMed]
- Gerstung, M.; Pellagatti, A.; Malcovati, L.; Giagounidis, A.; Porta, M.G.; Jadersten, M.; Dolatshad, H.; Verma, A.; Cross, N.C.; Vyas, P.; et al. Combining gene mutation with gene expression data improves outcome prediction in myelodysplastic syndromes. Nat. Commun. 2015, 6, 5901. [Google Scholar] [CrossRef]
- Dolatshad, H.; Pellagatti, A.; Liberante, F.G.; Llorian, M.; Repapi, E.; Steeples, V.; Roy, S.; Scifo, L.; Armstrong, R.N.; Shaw, J.; et al. Cryptic splicing events in the iron transporter ABCB7 and other key target genes in SF3B1-mutant myelodysplastic syndromes. Leukemia 2016, 30, 2322–2331. [Google Scholar] [CrossRef] [PubMed]
- Pellagatti, A.; Boultwood, J. The molecular pathogenesis of the myelodysplastic syndromes. Eur. J. Haematol. 2015, 95, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Dolatshad, H.; Pellagatti, A.; Fernandez-Mercado, M.; Yip, B.H.; Malcovati, L.; Attwood, M.; Przychodzen, B.; Sahgal, N.; Kanapin, A.A.; Lockstone, H.; et al. Disruption of SF3B1 results in deregulated expression and splicing of key genes and pathways in myelodysplastic syndrome hematopoietic stem and progenitor cells. Leukemia 2015, 29, 1092–1103. [Google Scholar] [CrossRef]
- Zhao, B.; Li, Z.; Qian, R.; Liu, G.; Fan, M.; Liang, Z.; Hu, X.; Wan, Y. Cancer-associated mutations in SF3B1 disrupt the interaction between SF3B1 and DDX42. J. Biochem. 2022, 172, 117–126. [Google Scholar] [CrossRef]
- Dalton, W.B.; Helmenstine, E.; Walsh, N.; Gondek, L.P.; Kelkar, D.S.; Read, A.; Natrajan, R.; Christenson, E.S.; Roman, B.; Das, S.; et al. Hotspot SF3B1 mutations induce metabolic reprogramming and vulnerability to serine deprivation. J. Clin. Investig. 2019, 129, 4708–4723. [Google Scholar] [CrossRef] [PubMed]
- Cazzola, M.; Rossi, M.; Malcovati, L.; Associazione Italiana per la Ricerca sul Cancro Gruppo Italiano Malattie Mieloproliferative. Biologic and clinical significance of somatic mutations of SF3B1 in myeloid and lymphoid neoplasms. Blood 2013, 121, 260–269. [Google Scholar] [CrossRef] [PubMed]
- Yoshimi, A.; Abdel-Wahab, O. Splicing factor mutations in MDS RARS and MDS/MPN-RS-T. Int. J. Hematol. 2017, 105, 720–731. [Google Scholar] [CrossRef]
- Giagounidis, A. Current treatment algorithm for the management of lower-risk MDS. Hematol. Am. Soc. Hematol. Educ. Program 2017, 2017, 453–459. [Google Scholar] [CrossRef]
- Fenaux, P.; Ades, L. How we treat lower-risk myelodysplastic syndromes. Blood 2013, 121, 4280–4286. [Google Scholar] [CrossRef]
- Platzbecker, U.; Germing, U.; Gotze, K.S.; Kiewe, P.; Mayer, K.; Chromik, J.; Radsak, M.; Wolff, T.; Zhang, X.; Laadem, A.; et al. Luspatercept for the treatment of anaemia in patients with lower-risk myelodysplastic syndromes (PACE-MDS): A multicentre, open-label phase 2 dose-finding study with long-term extension study. Lancet Oncol. 2017, 18, 1338–1347. [Google Scholar] [CrossRef]
- Fenaux, P.; Platzbecker, U.; Mufti, G.J.; Garcia-Manero, G.; Buckstein, R.; Santini, V.; Diez-Campelo, M.; Finelli, C.; Cazzola, M.; Ilhan, O.; et al. Luspatercept in Patients with Lower-Risk Myelodysplastic Syndromes. N. Engl. J. Med. 2020, 382, 140–151. [Google Scholar] [CrossRef]
- Della Porta, M.; Platzbecker, U.; Santini, V.; Garcia-Manero, G.; Komrokji, R.S.; Ito, R.; Fenaux, P. The Commands Trial: A Phase 3 Study of the Efficacy and Safety of Luspatercept Versus Epoetin Alfa for the Treatment of Anemia Due to IPSS-R Very Low-, Low-, or Intermediate-Risk MDS in Erythropoiesis Stimulating Agent-Naive Patients Who Require RBC Transfusions. Blood 2020, 136, 1–2. [Google Scholar] [CrossRef]
- Lasho, T.L.; Finke, C.M.; Hanson, C.A.; Jimma, T.; Knudson, R.A.; Ketterling, R.P.; Pardanani, A.; Tefferi, A. SF3B1 mutations in primary myelofibrosis: Clinical, histopathology and genetic correlates among 155 patients. Leukemia 2012, 26, 1135–1137. [Google Scholar] [CrossRef]
- Zhao, L.-P.; Daltro de Oliveira, R.; Marcault, C.; Soret, J.; Gauthier, N.; Verger, E.; Maslah, N.; Roux, B.; Parquet, N.; Dosquet, C.; et al. SF3B1 mutations in the Driver Clone Increase the Risk of Evolution to Myelofibrosis in Patients with Myeloproliferative Neoplasms (MPN). Blood 2020, 136, 1. [Google Scholar] [CrossRef]
- Lee, J.M.; Lee, H.; Eom, K.S.; Lee, S.E.; Kim, M.; Kim, Y. Impact of Integrated Genetic Information on Diagnosis and Prognostication for Myeloproliferative Neoplasms in the Next-Generation Sequencing Era. J. Clin. Med. 2021, 10, 1033. [Google Scholar] [CrossRef] [PubMed]
- Visconte, V.; Makishima, H.; Jankowska, A.; Szpurka, H.; Traina, F.; Jerez, A.; O’Keefe, C.; Rogers, H.J.; Sekeres, M.A.; Maciejewski, J.P.; et al. SF3B1, a splicing factor is frequently mutated in refractory anemia with ring sideroblasts. Leukemia 2012, 26, 542–545. [Google Scholar] [CrossRef] [PubMed]
- Boiocchi, L.; Hasserjian, R.P.; Pozdnyakova, O.; Wong, W.J.; Lennerz, J.K.; Le, L.P.; Dias-Santagata, D.; Iafrate, A.J.; Hobbs, G.S.; Nardi, V. Clinicopathological and molecular features of SF3B1-mutated myeloproliferative neoplasms. Hum. Pathol. 2019, 86, 1–11. [Google Scholar] [CrossRef]
- Senin, A.; Fernandez-Rodriguez, C.; Bellosillo, B.; Camacho, L.; Longaron, R.; Angona, A.; Besses, C.; Alvarez-Larran, A. Non-driver mutations in patients with JAK2V617F-mutated polycythemia vera or essential thrombocythemia with long-term molecular follow-up. Ann. Hematol. 2018, 97, 443–451. [Google Scholar] [CrossRef]
- Tefferi, A.; Lasho, T.L.; Guglielmelli, P.; Finke, C.M.; Rotunno, G.; Elala, Y.; Pacilli, A.; Hanson, C.A.; Pancrazzi, A.; Ketterling, R.P.; et al. Targeted deep sequencing in polycythemia vera and essential thrombocythemia. Blood Adv. 2016, 1, 21–30. [Google Scholar] [CrossRef]
- Loscocco, G.G.; Guglielmelli, P.; Mannelli, F.; Mora, B.; Mannarelli, C.; Rotunno, G.; Pancani, F.; Maccari, C.; Bartalucci, N.; Romagnoli, S.; et al. SF3B1 mutations in primary and secondary myelofibrosis: Clinical, molecular and prognostic correlates. Am. J. Hematol. 2022, 97, E347–E349. [Google Scholar] [CrossRef]
- Jeromin, S.; Haferlach, T.; Weissmann, S.; Meggendorfer, M.; Eder, C.; Nadarajah, N.; Alpermann, T.; Kohlmann, A.; Kern, W.; Haferlach, C.; et al. Refractory anemia with ring sideroblasts and marked thrombocytosis cases harbor mutations in SF3B1 or other spliceosome genes accompanied by JAK2V617F and ASXL1 mutations. Haematologica 2015, 100, e125–e127. [Google Scholar] [CrossRef] [PubMed]
- Inano, T.; Araki, M.; Morishita, S.; Imai, M.; Yasuda, H.; Nitta, H.; Ito, M.; Edahiro, Y.; Ochiai, T.; Misawa, K.; et al. JAK2 exon 12 mutation in myelodysplastic/myeloproliferative neoplasm with ring sideroblasts and thrombocytosis: Not an exclusive mutation to polycythaemia vera. Br. J. Haematol. 2019, 187, e27–e31. [Google Scholar] [CrossRef] [PubMed]
- Palomo, L.; Meggendorfer, M.; Hutter, S.; Twardziok, S.; Adema, V.; Fuhrmann, I.; Fuster-Tormo, F.; Xicoy, B.; Zamora, L.; Acha, P.; et al. Molecular landscape and clonal architecture of adult myelodysplastic/myeloproliferative neoplasms. Blood 2020, 136, 1851–1862. [Google Scholar] [CrossRef] [PubMed]
- Patnaik, M.M.; Barraco, D.; Lasho, T.L.; Finke, C.M.; Hanson, C.A.; Ketterling, R.P.; Gangat, N.; Tefferi, A. DNMT3A mutations are associated with inferior overall and leukemia-free survival in chronic myelomonocytic leukemia. Am. J. Hematol. 2017, 92, 56–61. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Sanada, M.; Shiraishi, Y.; Nowak, D.; Nagata, Y.; Yamamoto, R.; Sato, Y.; Sato-Otsubo, A.; Kon, A.; Nagasaki, M.; et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011, 478, 64–69. [Google Scholar] [CrossRef]
- Kar, S.A.; Jankowska, A.; Makishima, H.; Visconte, V.; Jerez, A.; Sugimoto, Y.; Muramatsu, H.; Traina, F.; Afable, M.; Guinta, K.; et al. Spliceosomal gene mutations are frequent events in the diverse mutational spectrum of chronic myelomonocytic leukemia but largely absent in juvenile myelomonocytic leukemia. Haematologica 2013, 98, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Patnaik, M.M.; Lasho, T.L.; Finke, C.M.; Hanson, C.A.; Hodnefield, J.M.; Knudson, R.A.; Ketterling, R.P.; Pardanani, A.; Tefferi, A. Spliceosome mutations involving SRSF2, SF3B1, and U2AF35 in chronic myelomonocytic leukemia: Prevalence, clinical correlates, and prognostic relevance. Am. J. Hematol. 2013, 88, 201–206. [Google Scholar] [CrossRef]
- Wassie, E.A.; Itzykson, R.; Lasho, T.L.; Kosmider, O.; Finke, C.M.; Hanson, C.A.; Ketterling, R.P.; Solary, E.; Tefferi, A.; Patnaik, M.M. Molecular and prognostic correlates of cytogenetic abnormalities in chronic myelomonocytic leukemia: A Mayo Clinic-French Consortium Study. Am. J. Hematol. 2014, 89, 1111–1115. [Google Scholar] [CrossRef]
- Wudhikarn, K.; Loghavi, S.; Mangaonkar, A.A.; Al-Kali, A.; Binder, M.; Carr, R.; Reichard, K.; Finke, C.; Howard, M.; Gangat, N.; et al. SF3B1-mutant CMML defines a predominantly dysplastic CMML subtype with a superior acute leukemia-free survival. Blood Adv. 2020, 4, 5716–5721. [Google Scholar] [CrossRef]
- Patnaik, M.M.; Padron, E.; LaBorde, R.R.; Lasho, T.L.; Finke, C.M.; Hanson, C.A.; Hodnefield, J.M.; Knudson, R.A.; Ketterling, R.P.; Al-kali, A.; et al. Mayo prognostic model for WHO-defined chronic myelomonocytic leukemia: ASXL1 and spliceosome component mutations and outcomes. Leukemia 2013, 27, 1504–1510. [Google Scholar] [CrossRef]
- Ferrara, F.; Schiffer, C.A. Acute myeloid leukaemia in adults. Lancet 2013, 381, 484–495. [Google Scholar] [CrossRef]
- Venable, E.R.; Chen, D.; Chen, C.P.; Bessonen, K.R.; Nguyen, P.L.; Oliveira, J.L.; Reichard, K.K.; Hoyer, J.D.; Althoff, S.D.; Roh, D.J.; et al. Pathologic Spectrum and Molecular Landscape of Myeloid Disorders Harboring SF3B1 Mutations. Am. J. Clin. Pathol. 2021, 156, 679–690. [Google Scholar] [CrossRef]
- Jeromin, S.; Bacher, U.; Bayer, K.; Dicker, F.; Eder, C.; Fasan, A.; Grossmann, V.; Kohlmann, A.; Kern, W.; Haferlach, C.; et al. SF3B1 Mutations Are Detectable in 48.9% of Acute Myeloid Leukemia with Normal Karyotype (AML-NK) and ≥15% Ring Sideroblasts and Are Closely Related to FLT3-ITD and RUNX1 Mutations. Blood 2012, 120, 406. [Google Scholar] [CrossRef]
- Berger, G.; Gerritsen, M.; Yi, G.; Koorenhof-Scheele, T.N.; Kroeze, L.I.; Stevens-Kroef, M.; Yoshida, K.; Shiraishi, Y.; van den Berg, E.; Schepers, H.; et al. Ring sideroblasts in AML are associated with adverse risk characteristics and have a distinct gene expression pattern. Blood Adv. 2019, 3, 3111–3122. [Google Scholar] [CrossRef]
- Zhang, H.; Nakauchi, Y.; Kohnke, T.; Stafford, M.; Bottomly, D.; Thomas, R.; Wilmot, B.; McWeeney, S.K.; Majeti, R.; Tyner, J.W. Integrated analysis of patient samples identifies biomarkers for venetoclax efficacy and combination strategies in acute myeloid leukemia. Nat. Cancer 2020, 1, 826–839. [Google Scholar] [CrossRef]
- Welch, J.S.; Petti, A.A.; Miller, C.A.; Fronick, C.C.; O’Laughlin, M.; Fulton, R.S.; Wilson, R.K.; Baty, J.D.; Duncavage, E.J.; Tandon, B.; et al. TP53 and Decitabine in Acute Myeloid Leukemia and Myelodysplastic Syndromes. N. Engl. J. Med. 2016, 375, 2023–2036. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lawrence, M.S.; Wan, Y.; Stojanov, P.; Sougnez, C.; Stevenson, K.; Werner, L.; Sivachenko, A.; DeLuca, D.S.; Zhang, L.; et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. N. Engl. J. Med. 2011, 365, 2497–2506. [Google Scholar] [CrossRef]
- Quesada, V.; Conde, L.; Villamor, N.; Ordonez, G.R.; Jares, P.; Bassaganyas, L.; Ramsay, A.J.; Bea, S.; Pinyol, M.; Martinez-Trillos, A.; et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nat. Genet. 2011, 44, 47–52. [Google Scholar] [CrossRef]
- Rossi, D.; Bruscaggin, A.; Spina, V.; Rasi, S.; Khiabanian, H.; Messina, M.; Fangazio, M.; Vaisitti, T.; Monti, S.; Chiaretti, S.; et al. Mutations of the SF3B1 splicing factor in chronic lymphocytic leukemia: Association with progression and fludarabine-refractoriness. Blood 2011, 118, 6904–6908. [Google Scholar] [CrossRef]
- Taus, P.; Pospisilova, S.; Plevova, K. Identification of Clinically Relevant Subgroups of Chronic Lymphocytic Leukemia Through Discovery of Abnormal Molecular Pathways. Front. Genet. 2021, 12, 627964. [Google Scholar] [CrossRef]
- Rosenquist, R.; Cortese, D.; Bhoi, S.; Mansouri, L.; Gunnarsson, R. Prognostic markers and their clinical applicability in chronic lymphocytic leukemia: Where do we stand? Leuk. Lymphoma 2013, 54, 2351–2364. [Google Scholar] [CrossRef]
- Rozovski, U.; Keating, M.; Estrov, Z. The significance of spliceosome mutations in chronic lymphocytic leukemia. Leuk Lymphoma 2013, 54, 1364–1366. [Google Scholar] [CrossRef] [PubMed]
- Woyach, J.A.; Furman, R.R.; Liu, T.M.; Ozer, H.G.; Zapatka, M.; Ruppert, A.S.; Xue, L.; Li, D.H.; Steggerda, S.M.; Versele, M.; et al. Resistance mechanisms for the Bruton’s tyrosine kinase inhibitor ibrutinib. N. Engl. J. Med. 2014, 370, 2286–2294. [Google Scholar] [CrossRef] [PubMed]
- Pozzo, F.; Bittolo, T.; Tissino, E.; Vit, F.; Vendramini, E.; Laurenti, L.; D’Arena, G.; Olivieri, J.; Pozzato, G.; Zaja, F.; et al. SF3B1-mutated chronic lymphocytic leukemia shows evidence of NOTCH1 pathway activation including CD20 downregulation. Haematologica 2021, 106, 3125–3135. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.J.; Ashe, S.L.; Leich, E.; Burek, C.; Barrans, S.; Fenton, J.A.; Jack, A.S.; Pulford, K.; Rosenwald, A.; Banham, A.H. Potentially oncogenic B-cell activation-induced smaller isoforms of FOXP1 are highly expressed in the activated B cell-like subtype of DLBCL. Blood 2008, 111, 2816–2824. [Google Scholar] [CrossRef]
- te Raa, D.; Derks, I.A.; Luijks, D.M.; van Laar, J.; Monsuur, H.; Oldreive, C.; Jethwa, A.; Hüllein, J.; Stankovic, T.; Zenz, T.; et al. SF3B1 Mutations in CLL Are Equivalent to p53/ATM Dysfunction and Cause Defective Puma Upregulation in Response to Chemotherapy. Blood 2012, 120, 711. [Google Scholar] [CrossRef]
- Kashyap, M.K.; Kumar, D.; Villa, R.; La Clair, J.J.; Benner, C.; Sasik, R.; Jones, H.; Ghia, E.M.; Rassenti, L.Z.; Kipps, T.J.; et al. Targeting the spliceosome in chronic lymphocytic leukemia with the macrolides FD-895 and pladienolide-B. Haematologica 2015, 100, 945–954. [Google Scholar] [CrossRef]
- Bernard, E.; Tuechler, H.; Greenberg, P.L.; Hasserjian, R.P.; Ossa, J.E.A.; Nannya, Y.; Devlin, S.M.; Creignou, M.; Pinel, P.; Monnier, L.; et al. Molecular International Prognostic Scoring System for Myelodysplastic Syndromes. NEJM Evidence 2022, 1, EVIDoa2200008. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cilloni, D.; Itri, F.; Bonuomo, V.; Petiti, J. SF3B1 Mutations in Hematological Malignancies. Cancers 2022, 14, 4927. https://doi.org/10.3390/cancers14194927
Cilloni D, Itri F, Bonuomo V, Petiti J. SF3B1 Mutations in Hematological Malignancies. Cancers. 2022; 14(19):4927. https://doi.org/10.3390/cancers14194927
Chicago/Turabian StyleCilloni, Daniela, Federico Itri, Valentina Bonuomo, and Jessica Petiti. 2022. "SF3B1 Mutations in Hematological Malignancies" Cancers 14, no. 19: 4927. https://doi.org/10.3390/cancers14194927
APA StyleCilloni, D., Itri, F., Bonuomo, V., & Petiti, J. (2022). SF3B1 Mutations in Hematological Malignancies. Cancers, 14(19), 4927. https://doi.org/10.3390/cancers14194927






