Single-Cell Identification of Melanoma Biomarkers in Circulating Tumor Cells
Abstract
Simple Summary
Abstract
1. Introduction
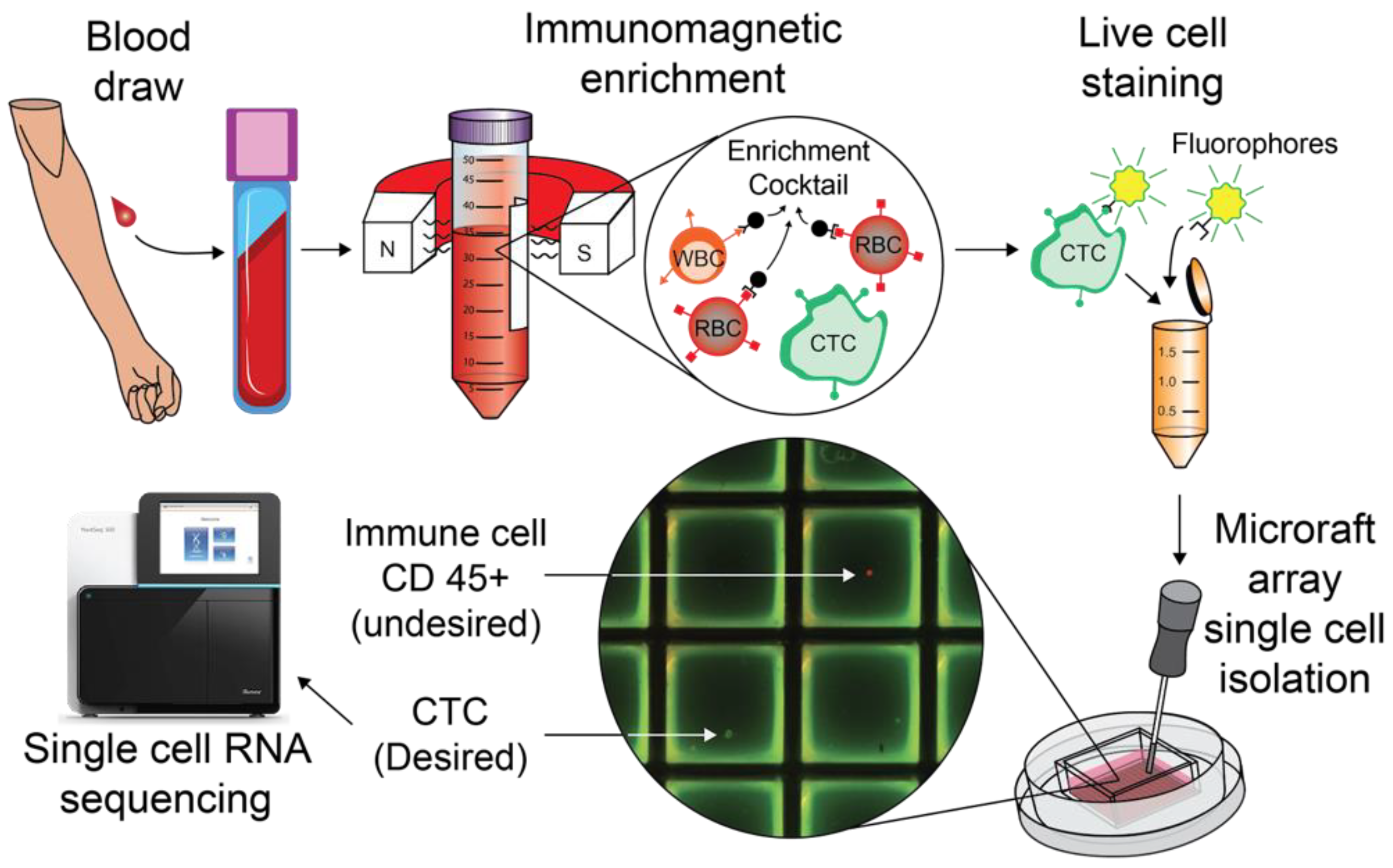
2. Materials and Methods
2.1. Patient Recruitment and Specimen Collection
2.2. Ficoll-Paque Density Gradient Separation for Isolation of Healthy Control PBMCs
2.3. CTC Enrichment with StemCell Technologies EasySep Direct Human CTC Enrichment Kit
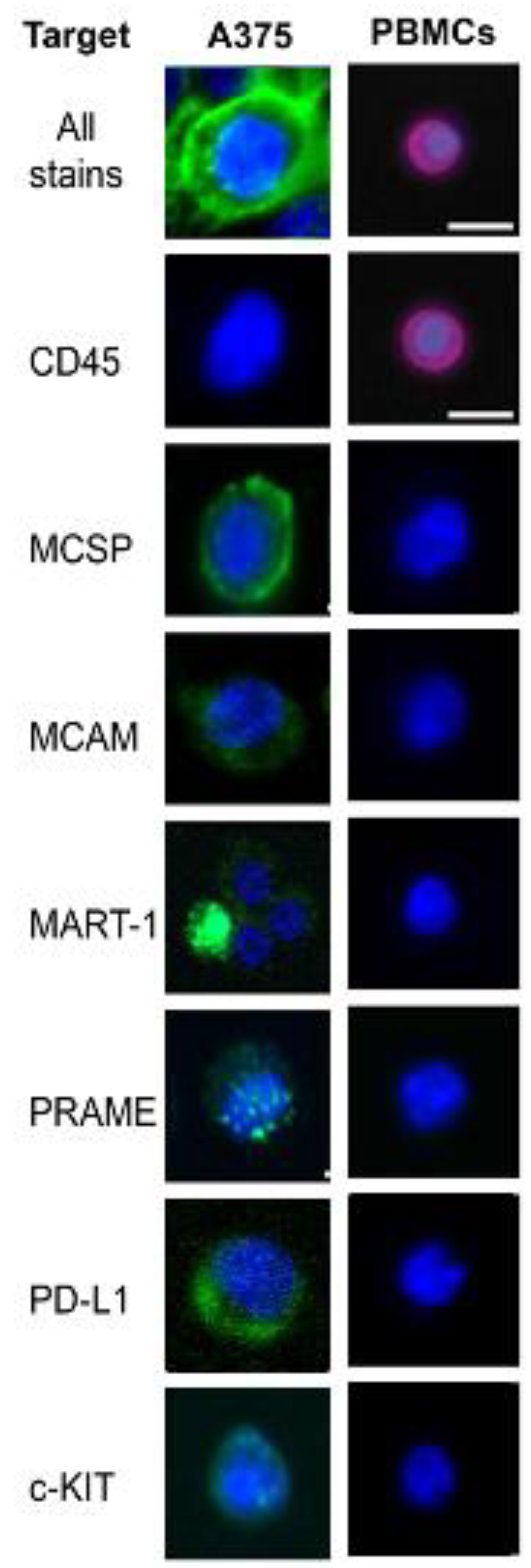
2.4. Live Cell Staining
2.5. Prepping Cell Microsystems CytoSort Arrays with Corning Cell-Tak Cell and Tissue Adhesive
2.6. CytoSort Array Picking
2.7. Single-Cell Library Preps
2.8. A375 Single Cell Preps
2.9. A375 Mini-Bulk Preparations
2.10. Data Analysis
- TrimGalore: 0.6.2
- fastqscreen: 0.14.0
- Seurat: 4.0.1
- Hisat: 2.0.4
- featureCounts=subread version 1.6.4
3. Results
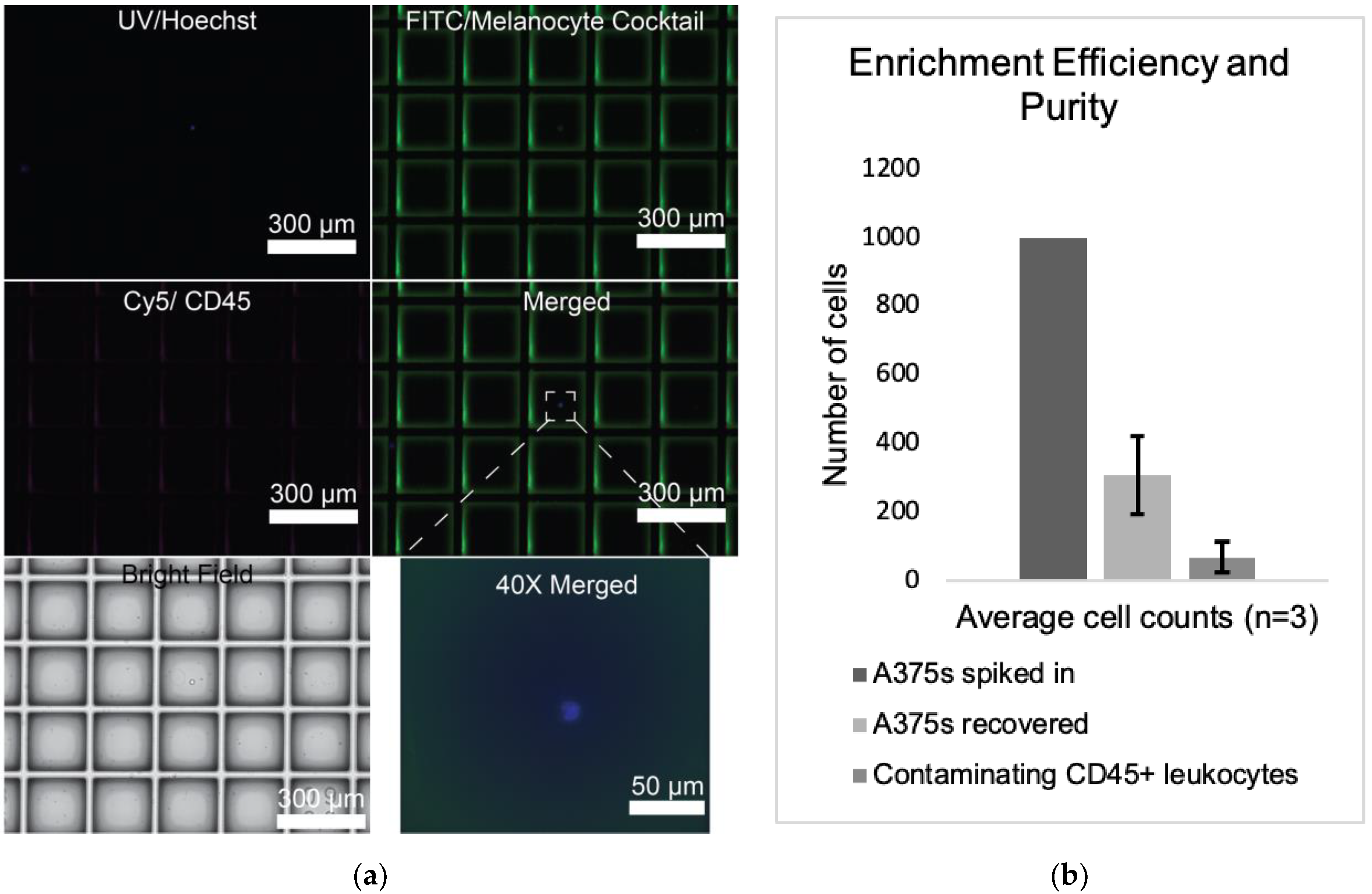
3.1. Negative Depletion Cocktail Yields High CTC Purity and Efficiency
3.2. CTCs Are Present in Many Advanced Melanoma Patients
3.3. A375 Validation of Smartseq-2 Single-Cell Analysis
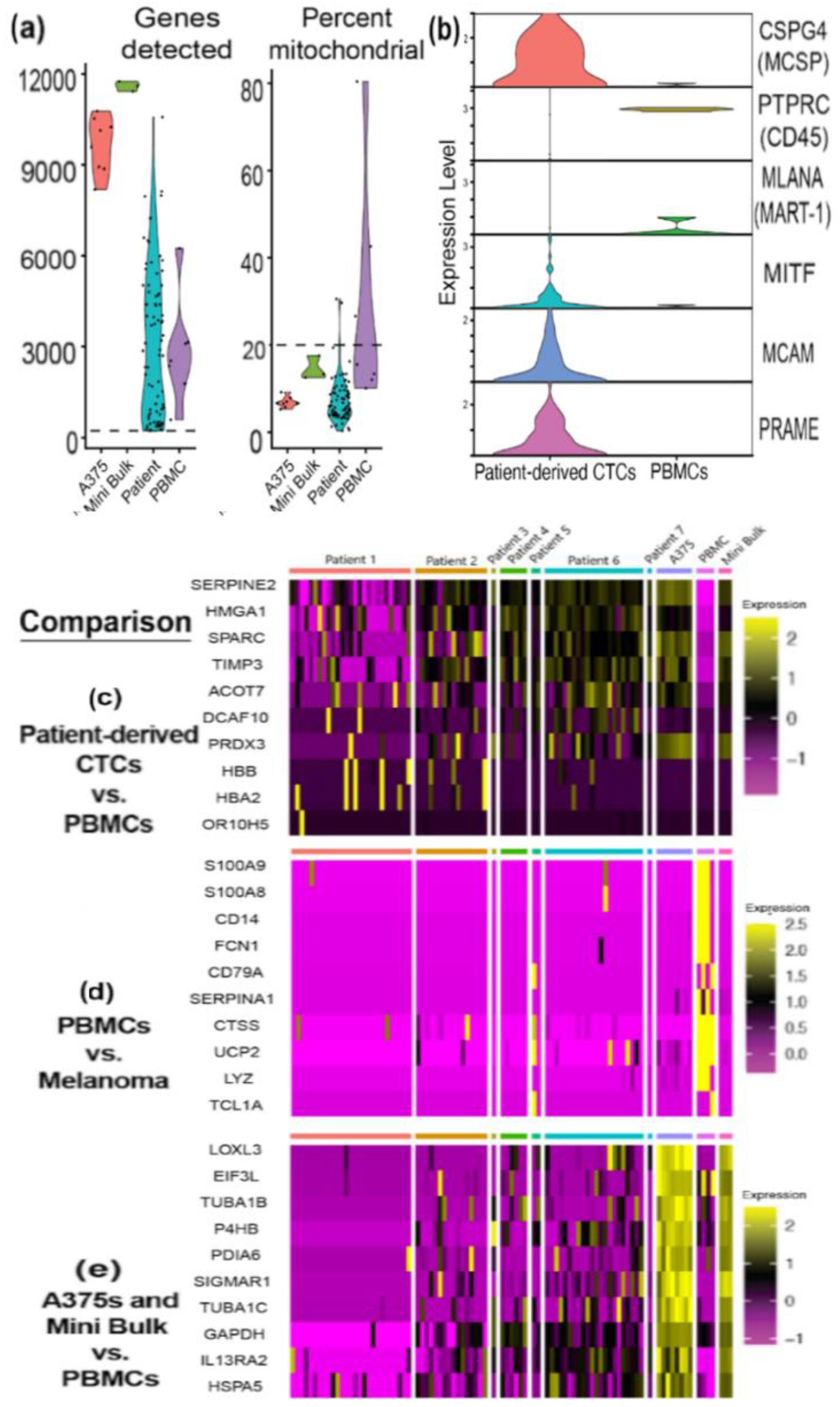
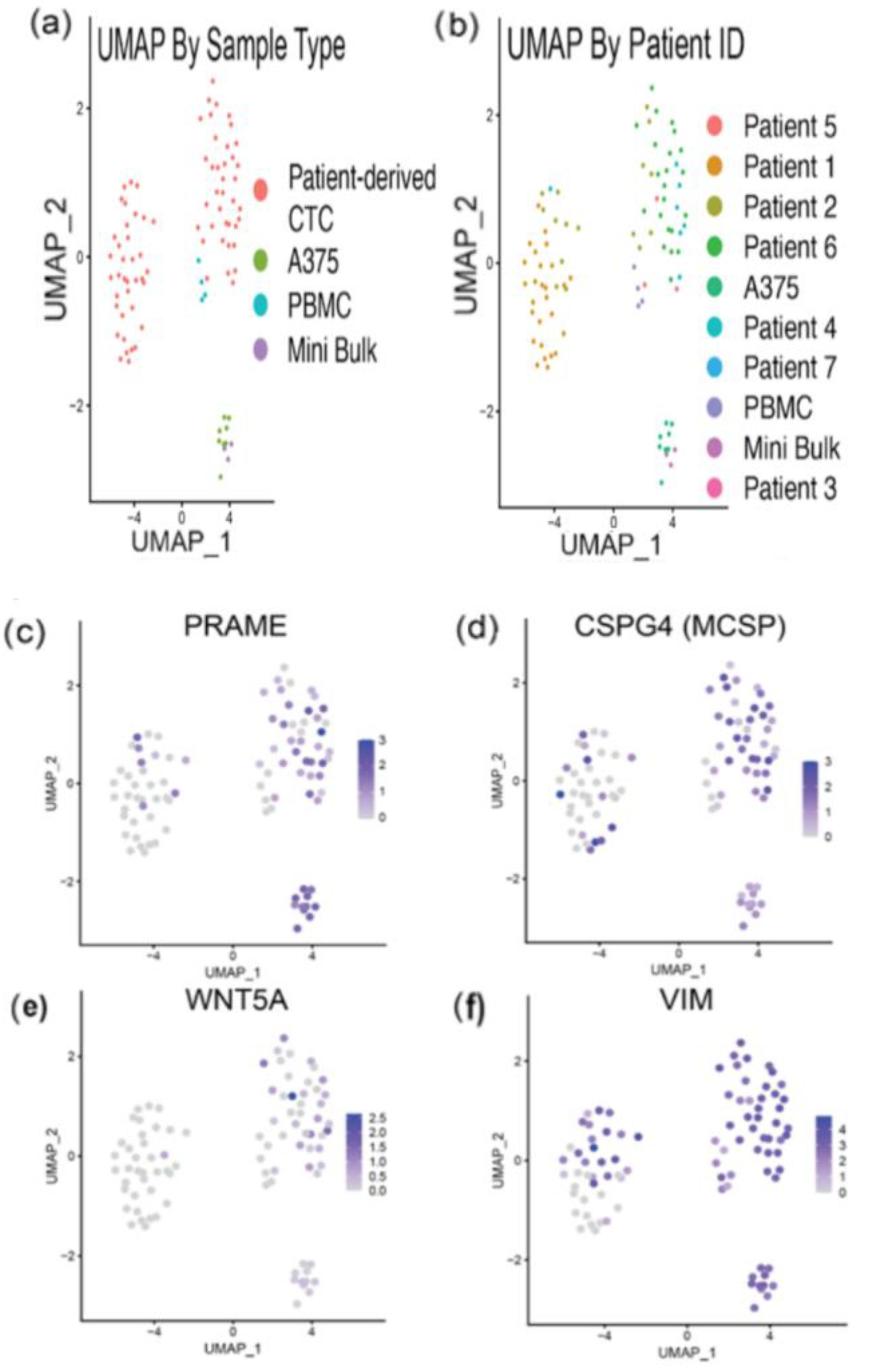
3.4. RNA Sequencing Helps Confirm Melanocytic Identity of Patient-Derived CTCs
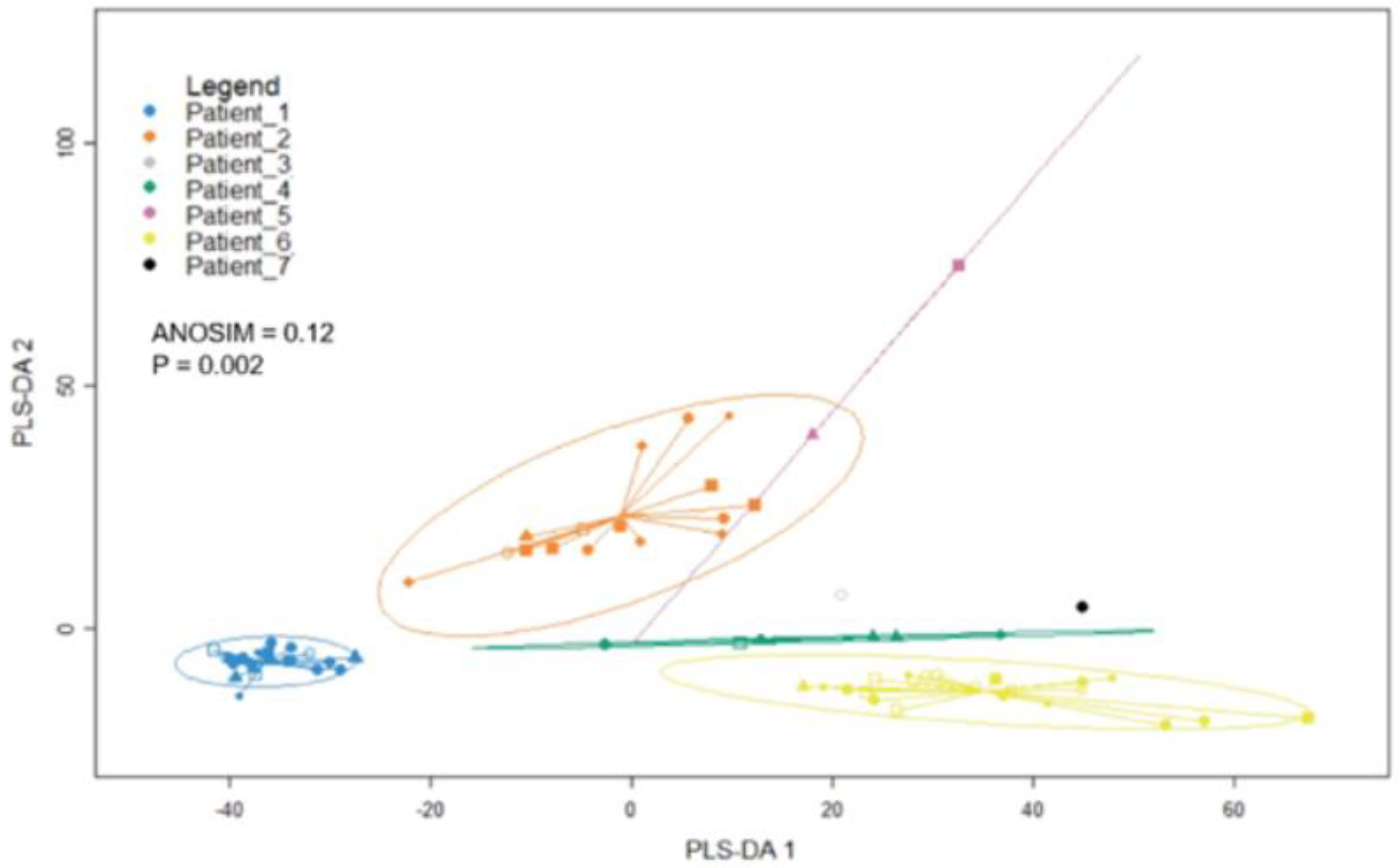
3.5. RNA Sequencing Analysis Reveals Expression of Melanoma-Related Genes in Melanoma CTCs and Heterogeneity of Gene Expression
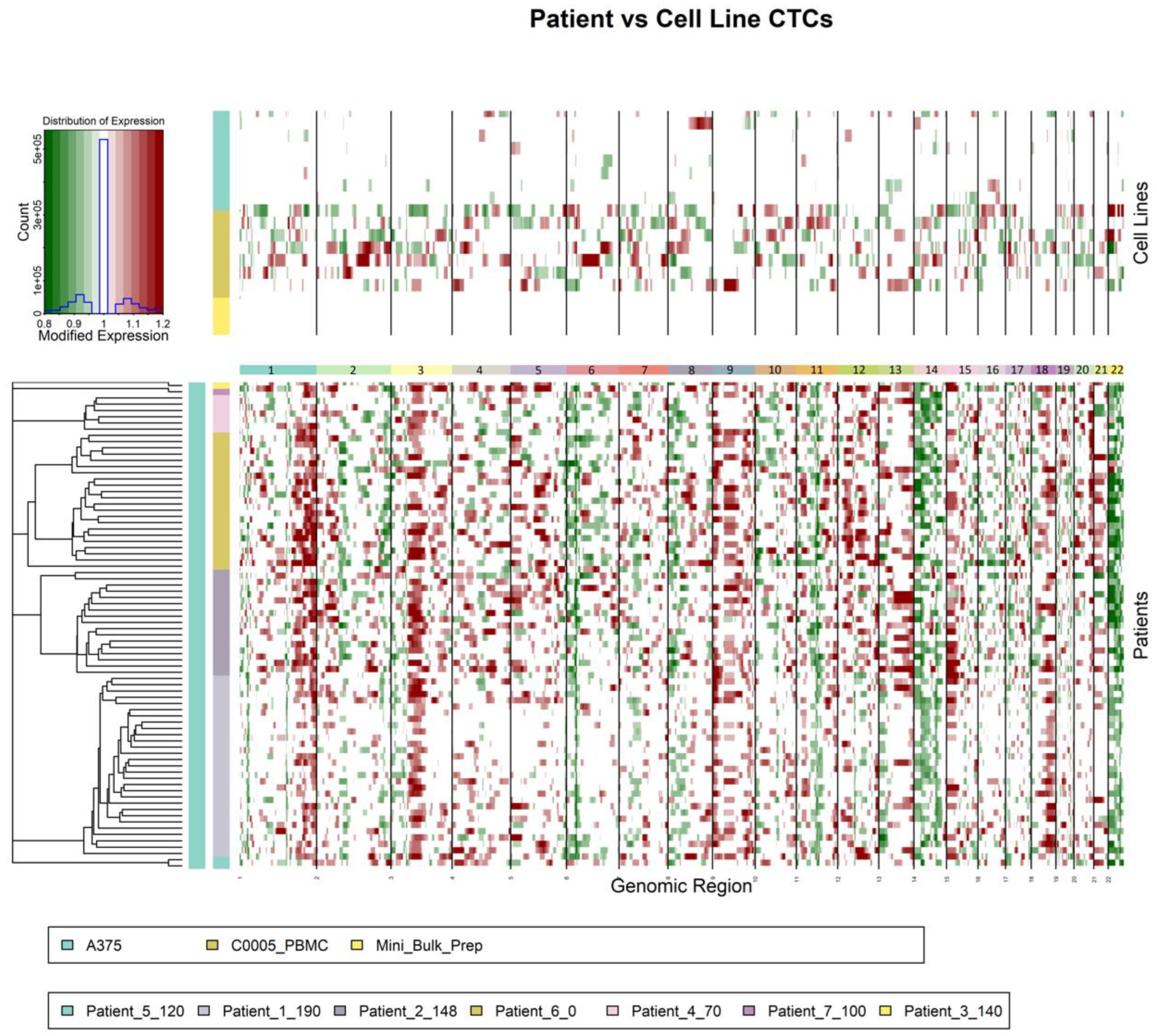
3.6. Copy Number Variation Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ashworth, T.R. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Aus. Med. J. 1869, 14, 146. [Google Scholar]
- Chaffer, C.L.; Weinberg, R.A. A Perspective on Cancer Cell Metastasis. Science 2011, 331, 1559–1564. [Google Scholar] [CrossRef] [PubMed]
- Mehlen, P.; Puisieux, A. Metastasis: A question of life or death. Nat. Cancer 2006, 6, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Dall’Olio, F.G.; Gelsomino, F.; Conci, N.; Marcolin, L.; De Giglio, A.; Grilli, G.; Sperandi, F.; Fontana, F.; Terracciano, M.; Fragomeno, B.; et al. PD-L1 Expression in Circulating Tumor Cells as a Promising Prognostic Biomarker in Advanced Non–small-cell Lung Cancer Treated with Immune Checkpoint Inhibitors. Clin. Lung Cancer 2021, 22, 423–431. [Google Scholar] [CrossRef]
- Dhar, M.; Wong, J.; Che, J.; Matsumoto, M.; Grogan, T.; Elashoff, D.; Garon, E.B.; Goldman, J.W.; Christen, E.S.; Di Carlo, D.; et al. Evaluation of PD-L1 expression on vortex-isolated circulating tumor cells in metastatic lung cancer. Sci. Rep. 2018, 8, 1–10. [Google Scholar] [CrossRef]
- Riethdorf, S.; Fritsche, H.; Müller, V.; Rau, T.; Schindlbeck, C.; Rack, B.; Janni, W.; Coith, C.; Beck, K.; Jänicke, F.; et al. Detection of Circulating Tumor Cells in Peripheral Blood of Patients with Metastatic Breast Cancer: A Validation Study of the CellSearch System. Clin. Cancer Res. 2007, 13, 920–928. [Google Scholar] [CrossRef]
- Cristofanilli, M.; Budd, G.T.; Mathew, J.E.; Alison, S.; Jeri, M.; Millar, J.C.; Reuben, J.M.; Doyle, G.V.; Allard, J.W.; Terstappen, L.W.M.M.; et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef]
- Nolé, F.; Munzone, E.; Zorzino, L.; Minchella, I.; Salvatici, M.; Botteri, E.; Medici, M.; Verri, E.; Adamoli, L.; Rotmensz, N.; et al. Variation of circulating tumor cell levels during treatment of metastatic breast cancer: Prognostic and therapeutic implications. Ann. Oncol. 2007, 19, 891–897. [Google Scholar] [CrossRef]
- Hoshimoto, S.; Shingai, T.; Morton, D.L.; Kuo, C.; Faries, M.B.; Chong, K.; Elashoff, D.; Wang, H.-J.; Elashoff, R.M.; Hoon, D.S.B. Association between circulating tumor cells and prognosis in patients with stage III melanoma with sentinel lymph node metastasis in a phase III international multicenter trial. J. Clin. Oncol. 2012, 30, 3819–3826. [Google Scholar] [CrossRef]
- Rushton, A.; Nteliopoulos, G.; Shaw, J.; Coombes, R. A Review of Circulating Tumour Cell Enrichment Technologies. Cancers 2021, 13, 970. [Google Scholar] [CrossRef]
- Alix-Panabières, C.; Schwarzenbach, H.; Pantel, K. Circulating Tumor Cells and Circulating Tumor DNA. Annu. Rev. Med. 2012, 63, 199–215. [Google Scholar] [CrossRef] [PubMed]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.J.; Uhr, J.W.; Terstappen, L.W.M.M. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef] [PubMed]
- Nagrath, S.; Sequist, L.V.; Maheswaran, S.; Bell, D.W.; Irimia, D.; Ulkus, L.; Smith, M.R.; Kwak, E.L.; Digumarthy, S.; Muzikansky, A.; et al. Isolation of rare circulating tumour cells in cancer patients by microchip technology. Nature 2007, 450, 1235–1239. [Google Scholar] [CrossRef] [PubMed]
- Joosse, S.A.; Gorges, T.M.; Pantel, K. Biology, detection, and clinical implications of circulating tumor cells. EMBO Mol. Med. 2014, 7, 1–11. [Google Scholar] [CrossRef]
- Williams, A.; Balic, M.; Datar, R.; Cote, R. Size-Based Enrichment Technologies for CTC Detection and Characterization. Recent Results Cancer Res. 2012, 195, 87–95. [Google Scholar] [CrossRef]
- Vona, G.; Sabile, A.; Louha, M.; Sitruk, V.; Romana, S.; Schutze, K.; Capron, F.; Franco, D.; Pazzagli, M.; Vekemans, M.; et al. Isolation by size of epithelial tumor cells: A new method for the immunomorphological and molecular charac-terization of circulatingtumor cells. Am. J. Pathol. 2000, 156, 57–63. [Google Scholar] [CrossRef]
- Sollier, E.; Go, D.E.; Che, J.; Gossett, D.R.; O’Byrne, S.; Weaver, W.M.; Kummer, N.; Rettig, M.; Goldman, J.; Nickols, N.; et al. Size-selective collection of circulating tumor cells using Vortex technology. Lab. Chip 2013, 14, 63–77. [Google Scholar] [CrossRef]
- Cho, E.H.; Wendel, M.; Luttgen, M.; Yoshioka, C.; Marrinucci, D.; Lazar, D.; Schram, E.; Nieva, J.; Bazhenova, L.; Morgan, A.; et al. Characterization of circulating tumor cell aggregates identified in patients with epithelial tumors. Phys. Biol. 2012, 9, 016001. [Google Scholar] [CrossRef]
- Ozkumur, E.; Shah, A.M.; Ciciliano, J.C.; Emmink, B.L.; Miyamoto, D.T.; Brachtel, E.; Yu, M.; Chen, P.-I.; Morgan, B.; Trautwein, J.; et al. Inertial Focusing for Tumor Antigen–Dependent and –Independent Sorting of Rare Circulating Tumor Cells. Sci. Transl. Med. 2013, 5, 179ra47. [Google Scholar] [CrossRef]
- Park, S.; Ang, R.R.; Duffy, S.P.; Bazov, J.; Chi, K.N.; Black, P.C.; Ma, H. Morphological Differences between Circulating Tumor Cells from Prostate Cancer Patients and Cultured Prostate Cancer Cells. PLoS ONE 2014, 9, e85264. [Google Scholar] [CrossRef]
- Gascoyne, P.R.C.; Shim, S. Isolation of Circulating Tumor Cells by Dielectrophoresis. Cancers 2014, 6, 545–579. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.; Jafferji, I.; Garza, M.; Melnikova, V.O.; Hasegawa, D.K.; Pethig, R.; Davis, D.W. ApoStream™, a new dielectrophoretic device for antibody independent isolation and recovery of viable cancer cells from blood. Biomicrofluidics 2012, 6, 024133. [Google Scholar] [CrossRef] [PubMed]
- Freeman, J.B.; Gray, E.S.; Millward, M.; Pearce, R.; Ziman, M. Evaluation of a multi-marker immunomagnetic enrichment assay for the quantification of circulating melanoma cells. J. Transl. Med. 2012, 10, 192. [Google Scholar] [CrossRef] [PubMed]
- Pantel, K.; Alix-Panabières, C.; Riethdorf, S. Cancer micrometastases. Nat. Rev. Clin. Oncol. 2009, 6, 339–351. [Google Scholar] [CrossRef]
- Marsavela, G.; Aya-Bonilla, C.A.; Warkiani, M.E.; Gray, E.S.; Ziman, M. Melanoma circulating tumor cells: Benefits and challenges required for clinical application. Cancer Lett. 2018, 424, 1–8. [Google Scholar] [CrossRef]
- Castro-Giner, F.; Scheidmann, M.C.; Aceto, N. Beyond Enumeration: Functional and Computational Analysis of Circulating Tumor Cells to Investigate Cancer Metastasis. Front. Med. 2018, 5, 34. [Google Scholar] [CrossRef]
- Vitale, I.; Shema, E.; Loi, S.; Galluzzi, L. Intratumoral heterogeneity in cancer progression and response to immunotherapy. Nat. Med. 2021, 27, 212–224. [Google Scholar] [CrossRef]
- Picelli, S.; Faridani, O.R.; Björklund, K.; Winberg, G.; Sagasser, S.; Sandberg, R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014, 9, 171–181. [Google Scholar] [CrossRef]
- Ziegenhain, C.; Vieth, B.; Parekh, S.; Reinius, B.; Guillaumet-Adkins, A.; Smets, M.; Leonhardt, H.; Heyn, H.; Hellmann, I.; Enard, W. Comparative Analysis of Single-Cell RNA Sequencing Methods. Mol. Cell 2017, 65, 631–643.e4. [Google Scholar] [CrossRef]
- Lezcano, C.; Jungbluth, A.A.; Nehal, K.S.; Hollmann, T.J.; Busam, K.J. PRAME Expression in Melanocytic Tumors. Am. J. Surg. Pathol. 2018, 42, 1456–1465. [Google Scholar] [CrossRef]
- Rapanotti, M.C.; Campione, E.; Spallone, G.; Orlandi, A.; Bernardini, S.; Bianchi, L. Minimal residual disease in melanoma: Circulating melanoma cells and predictive role of MCAM/MUC18/MelCAM/CD146. Cell Death Discov. 2017, 3, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Campoli, M.R.; Chang, C.C.; Kageshita, T.; Wang, X.; McCarthy, J.B.; Ferrone, S. Human high molecular weight-melanoma-associated antigen (HMW-MAA): A melanoma cell surface chondroitin sulfate proteoglycan (MSCP) with biological and clinical signifi-cance. Crit. Rev. Immunol. 2004, 24, 267–296. [Google Scholar] [CrossRef] [PubMed]
- Matsui, Y.; Zsebo, K.M.; Hogan, B.L. Embryonic expression of a haematopoietic growth factor encoded by the Sl locus and the ligand for c-kit. Nature 1990, 347, 667–669. [Google Scholar] [CrossRef]
- Pilloni, L.; Bianco, P.; DiFelice, E.; Cabras, S.; Castellanos, M.E.; Atzori, L.; Ferreli, C.; Mulas, P.; Nemolato, S.; Faa, G. The usefulness of c-Kit in the immunohistochemical assessment of melanocytic lesions. Eur. J. Histochem. 2011, 55, e20. [Google Scholar] [CrossRef] [PubMed]
- Murer, K.; Urosevic, M.; Willers, J.; Selvam, P.; Laine, E.; Burg, G.; Dummer, R. Expression of Melan-A/MART-1 in primary melanoma cell cultures has prognostic implication in metastatic melanoma patients. Melanoma Res. 2004, 14, 257–262. [Google Scholar] [CrossRef]
- Stott, S.L.; Lee, R.J.; Nagrath, S.; Yu, M.; Miyamoto, D.T.; Ulkus, L.; Inserra, E.J.; Ulman, M.; Springer, S.; Nakamura, Z.; et al. Isolation and Characterization of Circulating Tumor Cells from Patients with Localized and Metastatic Prostate Cancer. Sci. Transl. Med. 2010, 2, 25ra23. [Google Scholar] [CrossRef]
- Ting, D.T.; Wittner, B.S.; Ligorio, M.; Jordan, N.V.; Shah, A.M.; Miyamoto, D.T.; Aceto, N.; Bersani, F.; Brannigan, B.W.; Xega, K.; et al. Single-Cell RNA Sequencing Identifies Extracellular Matrix Gene Expression by Pancreatic Circulating Tumor Cells. Cell Rep. 2015, 8, 1905–1918. [Google Scholar] [CrossRef]
- Miyamoto, D.T.; Zheng, Y.; Wittner, B.S.; Lee, R.J.; Zhu, H.; Broderick, K.T.; Desai, R.; Fox, D.S.; Brannigan, B.W.; Trautwein, J.; et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen re-sistance. Science 2015, 349, 1351–1356. [Google Scholar] [CrossRef]
- Lee, A.; Park, J.; Lim, M.; Sunkara, V.; Kim, S.Y.; Kim, G.H.; Kim, M.; Cho, Y. All-in-one centrifugal microfluidic device for size-selective circulating tumor cell isolation with high purity. Anal. Chem. 2014, 86, 11349–11356. [Google Scholar] [CrossRef]
- Perego, M.; Maurer, M.; Wang, J.X.; Shaffer, S.; Muller, A.C.; Parapatics, K.; Li, L.; Hrisova, D.; Shin, S.; Keeny, F.; et al. A slow-cycling subpopulation of melanoma cells with highly invasive properties. Oncogene 2017, 37, 302–312. [Google Scholar] [CrossRef] [PubMed]
- Sumter, T.; Xian, L.; Huso, T.; Koo, M.; Chang, Y.-T.; Almasri, T.; Chia, L.; Inglis, C.; Reid, D.; Resar, L. The High Mobility Group A1 (HMGA1) Transcriptome in Cancer and Development. Curr. Mol. Med. 2016, 16, 353–393. [Google Scholar] [CrossRef] [PubMed]
- Salvatierra, E.; Alvarez, M.J.; Leishman, C.C.; Baquero, E.R.; Lutzky, V.P.; Chuluyan, H.E.; Podhajcer, O.L. SPARC Controls Melanoma Cell Plasticity through Rac1. PLoS ONE 2015, 10, e0134714. [Google Scholar] [CrossRef] [PubMed]
- Girotti, M.R.; Fernández, M.; López, J.A.; Camafeita, E.; Fernández, E.A.; Albar, J.P.; Benedetti, L.G.; Valacco, M.P.; Brekken, R.A.; Podhajcer, O.L.; et al. SPARC Promotes Cathepsin B-Mediated Melanoma Invasiveness through a Collagen I/α2β1 Integrin Axis. J. Investig. Dermatol. 2011, 131, 2438–2447. [Google Scholar] [CrossRef]
- Das, A.M.; Bolkestein, M.; van der Klok, T.; Ophuis, C.M.O.; Vermeulen, C.E.; Rens, J.A.; Dinjens, W.N.; Atmodimedjo, P.N.; Verhoef, C.; Koljenović, S.; et al. Tissue inhibitor of metalloproteinase-3 (TIMP3) expression decreases during melanoma progression and inhibits melanoma cell migration. Eur. J. Cancer 2016, 66, 34–46. [Google Scholar] [CrossRef]
- Ramasamy, P.; Larkin, A.-M.; Linge, A.; Tiernan, D.; McAree, F.; Horgan, N.; Moriarty, P.; Beatty, S.; Murphy, C.; Clynes, M.; et al. PRDX3 is associated with metastasis and poor survival in uveal melanoma. J. Clin. Pathol. 2019, 73, 408–412. [Google Scholar] [CrossRef]
- Santamaría, P.G.; Floristán, A.; Fontanals-Cirera, B.; Vázquez-Naharro, A.; Santos, V.; Morales, S.; Yuste, L.; Peinado, H.; García-Gómez, A.; Portillo, F.; et al. Lysyl oxidase-like 3 is required for melanoma cell survival by maintaining genomic stability. Cell Death Differ. 2017, 25, 935–950. [Google Scholar] [CrossRef]
- Kioi, M.; Kawakami, M.; Shimamura, T.; Husain, S.R.; Puri, R.K. Interleukin-13 receptor alpha2 chain: A potential biomarker and molecular target for ovarian cancer therapy. Cancer 2006, 107, 1407–1418. [Google Scholar] [CrossRef]
- Beard, R.E.; Abate-Daga, D.; Rosati, S.F.; Zheng, Z.; Wunderlich, J.R.; Rosenberg, S.A.; Morgan, R.A. Gene Expression Profiling using Nanostring Digital RNA Counting to Identify Potential Target Antigens for Melanoma Immunotherapy. Clin. Cancer Res. 2013, 19, 4941–4950. [Google Scholar] [CrossRef]
- Guo, C.; Liu, S.; Wang, J.; Sun, M.-Z.; Greenaway, F.T. ACTB in cancer. Clin. Chim. Acta 2013, 417, 39–44. [Google Scholar] [CrossRef]
- Xue, G.; Romano, E.; Massi, D.; Mandalà, M. Wnt/β-catenin signaling in melanoma: Preclinical rationale and novel therapeutic insights. Cancer Treat. Rev. 2016, 49, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Webster, M.R.; Kugel, C.H., 3rd; Weeraratna, A.T. The Wnts of change: How Wnts regulate phenotype switching in melanoma. Biochim. Biophys. Acta 2015, 1856, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Dissanayake, S.K.; Wade, M.; Johnson, C.E.; O’Connell, M.P.; Leotlela, P.D.; French, A.D.; Shah, K.V.; Hewitt, K.J.; Rosenthal, D.T.; Indig, F.E.; et al. The Wnt5A/Protein Kinase C Pathway Mediates Motility in Melanoma Cells via the Inhibition of Metastasis Suppressors and Initiation of an Epithelial to Mesenchymal Transition. J. Biol. Chem. 2007, 282, 17259–17271. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, R.S.; Kulej, K.; Kathayat, R.S.; Garcis, B.A.; Dickinson, B.C.; Brandy, D.C.; Witze, E.A. Wnt5a signaling induced phosphorylation increases APT1 activity and promotes melanoma met-astatic behavior. Elife 2018, 7, e34362. [Google Scholar] [CrossRef]
- Da Forno, P.D.; Pringle, J.H.; Hutchinson, P.; Osborn, J.; Huang, Q.; Potter, L.; Hancox, R.A.; Fletcher, A.; Saldanha, G.S. WNT5A Expression Increases during Melanoma Progression and Correlates with Outcome. Clin. Cancer Res. 2008, 14, 5825–5832. [Google Scholar] [CrossRef]
- Holtzhausen, A.; Zhao, F.; Evans, K.S.; Tsutsui, M.; Orabona, C.; Tyler, D.S.; Hanks, B.A. Melanoma-Derived Wnt5a Promotes Local Dendritic-Cell Expression of IDO and Immunotol-erance: Opportunities for Pharmacologic Enhancement of Immunotherapy. Cancer Immunol. Res. 2015, 3, 1082–1095. [Google Scholar] [CrossRef]
- Spranger, S.; Bao, R.; Gajewski, T.F. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature 2015, 523, 231–235. [Google Scholar] [CrossRef]
- Satelli, A.; Li, S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Exp. Mol. Life Sci. 2011, 68, 3033–3046. [Google Scholar] [CrossRef]
- Fane, M.E.; Chhabra, Y.; Alicea, G.M.; Maranto, D.A.; Douglass, S.M.; Webster, M.R.; Rebecca, V.W.; Marino, G.E.; Almeida, F.; Ecker, B.L.; et al. Stromal changes in the aged lung induce an emergence from melanoma dormancy. Nature 2022, 606, 396–405. [Google Scholar] [CrossRef]
- Vogel, C.; Marcotte, E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 2012, 13, 227–232. [Google Scholar] [CrossRef]
- de Sousa Abreu, R.; Penalva, L.O.; Marcotte, E.M.; Vogel, C. Global signatures of protein and mRNA expression levels. Mol. Biosyst. 2009, 5, 1512–1526. [Google Scholar] [CrossRef] [PubMed]
- Galanzha, E.I.; Zharov, V.P. Circulating Tumor Cell Detection and Capture by Photoacoustic Flow Cytometry in Vivo and ex Vivo. Cancers 2013, 5, 1691–1738. [Google Scholar] [CrossRef] [PubMed]
- Klinac, D.; Gray, E.S.; Freeman, J.B.; Reid, A.; Bowyer, S.; Millward, M.; Ziman, M. Monitoring changes in circulating tumour cells as a prognostic indicator of overall survival and treatment response in patients with metastatic melanoma. BMC Cancer 2014, 14, 423. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fankhauser, R.; Chang, M.; Garrison, Z.; Berryman, R.; Lucero, O.M.; Fuiten, A.; DePatie, N.; Seifert, H.; Kulkarni, R.P. Single-Cell Identification of Melanoma Biomarkers in Circulating Tumor Cells. Cancers 2022, 14, 4921. https://doi.org/10.3390/cancers14194921
Fankhauser R, Chang M, Garrison Z, Berryman R, Lucero OM, Fuiten A, DePatie N, Seifert H, Kulkarni RP. Single-Cell Identification of Melanoma Biomarkers in Circulating Tumor Cells. Cancers. 2022; 14(19):4921. https://doi.org/10.3390/cancers14194921
Chicago/Turabian StyleFankhauser, Reilly, Matthew Chang, Zachary Garrison, Rachel Berryman, Olivia M. Lucero, Allison Fuiten, Nicholas DePatie, Hilary Seifert, and Rajan P. Kulkarni. 2022. "Single-Cell Identification of Melanoma Biomarkers in Circulating Tumor Cells" Cancers 14, no. 19: 4921. https://doi.org/10.3390/cancers14194921
APA StyleFankhauser, R., Chang, M., Garrison, Z., Berryman, R., Lucero, O. M., Fuiten, A., DePatie, N., Seifert, H., & Kulkarni, R. P. (2022). Single-Cell Identification of Melanoma Biomarkers in Circulating Tumor Cells. Cancers, 14(19), 4921. https://doi.org/10.3390/cancers14194921





