Multiplexed In Situ Spatial Protein Profiling in the Pursuit of Precision Immuno-Oncology for Patients with Breast Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Antigenicity
2.1. Tumor-Specific Antigens
2.2. Tumor-Associated Antigens
3. Adjuvanticity
4. Patterns of Resistance to Innate Immunity
4.1. Dendritic Cells
4.2. Macrophages
4.3. NK Cells
5. Homing and Migration
5.1. Endothelial Cells
5.2. Cancer-Associated Fibroblasts
5.3. Chemokines
6. Recognition and Killing
6.1. TILs Density
6.2. Spatial Organization
6.3. T Cell States and Trajectories
6.3.1. CD8+ T Cells
6.3.2. CD4+ T Cells
6.3.3. TIL-B and TLS
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Karasaki, T.; Nagayama, K.; Kuwano, H.; Nitadori, J.-I.; Sato, M.; Anraku, M.; Hosoi, A.; Matsushita, H.; Morishita, Y.; Kashiwabara, K.; et al. An Immunogram for the Cancer-Immunity Cycle: Towards Personalized Immunotherapy of Lung Cancer. J. Thorac. Oncol. 2017, 12, 791–803. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.S.; Mellman, I. Oncology Meets Immunology: The Cancer-Immunity Cycle. Immunity 2013, 39, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Boumahdi, S.; de Sauvage, F.J. The Great Escape: Tumour Cell Plasticity in Resistance to Targeted Therapy. Nat. Rev. Drug. Discov. 2020, 19, 39–56. [Google Scholar] [CrossRef]
- Galluzzi, L.; Petroni, G.; Kroemer, G. Immunogenicity of cell death driven by immune effectors. J. Immunother. Cancer 2019, 8, e000802. [Google Scholar] [CrossRef]
- Somarelli, J.A. The Hallmarks of Cancer as Ecologically Driven Phenotypes. Front. Ecol. Evol. 2021, 9. [Google Scholar] [CrossRef]
- Turajlic, S.; Sottoriva, A.; Graham, T.; Swanton, C. Resolving genetic heterogeneity in cancer. Nat. Rev. Genet. 2019, 20, 404–416. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Miglietta, F.; Bottosso, M.; Griguolo, G.; Dieci, M.; Guarneri, V. Major advancements in metastatic breast cancer treatment: When expanding options means prolonging survival. ESMO Open 2022, 7. [Google Scholar] [CrossRef]
- Schmid, P.; Adams, S.; Rugo, H.S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Diéras, V.; Hegg, R.; Im, S.-A.; Shaw Wright, G.; et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N. Engl. J. Med. 2018, 379, 2108–2121. [Google Scholar] [CrossRef]
- Cortes, J.; Rugo, H.S.; Cescon, D.W.; Im, S.-A.; Yusof, M.M.; Gallardo, C.; Lipatov, O.; Barrios, C.H.; Perez-Garcia, J.; Iwata, H.; et al. Pembrolizumab plus Chemotherapy in Advanced Triple-Negative Breast Cancer. N. Engl. J. Med. 2022, 387, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Schmid, P.; Cortes, J.; Dent, R.; Pusztai, L.; McArthur, H.; Kümmel, S.; Bergh, J.; Denkert, C.; Park, Y.H.; Hui, R.; et al. Event-free Survival with Pembrolizumab in Early Triple-Negative Breast Cancer. N. Engl. J. Med. 2022, 386, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Tarantino, P.; Gandini, S.; Trapani, D.; Criscitiello, C.; Curigliano, G. Immunotherapy addition to neoadjuvant chemotherapy for early triple negative breast cancer: A systematic review and meta-analysis of randomized clinical trials. Crit. Rev. Oncol. 2021, 159, 103223. [Google Scholar] [CrossRef] [PubMed]
- Loibl, S.; Schneeweiss, A.; Huober, J.B.; Braun, M.; Rey, J.; Blohmer, J.U.; Furlanetto, J.; Zahm, D.M.; Hanusch, C.; Thomalla, J.; et al. Durvalumab improves long-term outcome in TNBC: Results from the phase II randomized GeparNUEVO study investigating neodjuvant durvalumab in addition to an anthracycline/taxane based neoadjuvant chemotherapy in early triple-negative breast cancer (TNBC). J. Clin. Oncol. 2021, 39, 506. [Google Scholar] [CrossRef]
- Roelofsen, L.; Kaptein, P.; Thommen, D. Multimodal predictors for precision immunotherapy. Immuno-Oncol. Technol. 2022, 14, 100071. [Google Scholar] [CrossRef]
- Sharma, P.; Siddiqui, B.A.; Anandhan, S.; Yadav, S.S.; Subudhi, S.K.; Gao, J.; Goswami, S.; Allison, J.P. The Next Decade of Immune Checkpoint Therapy. Cancer Discov. 2021, 11, 838–857. [Google Scholar] [CrossRef]
- Sharma, P.; Hu-Lieskovan, S.; Wargo, J.A.; Ribas, A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 2017, 168, 707–723. [Google Scholar] [CrossRef]
- Karasarides, M.; Cogdill, A.P.; Robbins, P.B.; Bowden, M.; Burton, E.M.; Butterfield, L.H.; Cesano, A.; Hammer, C.; Haymaker, C.L.; Horak, C.E.; et al. Hallmarks of Resistance to Immune-Checkpoint Inhibitors. Cancer Immunol. Res. 2022, 10, 372–383. [Google Scholar] [CrossRef]
- Giugliano, F.; Antonarelli, G.; Tarantino, P.; Cortes, J.; Rugo, H.S.; Curigliano, G. Harmonizing PD-L1 testing in metastatic triple negative breast cancer. Expert Opin. Biol. Ther. 2021, 22, 345–348. [Google Scholar] [CrossRef] [PubMed]
- Rugo, H.S.; Loi, S.; Adams, S.; Schmid, P.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Diéras, V.; Winer, E.P.; Kockx, M.M.; et al. PD-L1 Immunohistochemistry Assay Comparison in Atezolizumab Plus nab-Paclitaxel–Treated Advanced Triple-Negative Breast Cancer. JNCI J. Natl. Cancer Inst. 2021, 113, 1733–1743. [Google Scholar] [CrossRef]
- Miglietta, F.; Griguolo, G.; Guarneri, V.; Dieci, M.V. Programmed Cell Death Ligand 1 in Breast Cancer: Technical Aspects, Prognostic Implications, and Predictive Value. Oncologist 2019, 24, e1055–e1069. [Google Scholar] [CrossRef]
- Keren, L.; Bosse, M.; Marquez, D.; Angoshtari, R.; Jain, S.; Varma, S.; Yang, S.-R.; Kurian, A.; Van Valen, D.; West, R.; et al. A Structured Tumor-Immune Microenvironment in Triple Negative Breast Cancer Revealed by Multiplexed Ion Beam Imaging. Cell 2018, 174, 1373–1387.e19. [Google Scholar] [CrossRef] [PubMed]
- Dieci, M.V.; Guarneri, V.; Tosi, A.; Bisagni, G.; Musolino, A.; Spazzapan, S.; Moretti, G.; Vernaci, G.M.; Griguolo, G.; Giarratano, T.; et al. Neoadjuvant Chemotherapy and Immunotherapy in Luminal B-like Breast Cancer: Results of the Phase II GIADA Trial. Clin. Cancer Res. 2022, 28, 308–317. [Google Scholar] [CrossRef] [PubMed]
- El Bairi, K.; Haynes, H.R.; Blackley, E.; Fineberg, S.; Shear, J.; Turner, S.; de Freitas, J.R.; Sur, D.; Amendola, L.C.; Gharib, M.; et al. The Tale of TILs in Breast Cancer: A Report from The International Immuno-Oncology Biomarker Working Group. NPJ Breast Cancer 2021, 7, 150. [Google Scholar] [CrossRef]
- Schmid, P.; Salgado, R.; Park, Y.H.; Muñoz-Couselo, E.; Kim, S.B.; Sohn, J.; Im, S.A.; Foukakis, T.; Kuemmel, S.; Dent, R.; et al. Pembrolizumab plus Chemotherapy as Neoadjuvant Treatment of High-Risk, Early-Stage Tri-ple-Negative Breast Cancer: Results from the Phase 1b Open-Label, Multicohort KEYNOTE-173 Study. Ann. Oncol. 2020, 31, 569–581. [Google Scholar] [CrossRef]
- Winer, E.P.; Lipatov, O.; Im, S.-A.; Goncalves, A.; Muñoz-Couselo, E.; Lee, K.S.; Schmid, P.; Tamura, K.; Testa, L.; Witzel, I.; et al. Pembrolizumab versus investigator-choice chemotherapy for metastatic triple-negative breast cancer (KEYNOTE-119): A randomised, open-label, phase 3 trial. Lancet Oncol. 2021, 22, 499–511. [Google Scholar] [CrossRef]
- O’Meara, T.A.; Tolaney, S.M. Tumor mutational burden as a predictor of immunotherapy response in breast cancer. Oncotarget 2021, 12, 394–400. [Google Scholar] [CrossRef]
- Barroso-Sousa, R.; Li, T.; Reddy, S.; Emens, L.A.; Overmoyer, B.; Lange, P.; Dilullo, M.K.; Attaya, V.; Kimmel, J.; Winer, E.P.; et al. Abstract GS2-10: Nimbus: A phase 2 trial of nivolumab plus ipilimumab for patients with hypermutated her2-negative metastatic breast cancer (MBC). Cancer Res. 2022, 82, GS2-10. [Google Scholar] [CrossRef]
- Horimoto, Y.; Hlaing, M.T.; Saeki, H.; Kitano, S.; Nakai, K.; Sasaki, R.; Kurisaki-Arakawa, A.; Arakawa, A.; Otsuji, N.; Matsuoka, S.; et al. Microsatellite instability and mismatch repair protein expressions in lymphocyte-predominant breast cancer. Cancer Sci. 2020, 111, 2647–2654. [Google Scholar] [CrossRef]
- Barroso-Sousa, R.; Jain, E.; Cohen, O.; Kim, D.; Buendia-Buendia, J.; Winer, E.; Lin, N.; Tolaney, S.; Wagle, N. Prevalence and mutational determinants of high tumor mutation burden in breast cancer. Ann. Oncol. 2020, 31, 387–394. [Google Scholar] [CrossRef] [PubMed]
- Kossai, M.; Radosevic-Robin, N.; Penault-Llorca, F. Refining patient selection for breast cancer immunotherapy: Beyond PD-L1. ESMO Open 2021, 6. [Google Scholar] [CrossRef]
- McGrail, D.; Pilié, P.; Rashid, N.; Voorwerk, L.; Slagter, M.; Kok, M.; Jonasch, E.; Khasraw, M.; Heimberger, A.; Lim, B.; et al. High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann. Oncol. 2021, 32, 661–672. [Google Scholar] [CrossRef]
- Dieci, M.V.; Tsvetkova, V.; Griguolo, G.; Miglietta, F.; Tasca, G.; Giorgi, C.A.; Cumerlato, E.; Massa, D.; Mele, M.L.; Orvieto, E.; et al. Integration of tumour infiltrating lymphocytes, programmed cell-death ligand-1, CD8 and FOXP3 in prognostic models for triple-negative breast cancer: Analysis of 244 stage I–III patients treated with standard therapy. Eur. J. Cancer 2020, 136, 7–15. [Google Scholar] [CrossRef]
- Cristescu, R.; Mogg, R.; Ayers, M.; Albright, A.; Murphy, E.; Yearley, J.; Sher, X.; Liu, X.Q.; Lu, H.; Nebozhyn, M.; et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade–based immunotherapy. Science 2018, 362, eaar3593. [Google Scholar] [CrossRef]
- Montesion, M.; Murugesan, K.; Jin, D.X.; Sharaf, R.; Sanchez, N.; Guria, A.; Minker, M.; Li, G.; Fisher, V.; Sokol, E.S.; et al. Somatic HLA Class I Loss Is a Widespread Mechanism of Immune Evasion Which Refines the Use of Tumor Mutational Burden as a Biomarker of Checkpoint Inhibitor Response. Cancer Discov. 2021, 11, 282–292. [Google Scholar] [CrossRef] [PubMed]
- Loi, S.; Adams, S.; Schmid, P.; Cortés, J.; Cescon, D.W.; Winer, E.P.; Toppmeyer, D.L.; Rugo, H.S.; De Laurentiis, M.; Nanda, R.; et al. Relationship between Tumor Infiltrating Lymphocyte (TIL) Levels and Response to Pembrolizumab (Pembro) in Metastatic Triple-Negative Breast Cancer (mTNBC): Results from KEYNOTE-086. In Proceedings of the ESMO Annul Meeting, Madrid, Spain, 8–12 September 2017; Elsevier Inc.: Amsterdam, The Netherlands, 2017. [Google Scholar]
- Jackson, H.; Fischer, J.R.; Zanotelli, V.R.T.; Ali, H.R.; Mechera, R.; Soysal, S.D.; Moch, H.; Muenst, S.; Varga, Z.; Weber, W.P.; et al. The single-cell pathology landscape of breast cancer. Nature 2020, 578, 615–620. [Google Scholar] [CrossRef]
- Wu, S.Z.; Al-Eryani, G.; Roden, D.L.; Junankar, S.; Harvey, K.; Andersson, A.; Thennavan, A.; Wang, C.; Torpy, J.R.; Bartonicek, N.; et al. A single-cell and spatially resolved atlas of human breast cancers. Nat. Genet. 2021, 53, 1334–1347. [Google Scholar] [CrossRef]
- Azizi, E.; Carr, A.J.; Plitas, G.; Cornish, A.E.; Konopacki, C.; Prabhakaran, S.; Nainys, J.; Wu, K.; Kiseliovas, V.; Setty, M.; et al. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell 2018, 174, 1293–1308.e36. [Google Scholar] [CrossRef] [PubMed]
- Kieffer, Y.; Hocine, H.R.; Gentric, G.; Pelon, F.; Bernard, C.; Bourachot, B.; Lameiras, S.; Albergante, L.; Bonneau, C.; Guyard, A.; et al. Single-Cell Analysis Reveals Fibroblast Clusters Linked to Immunotherapy Resistance in Cancer. Cancer Discov. 2020, 10, 1330–1351. [Google Scholar] [CrossRef] [PubMed]
- Savas, P.; Virassamy, B.; Ye, C.; Salim, A.; Mintoff, C.P.; Caramia, F.; Salgado, R.; Byrne, D.J.; Teo, Z.L.; Dushyanthen, S.; et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat. Med. 2018, 24, 986–993. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Hong, Y.; Qi, P.; Lu, G.; Mai, X.; Xu, S.; He, X.; Guo, Y.; Gao, L.; Jing, Z.; et al. Atlas of breast cancer infiltrated B-lymphocytes revealed by paired single-cell RNA-sequencing and antigen receptor profiling. Nat. Commun. 2021, 12, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.R.; Jackson, H.W.; Zanotelli, V.R.T.; Danenberg, E.; Fischer, J.R.; Bardwell, H.; Provenzano, E.; Cruk Imaxt Grand Challenge Team; Rueda, O.M.; Chin, S.-F.; et al. Imaging mass cytometry and multiplatform genomics define the phenogenomic landscape of breast cancer. Nat. Cancer 2020, 1, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Barkley, D.; Moncada, R.; Pour, M.; Liberman, D.A.; Dryg, I.; Werba, G.; Wang, W.; Baron, M.; Rao, A.; Xia, B.; et al. Cancer cell states recur across tumor types and form specific interactions with the tumor microenvironment. Nat. Genet. 2022, 54, 1192–1201. [Google Scholar] [CrossRef] [PubMed]
- Lewis, S.M.; Asselin-Labat, M.-L.; Nguyen, Q.; Berthelet, J.; Tan, X.; Wimmer, V.C.; Merino, D.; Rogers, K.L.; Naik, S.H. Spatial omics and multiplexed imaging to explore cancer biology. Nat. Methods 2021, 18, 997–1012. [Google Scholar] [CrossRef] [PubMed]
- Zahir, N.; Sun, R.; Gallahan, D.; Gatenby, R.A.; Curtis, C. Characterizing the ecological and evolutionary dynamics of cancer. Nat. Genet. 2020, 52, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Gruosso, T.; Gigoux, M.; Manem, V.S.K.; Bertos, N.; Zuo, D.; Perlitch, I.; Saleh, S.M.I.; Zhao, H.; Souleimanova, M.; Johnson, R.M.; et al. Spatially distinct tumor immune microenvironments stratify triple-negative breast cancers. J. Clin. Investig. 2019, 129, 1785–1800. [Google Scholar] [CrossRef]
- Hammerl, D.; Martens, J.W.M.; Timmermans, M.; Smid, M.; Trapman-Jansen, A.M.; Foekens, R.; Isaeva, O.I.; Voorwerk, L.; Balcioglu, H.E.; Wijers, R.; et al. Spatial immunophenotypes predict response to anti-PD1 treatment and capture distinct paths of T cell evasion in triple negative breast cancer. Nat. Commun. 2021, 12, 1–13. [Google Scholar] [CrossRef]
- Baertsch, M.-A.; Nolan, G.P.; Hickey, J.W. Multicellular modules as clinical diagnostic and therapeutic targets. Trends Cancer 2021, 8, 164–173. [Google Scholar] [CrossRef]
- Albers, J.J.; Pelka, K. Listening in on Multicellular Communication in Human Tissue Immunology. Front. Immunol. 2022, 13. [Google Scholar] [CrossRef]
- Fu, T.; Dai, L.-J.; Wu, S.-Y.; Xiao, Y.; Ma, D.; Jiang, Y.-Z.; Shao, Z.-M. Spatial architecture of the immune microenvironment orchestrates tumor immunity and therapeutic response. J. Hematol. Oncol. 2021, 14, 1–25. [Google Scholar] [CrossRef]
- Hickey, J.W.; Neumann, E.K.; Radtke, A.J.; Camarillo, J.M.; Beuschel, R.T.; Albanese, A.; McDonough, E.; Hatler, J.; Wiblin, A.E.; Fisher, J.; et al. Spatial mapping of protein composition and tissue organization: A primer for multiplexed antibody-based imaging. Nat. Methods 2021, 19, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Tzoras, E.; Zerdes, I.; Tsiknakis, N.; Manikis, G.C.; Mezheyeuski, A.; Bergh, J.; Matikas, A.; Foukakis, T. Dissecting Tumor-Immune Microenvironment in Breast Cancer at a Spatial and Multiplex Resolution. Cancers 2022, 14, 1999. [Google Scholar] [CrossRef] [PubMed]
- Taube, J.M.; Akturk, G.; Angelo, M.; Engle, E.L.; Gnjatic, S.; Greenbaum, S.; Greenwald, N.F.; Hedvat, C.V.; Hollmann, T.J.; Juco, J.; et al. The Society for Immunotherapy of Cancer statement on best practices for multiplex immunohistochemistry (IHC) and immunofluorescence (IF) staining and validation. J. Immunother. Cancer 2020, 8, e000155. [Google Scholar] [CrossRef] [PubMed]
- Berry, S.; Giraldo, N.A.; Green, B.F.; Cottrell, T.R.; Stein, J.E.; Engle, E.L.; Xu, H.; Ogurtsova, A.; Roberts, C.; Wang, D.; et al. Analysis of multispectral imaging with the AstroPath platform informs efficacy of PD-1 blockade. Science 2021, 372. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Stein, J.E.; Rimm, D.L.; Wang, D.W.; Bell, J.M.; Johnson, D.B.; Sosman, J.A.; Schalper, K.A.; Anders, R.A.; Wang, H.; et al. Comparison of Biomarker Modalities for Predicting Response to PD-1/PD-L1 Checkpoint Blockade: A Systematic Review and Meta-Analysis. JAMA Oncol. 2019, 5, 1195–1204. [Google Scholar] [CrossRef] [PubMed]
- Ogbeide, S.; Giannese, F.; Mincarelli, L.; Macaulay, I.C. Into the multiverse: Advances in single-cell multiomic profiling. Trends Genet. 2022, 38, 831–843. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Cheng, Y.; Wang, X.; Fan, J.; Gao, Q. Spatial omics: Navigating to the golden era of cancer research. Clin. Transl. Med. 2022, 12, e696. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Leone, P.; Shin, E.-C.; Perosa, F.; Vacca, A.; Dammacco, F.; Racanelli, V. MHC Class I Antigen Processing and Presenting Machinery: Organization, Function, and Defects in Tumor Cells. JNCI J. Natl. Cancer Inst. 2013, 105, 1172–1187. [Google Scholar] [CrossRef]
- Dhatchinamoorthy, K.; Colbert, J.D.; Rock, K.L. Cancer Immune Evasion Through Loss of MHC Class I Antigen Presentation. Front. Immunol. 2021, 12. [Google Scholar] [CrossRef]
- Maleno, I.; Aptsiauri, N.; Cabrera, T.; Gallego, A.; Paschen, A.; López-Nevot, M.A.; Garrido, F. Frequent loss of heterozygosity in the β2-microglobulin region of chromosome 15 in primary human tumors. Immunogenetics 2010, 63, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Inoue, M.; Mimura, K.; Izawa, S.; Shiraishi, K.; Inoue, A.; Shiba, S.; Watanabe, M.; Maruyama, T.; Kawaguchi, Y.; Inoue, S.; et al. Expression of MHC Class I on breast cancer cells correlates inversely with HER2 expression. OncoImmunology 2012, 1, 1104–1110. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Shklovskaya, E.; Lim, S.Y.; Carlino, M.S.; Menzies, A.M.; Stewart, A.; Pedersen, B.; Irvine, M.; Alavi, S.; Yang, J.Y.H.; et al. Transcriptional downregulation of MHC class I and melanoma de- differentiation in resistance to PD-1 inhibition. Nat. Commun. 2020, 11, 1–12. [Google Scholar] [CrossRef]
- Berglund, A.K.; Long, J.M.; Schnabel, L.V. TGF-β Downregulates MHC I Surface Expression through a Smad3-Dependent Mechanism. J. Immunol. 2020, 204, 140.2. [Google Scholar]
- Lehmann, B.D.; Colaprico, A.; Silva, T.C.; Chen, J.; An, H.; Ban, Y.; Huang, H.; Wang, L.; James, J.L.; Balko, J.M.; et al. Multi-omics analysis identifies therapeutic vulnerabilities in triple-negative breast cancer subtypes. Nat. Commun. 2021, 12, 1–18. [Google Scholar] [CrossRef]
- Fang, Y.; Wang, L.; Wan, C.; Sun, Y.; Van der Jeught, K.; Zhou, Z.; Dong, T.; So, K.M.; Yu, T.; Li, Y.; et al. MAL2 drives immune evasion in breast cancer by suppressing tumor antigen presentation. J. Clin. Investig. 2021, 131. [Google Scholar] [CrossRef]
- Hazini, A.; Fisher, K.; Seymour, L. Deregulation of HLA-I in cancer and its central importance for immunotherapy. J. Immunother. Cancer 2021, 9, e002899. [Google Scholar] [CrossRef]
- Park, I.A.; Hwang, S.-H.; Song, I.H.; Heo, S.-H.; Kim, Y.-A.; Bang, W.S.; Park, H.S.; Lee, M.; Gong, G.; Lee, H.J. Expression of the MHC class II in triple-negative breast cancer is associated with tumor-infiltrating lymphocytes and interferon signaling. PLoS ONE 2017, 12, e0182786. [Google Scholar] [CrossRef]
- Sade-Feldman, M.; Jiao, Y.J.; Chen, J.H.; Rooney, M.S.; Barzily-Rokni, M.; Eliane, J.-P.; Bjorgaard, S.L.; Hammond, M.R.; Vitzthum, H.; Blackmon, S.M.; et al. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat. Commun. 2017, 8, 1–11. [Google Scholar] [CrossRef]
- Sinn, B.V.; Weber, K.E.; Schmitt, W.; Fasching, P.A.; Symmans, W.F.; Blohmer, J.-U.; Karn, T.; Taube, E.T.; Klauschen, F.; Marmé, F.; et al. Human leucocyte antigen class I in hormone receptor-positive, HER2-negative breast cancer: Association with response and survival after neoadjuvant chemotherapy. Breast Cancer Res. 2019, 21, 1–9. [Google Scholar] [CrossRef]
- Muntasell, A.; Rojo, F.; Servitja, S.; Rubio-Perez, C.; Cabo, M.; Tamborero, D.; Costa-García, M.; Martínez-Garcia, M.; Menéndez, S.; Vazquez, I.; et al. NK Cell Infiltrates and HLA Class I Expression in Primary HER2+ Breast Cancer Predict and Uncouple Pathological Response and Disease-free Survival. Clin. Cancer Res. 2019, 25, 1535–1545. [Google Scholar] [CrossRef] [PubMed]
- Tsang, J.Y.; Ho, C.-S.; Ni, Y.-B.; Shao, Y.; Poon, I.K.; Chan, S.-K.; Cheung, S.-Y.; Shea, K.-H.; Marabi, M.; Tse, G.M. Co-expression of HLA-I loci improved prognostication in HER2+ breast cancers. Cancer Immunol. Immunother. 2020, 69, 799–811. [Google Scholar] [CrossRef]
- Bagaev, A.; Kotlov, N.; Nomie, K.; Svekolkin, V.; Gafurov, A.; Isaeva, O.; Osokin, N.; Kozlov, I.; Frenkel, F.; Gancharova, O.; et al. Conserved pan-cancer microenvironment subtypes predict response to immunotherapy. Cancer Cell 2021, 39, 845–865.e7. [Google Scholar] [CrossRef] [PubMed]
- Patwa, A.; Yamashita, R.; Long, J.; Risom, T.; Angelo, M.; Keren, L.; Rubin, D.L. Multiplexed imaging analysis of the tumor-immune microenvironment reveals predictors of outcome in triple-negative breast cancer. Commun. Biol. 2021, 4, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Axelrod, M.L.; Cook, R.S.; Johnson, D.B.; Balko, J.M. Biological Consequences of MHC-II Expression by Tumor Cells in Cancer. Clin. Cancer Res. 2019, 25, 2392–2402. [Google Scholar] [CrossRef]
- Asleh, K.; Negri, G.L.; Miko, S.E.S.; Colborne, S.; Hughes, C.S.; Wang, X.Q.; Gao, D.; Gilks, C.B.; Chia, S.K.L.; Nielsen, T.O.; et al. Proteomic analysis of archival breast cancer clinical specimens identifies biological subtypes with distinct survival outcomes. Nat. Commun. 2022, 13, 1–19. [Google Scholar] [CrossRef]
- Datar, I.J.; Hauc, S.C.; Desai, S.S.; Gianino, N.; Henick, B.S.; Liu, Y.; Syrigos, K.N.; Rimm, D.L.; Kavathas, P.; Ferrone, S.; et al. Spatial Analysis and Clinical Significance of HLA Class-I and Class-II Subunit Expression in Non–Small Cell Lung Cancer. Clin. Cancer Res. 2021, 27, 2837–2847. [Google Scholar] [CrossRef]
- Rodig, S.J.; Gusenleitner, D.; Jackson, D.G.; Gjini, E.; Giobbie-Hurder, A.; Jin, C.; Chang, H.; Lovitch, S.B.; Horak, C.; Weber, J.S.; et al. MHC proteins confer differential sensitivity to CTLA-4 and PD-1 blockade in untreated metastatic melanoma. Sci. Transl. Med. 2018, 10, eaar3342. [Google Scholar] [CrossRef]
- Johnson, D.B.; Estrada, M.V.; Salgado, R.; Sanchez, V.; Doxie, D.; Opalenik, S.R.; Vilgelm, A.; Feld, E.; Johnson, A.S.; Greenplate, A.R.; et al. Melanoma-specific MHC-II expression represents a tumour-autonomous phenotype and predicts response to anti-PD-1/PD-L1 therapy. Nat. Commun. 2016, 7, 10582. [Google Scholar] [CrossRef]
- Johnson, A.M.; Boland, J.M.; Wrobel, J.; Klezcko, E.K.; Weiser-Evans, M.; Hopp, K.; Heasley, L.; Clambey, E.T.; Jordan, K.; Nemenoff, R.A.; et al. Cancer Cell-Specific Major Histocompatibility Complex II Expression as a Determinant of the Immune Infiltrate Organization and Function in the NSCLC Tumor Microenvironment. J. Thorac. Oncol. 2021, 16, 1694–1704. [Google Scholar] [CrossRef]
- Gonzalez-Ericsson, P.I.; Wulfkhule, J.D.; Gallagher, R.I.; Sun, X.; Axelrod, M.L.; Sheng, Q.; Luo, N.; Gomez, H.; Sanchez, V.; Sanders, M.; et al. Tumor-Specific Major Histocompatibility-II Expression Predicts Benefit to Anti–PD-1/L1 Therapy in Patients With HER2-Negative Primary Breast Cancer. Clin. Cancer Res. 2021, 27, 5299–5306. [Google Scholar] [CrossRef] [PubMed]
- Bianchini, G.; Wang, X.Q.; Danenberg, E.; Huang, C.-S.; Egle, D.; Callari, M.; Bermejo, B.; Zamagni, C.; Thill, M.; Anton, A.; et al. Abstract GS1-00: Single-cell spatial analysis by imaging mass cytometry and immunotherapy response in triple-negative breast cancer (TNBC) in the NeoTRIPaPDL1 trial. Cancer Res. 2022, 82, GS1-00. [Google Scholar] [CrossRef]
- Mostafa, A.; Codner, D.; Hirasawa, K.; Komatsu, Y.; Young, M.N.; Steimle, V.; Drover, S. Activation of ERα Signaling Differentially Modulates IFN-γ Induced HLA-Class II Expression in Breast Cancer Cells. PLoS ONE 2014, 9, e87377. [Google Scholar] [CrossRef] [PubMed]
- Dieci, M.V.; Griguolo, G.; Miglietta, F.; Guarneri, V. The Immune System and Hormone-Receptor Positive Breast Cancer: Is It Really a Dead End? Cancer Treat. Rev. 2016, 46, 9–19. [Google Scholar] [CrossRef]
- Johnson, D.B.; Nixon, M.J.; Wang, Y.; Wang, D.Y.; Castellanos, E.; Estrada, M.V.; Ericsson-Gonzalez, P.I.; Cote, C.H.; Salgado, R.; Sanchez, V.; et al. Tumor-specific MHC-II expression drives a unique pattern of resistance to immunotherapy via LAG-3/FCRL6 engagement. JCI Insight 2018, 3. [Google Scholar] [CrossRef]
- Alspach, E.; Lussier, D.M.; Miceli, A.P.; Kizhvatov, I.; DuPage, M.; Luoma, A.M.; Meng, W.; Lichti, C.F.; Esaulova, E.; Vomund, A.N.; et al. MHC-II neoantigens shape tumour immunity and response to immunotherapy. Nature 2019, 574, 696–701. [Google Scholar] [CrossRef]
- Cachot, A.; Bilous, M.; Liu, Y.-C.; Li, X.; Saillard, M.; Cenerenti, M.; Rockinger, G.A.; Wyss, T.; Guillaume, P.; Schmidt, J.; et al. Tumor-specific cytolytic CD4 T cells mediate immunity against human cancer. Sci. Adv. 2021, 7, eabe3348. [Google Scholar] [CrossRef]
- Quezada, S.; Simpson, T.R.; Peggs, K.S.; Merghoub, T.; Vider, J.; Fan, X.; Blasberg, R.; Yagita, H.; Muranski, P.; Antony, P.A.; et al. Tumor-reactive CD4+ T cells develop cytotoxic activity and eradicate large established melanoma after transfer into lymphopenic hosts. J. Exp. Med. 2010, 207, 637–650. [Google Scholar] [CrossRef]
- Forero, A.; Li, Y.; Chen, D.; Grizzle, W.E.; Updike, K.L.; Merz, N.D.; Downs-Kelly, E.; Burwell, T.C.; Vaklavas, C.; Buchsbaum, D.J.; et al. Expression of the MHC Class II Pathway in Triple-Negative Breast Cancer Tumor Cells Is Associated with a Good Prognosis and Infiltrating Lymphocytes. Cancer Immunol. Res. 2016, 4, 390–399. [Google Scholar] [CrossRef]
- Loi, S.; Dushyanthen, S.; Beavis, P.A.; Salgado, R.; Denkert, C.; Savas, P.; Combs, S.; Rimm, D.L.; Giltnane, J.M.; Estrada, M.V.; et al. RAS/MAPK Activation Is Associated with Reduced Tumor-Infiltrating Lymphocytes in Triple-Negative Breast Cancer: Therapeutic Cooperation Between MEK and PD-1/PD-L1 Immune Checkpoint Inhibitors. Clin. Cancer Res. 2016, 22, 1499–1509. [Google Scholar] [CrossRef]
- Carter, J.M.; Polley, M.Y.C.; Leon-Ferre, R.A.; Sinnwell, J.; Thompson, K.J.; Wang, X.; Ma, Y.; Zahrieh, D.; Ka-chergus, J.M.; Solanki, M.; et al. Characteristics and Spatially Defined Immune (Micro)Landscapes of Ear-ly-Stage PD-L1–Positive Triple-Negative Breast Cancer. Clin. Cancer Res. 2021, 27, 5628–5637. [Google Scholar] [CrossRef]
- Corti, C.; Antonarelli, G.; Valenza, C.; Nicolò, E.; Rugo, H.; Cortés, J.; Harbeck, N.; Carey, L.A.; Criscitiello, C.; Curigliano, G. Histology-Agnostic Approvals for Antibody–Drug Conjugates in Solid Tumours: Is the Time Ripe? Eur. J. Cancer 2022, 171, 25–42. [Google Scholar] [CrossRef]
- Modi, S.; Jacot, W.; Yamashita, T.; Sohn, J.; Vidal, M.; Tokunaga, E.; Tsurutani, J.; Ueno, N.T.; Prat, A.; Chae, Y.S.; et al. Trastuzumab Deruxtecan in Previously Treated HER2-Low Advanced Breast Cancer. N. Engl. J. Med. 2022, 387, 9–20. [Google Scholar] [CrossRef]
- Mosele, M.F.; Lusque, A.; Dierasa, V.; Deluche, E. Unraveling the Mechanism of Action and Resistance to Trastuzumab Deruxtecan (T-DXd): Biomarker Analyses from Patients from DAISY Trial. Ann. Oncol. 2022, 33 (Suppl. S3), S123–S147. [Google Scholar] [CrossRef]
- Rosenberg, J.E.; O’donnell, P.H.; Balar, A.V.; Mcgregor, B.A.; Heath, E.I.; Yu, E.Y.; Galsky, M.D.; Hahn, N.M.; Gartner, E.M.; Pinelli, J.M.; et al. Pivotal Trial of Enfortumab Vedotin in Urothelial Carcinoma After Platinum and Anti-Programmed Death 1/Programmed Death Ligand 1 Therapy. J. Clin. Oncol. 2019, 37, 2592–2600. [Google Scholar] [CrossRef]
- Bardia, A.; Tolaney, S.M.; Punie, K.; Loirat, D.; Oliveira, M.; Kalinsky, K.; Zelnak, A.; Aftimos, P.; Dalenc, F.; Sardesai, S.; et al. Biomarker analyses in the phase III ASCENT study of sacituzumab govitecan versus chemotherapy in patients with metastatic triple-negative breast cancer. Ann. Oncol. 2021, 32, 1148–1156. [Google Scholar] [CrossRef]
- Clark, A.S.; Yau, C.; Wolf, D.M.; Petricoin, E.F.; Veer, L.J.V.; Yee, D.; Moulder, S.L.; Wallace, A.M.; Chien, A.J.; Isaacs, C.; et al. Neoadjuvant T-DM1/pertuzumab and paclitaxel/trastuzumab/pertuzumab for HER2+ breast cancer in the adaptively randomized I-SPY2 trial. Nat. Commun. 2021, 12, 1–11. [Google Scholar] [CrossRef]
- Filho, O.M.; Viale, G.; Trippa, L.; Li, T.; Yardley, D.A.; Mayer, I.A.; Abramson, V.G.; Arteaga, C.L.; Spring, L.; Waks, A.G.; et al. HER2 heterogeneity as a predictor of response to neoadjuvant T-DM1 plus pertuzumab: Results from a prospective clinical trial. J. Clin. Oncol. 2019, 37, 502. [Google Scholar] [CrossRef]
- Moutafi, M.; Robbins, C.J.; Yaghoobi, V.; Fernandez, A.I.; Martinez-Morilla, S.; Xirou, V.; Bai, Y.; Song, Y.; Gaule, P.; Krueger, J.; et al. Quantitative measurement of HER2 expression to subclassify ERBB2 unamplified breast cancer. Lab. Investig. 2022, 1–8. [Google Scholar] [CrossRef]
- Drago, J.Z.; Modi, S.; Chandarlapaty, S. Unlocking the potential of antibody–drug conjugates for cancer therapy. Nat. Rev. Clin. Oncol. 2021, 18, 327–344. [Google Scholar] [CrossRef] [PubMed]
- Musolino, A.; Gradishar, W.J.; Rugo, H.S.; Nordstrom, J.L.; Rock, E.P.; Arnaldez, F.; Pegram, M.D. Role of Fcγ receptors in HER2-targeted breast cancer therapy. J. Immunother. Cancer 2022, 10, e003171. [Google Scholar] [CrossRef] [PubMed]
- Schmid, P.; Im, S.-A.; Armstrong, A.; Park, Y.H.; Chung, W.-P.; Nowecki, Z.; Lord, S.; Wysocki, P.J.; Lu, Y.-S.; Dry, H.; et al. BEGONIA: Phase 1b/2 study of durvalumab (D) combinations in locally advanced/metastatic triple-negative breast cancer (TNBC)—Initial results from arm 1, d+paclitaxel (P), and arm 6, d+trastuzumab deruxtecan (T-DXd). J. Clin. Oncol. 2021, 39, 1023. [Google Scholar] [CrossRef]
- Schmid, P.; Jung, K.; Wysocki, P.; Jassem, J.; Ma, C.; Fernandes, R.; Huisden, R.; Stewart, R.; Vukovic, P.; Nunes, A.T.; et al. 166MO Datopotamab deruxtecan (Dato-DXd) + durvalumab (D) as first-line (1L) treatment for unresectable locally advanced/metastatic triple-negative breast cancer (a/mTNBC): Initial results from BEGONIA, a phase Ib/II study. Ann. Oncol. 2022, 33, S199. [Google Scholar] [CrossRef]
- Philip, M.; Schietinger, A. CD8+ T cell differentiation and dysfunction in cancer. Nat. Rev. Immunol. 2021, 22, 209–223. [Google Scholar] [CrossRef]
- Kroemer, G.; Galluzzi, L.; Kepp, O.; Zitvogel, L. Immunogenic Cell Death in Cancer Therapy. Annu. Rev. Immunol. 2013, 31, 51–72. [Google Scholar] [CrossRef]
- Zitvogel, L.; Kepp, O.; Kroemer, G. Immune parameters affecting the efficacy of chemotherapeutic regimens. Nat. Rev. Clin. Oncol. 2011, 8, 151–160. [Google Scholar] [CrossRef]
- Galluzzi, L.; Zitvogel, L.; Kroemer, G. Immunological Mechanisms Underneath the Efficacy of Cancer Therapy. Cancer Immunol. Res. 2016, 4, 895–902. [Google Scholar] [CrossRef] [PubMed]
- Voorwerk, L.; Slagter, M.; Horlings, H.M.; Sikorska, K.; Van De Vijver, K.K.; De Maaker, M.; Nederlof, I.; Kluin, R.J.C.; Warren, S.; Ong, S.; et al. Immune induction strategies in metastatic triple-negative breast cancer to enhance the sensitivity to PD-1 blockade: The TONIC trial. Nat. Med. 2019, 25, 920–928. [Google Scholar] [CrossRef] [PubMed]
- Killock, D. Chemotherapy as a TONIC to invigorate PD-1 inhibition in TNBC. Nat. Rev. Clin. Oncol. 2019, 16, 464. [Google Scholar] [CrossRef]
- Parkes, E.E.; Humphries, M.P.; Gilmore, E.; Sidi, F.A.; Bingham, V.; Phyu, S.M.; Craig, S.; Graham, C.; Miller, J.; Griffin, D.; et al. The clinical and molecular significance associated with STING signaling in breast cancer. NPJ Breast Cancer 2021, 7, 1–11. [Google Scholar] [CrossRef]
- Kroemer, G.; Galassi, C.; Zitvogel, L.; Galluzzi, L. Immunogenic cell stress and death. Nat. Immunol. 2022, 23, 487–500. [Google Scholar] [CrossRef]
- Reis, E.S.; Mastellos, D.; Ricklin, D.; Mantovani, A.; Lambris, J.D. Complement in cancer: Untangling an intricate relationship. Nat. Rev. Immunol. 2017, 18, 5–18. [Google Scholar] [CrossRef]
- Lu, Y.; Zhao, Q.; Liao, J.-Y.; Song, E.; Xia, Q.; Pan, J.; Li, Y.; Li, J.; Zhou, B.; Ye, Y.; et al. Complement Signals Determine Opposite Effects of B Cells in Chemotherapy-Induced Immunity. Cell 2020, 180, 1081–1097.e24. [Google Scholar] [CrossRef]
- Chen, S.; Lai, S.W.T.; Brown, C.E.; Feng, M. Harnessing and Enhancing Macrophage Phagocytosis for Cancer Therapy. Front. Immunol. 2021, 12. [Google Scholar] [CrossRef]
- Samanta, D.; Park, Y.; Ni, X.; Li, H.; Zahnow, C.A.; Gabrielson, E.; Pan, F.; Semenza, G.L. Chemotherapy induces enrichment of CD47+/CD73+/PDL1+ immune evasive triple-negative breast cancer cells. Proc. Natl. Acad. Sci. USA 2018, 115, E1239–E1248. [Google Scholar] [CrossRef]
- Tsao, L.-C.; Crosby, E.J.; Trotter, T.N.; Agarwal, P.; Hwang, B.-J.; Acharya, C.; Shuptrine, C.W.; Wang, T.; Wei, J.; Yang, X.; et al. CD47 blockade augmentation of trastuzumab antitumor efficacy dependent on antibody-dependent cellular phagocytosis. JCI Insight 2019, 4. [Google Scholar] [CrossRef]
- Candas-Green, D.; Xie, B.; Huang, J.; Fan, M.; Wang, A.; Menaa, C.; Zhang, Y.; Zhang, L.; Jing, D.; Azghadi, S.; et al. Dual blockade of CD47 and HER2 eliminates radioresistant breast cancer cells. Nat. Commun. 2020, 11, 1–15. [Google Scholar] [CrossRef]
- Yuan, J.; He, H.; Chen, C.; Wu, J.; Rao, J.; Yan, H. Combined high expression of CD47 and CD68 is a novel prognostic factor for breast cancer patients. Cancer Cell Int. 2019, 19, 1–12. [Google Scholar] [CrossRef]
- Jiang, T.; Xu, X.; Qiao, M.; Li, X.; Zhao, C.; Zhou, F.; Gao, G.; Wu, F.; Chen, X.; Su, C.; et al. Comprehensive evaluation of NT5E/CD73 expression and its prognostic significance in distinct types of cancers. BMC Cancer 2018, 18, 1–10. [Google Scholar] [CrossRef]
- Petruk, N.; Tuominen, S.; Åkerfelt, M.; Mattsson, J.; Sandholm, J.; Nees, M.; Yegutkin, G.G.; Jukkola, A.; Tuomela, J.; Selander, K.S. CD73 facilitates EMT progression and promotes lung metastases in triple-negative breast cancer. Sci. Rep. 2021, 11, 1–13. [Google Scholar] [CrossRef]
- Buisseret, L.; Pommey, S.; Allard, B.; Garaud, S.; Bergeron, M.; Cousineau, I.; Ameye, L.; Bareche, Y.; Paesmans, M.; Crown, J.; et al. Clinical significance of CD73 in triple-negative breast cancer: Multiplex analysis of a phase III clinical trial. Ann. Oncol. 2017, 29, 1056–1062. [Google Scholar] [CrossRef]
- Cerbelli, B.; Botticelli, A.; Pisano, A.; Pernazza, A.; Campagna, D.; De Luca, A.; Ascierto, P.A.; Pignataro, M.G.; Pelullo, M.; Della Rocca, C.; et al. CD73 expression and pathologic response to neoadjuvant chemotherapy in triple negative breast cancer. Virchows Arch. 2019, 476, 569–576. [Google Scholar] [CrossRef]
- Turcotte, M.; Allard, D.; Mittal, D.; Bareche, Y.; Buisseret, L.; José, V.; Pommey, S.; Delisle, V.; Loi, S.; Joensuu, H.; et al. CD73 Promotes Resistance to HER2/ErbB2 Antibody Therapy. Cancer Res. 2017, 77, 5652–5663. [Google Scholar] [CrossRef]
- Wennerberg, E.; Spada, S.; Rudqvist, N.-P.; Lhuillier, C.; Gruber, S.; Chen, Q.; Zhang, F.; Zhou, X.K.; Gross, S.S.; Formenti, S.C.; et al. CD73 Blockade Promotes Dendritic Cell Infiltration of Irradiated Tumors and Tumor Rejection. Cancer Immunol. Res. 2020, 8, 465–478. [Google Scholar] [CrossRef]
- Roberts, E.W.; Broz, M.L.; Binnewies, M.; Headley, M.B.; Nelson, A.E.; Wolf, D.M.; Kaisho, T.; Bogunovic, D.; Bhardwaj, N.; Krummel, M.F. Critical Role for CD103(+)/CD141(+) Dendritic Cells Bearing CCR7 for Tumor Antigen Trafficking and Priming of T Cell Immunity in Melanoma. Cancer Cell 2016, 30, 324–336. [Google Scholar] [CrossRef]
- Spranger, S.; Dai, D.; Horton, B.; Gajewski, T.F. Tumor-Residing Batf3 Dendritic Cells Are Required for Effector T Cell Trafficking and Adoptive T Cell Therapy. Cancer Cell 2017, 31, 711–723.e4. [Google Scholar] [CrossRef]
- Eisenbarth, S.C. Dendritic cell subsets in T cell programming: Location dictates function. Nat. Rev. Immunol. 2018, 19, 89–103. [Google Scholar] [CrossRef]
- Böttcher, J.P.; Bonavita, E.; Chakravarty, P.; Blees, H.; Cabeza-Cabrerizo, M.; Sammicheli, S.; Rogers, N.C.; Sahai, E.; Zelenay, S.; e Sousa, C.R. NK Cells Stimulate Recruitment of cDC1 into the Tumor Microenvironment Promoting Cancer Immune Control. Cell 2018, 172, 1022–1037.e14. [Google Scholar] [CrossRef]
- Oh, S.A.; Wu, D.-C.; Cheung, J.; Navarro, A.; Xiong, H.; Cubas, R.; Totpal, K.; Chiu, H.; Wu, Y.; Comps-Agrar, L.; et al. PD-L1 expression by dendritic cells is a key regulator of T-cell immunity in cancer. Nat. Cancer 2020, 1, 681–691. [Google Scholar] [CrossRef]
- Barry, K.C.; Hsu, J.; Broz, M.L.; Cueto, F.J.; Binnewies, M.; Combes, A.J.; Nelson, A.E.; Loo, K.; Kumar, R.; Rosenblum, M.D.; et al. A natural killer–dendritic cell axis defines checkpoint therapy–responsive tumor microenvironments. Nat. Med. 2018, 24, 1178–1191. [Google Scholar] [CrossRef]
- Ovcinnikovs, V.; Ross, E.M.; Petersone, L.; Edner, N.M.; Heuts, F.; Ntavli, E.; Kogimtzis, A.; Kennedy, A.; Wang, C.J.; Bennett, C.L.; et al. CTLA-4–mediated transendocytosis of costimulatory molecules primarily targets migratory dendritic cells. Sci. Immunol. 2019, 4, e124716. [Google Scholar] [CrossRef]
- Schenkel, J.M.; Herbst, R.H.; Canner, D.; Li, A.; Hillman, M.; Shanahan, S.-L.; Gibbons, G.; Smith, O.C.; Kim, J.Y.; Westcott, P.; et al. Conventional type I dendritic cells maintain a reservoir of proliferative tumor-antigen specific TCF-1+ CD8+ T cells in tumor-draining lymph nodes. Immunity 2021, 54, 2338–2353.e6. [Google Scholar] [CrossRef]
- Connolly, K.A.; Kuchroo, M.; Venkat, A.; Khatun, A.; Wang, J.; William, I.; Hornick, N.I.; Fitzgerald, B.L.; Damo, M.; Kasmani, M.Y.; et al. A reservoir of stem-like CD8+ T cells in the tumor-draining lymph node preserves the ongoing antitumor immune response. Sci. Immunol. 2021, 6, eabg7836. [Google Scholar] [CrossRef]
- Dammeijer, F.; van Gulijk, M.; Mulder, E.E.; Lukkes, M.; Klaase, L.; Bosch, T.V.D.; van Nimwegen, M.; Lau, S.P.; Latupeirissa, K.; Schetters, S.; et al. The PD-1/PD-L1-Checkpoint Restrains T cell Immunity in Tumor-Draining Lymph Nodes. Cancer Cell 2020, 38, 685–700.e8. [Google Scholar] [CrossRef]
- Noël, G.; Fontsa, M.L.; Garaud, S.; De Silva, P.; de Wind, A.; Eynden, G.G.V.D.; Salgado, R.; Boisson, A.; Locy, H.; Thomas, N.; et al. Functional Th1-oriented T follicular helper cells that infiltrate human breast cancer promote effective adaptive immunity. J. Clin. Investig. 2021, 131. [Google Scholar] [CrossRef]
- Trüb, M.; Zippelius, A. Tertiary Lymphoid Structures as a Predictive Biomarker of Response to Cancer Immunotherapies. Front Immunol 2021, 12, 1432. [Google Scholar] [CrossRef]
- Duraiswamy, J.; Turrini, R.; Minasyan, A.; Barras, D.; Crespo, I.; Grimm, A.J.; Casado, J.; Genolet, R.; Benedetti, F.; Wicky, A.; et al. Myeloid antigen-presenting cell niches sustain antitumor T cells and license PD-1 blockade via CD28 costimulation. Cancer Cell 2021, 39, 1623–1642.e20. [Google Scholar] [CrossRef]
- Yost, K.E.; Chang, H.Y.; Satpathy, A.T. Recruiting T cells in cancer immunotherapy. Science 2021, 372, 130–131. [Google Scholar] [CrossRef]
- Lam, A.; Heit, B. Having an Old Friend for Dinner: The Interplay between Apoptotic Cells and Efferocytes. Cells 2021, 10, 1265. [Google Scholar] [CrossRef]
- Jamiyan, T.; Kuroda, H.; Yamaguchi, R.; Abe, A.; Hayashi, M. CD68- and CD163-positive tumor-associated macrophages in triple negative cancer of the breast. Virchows Arch. 2020, 477, 767–775. [Google Scholar] [CrossRef]
- Sousa, S.; Brion, R.; Lintunen, M.; Kronqvist, P.; Sandholm, J.; Mönkkönen, J.; Kellokumpu-Lehtinen, P.-L.; Lauttia, S.; Tynninen, O.; Joensuu, H.; et al. Human breast cancer cells educate macrophages toward the M2 activation status. Breast Cancer Res. 2015, 17, 1–14. [Google Scholar] [CrossRef]
- A Mahmoud, S.M.; Lee, A.H.S.; Paish, E.C.; Macmillan, R.D.; O Ellis, I.; Green, A.R. Tumour-infiltrating macrophages and clinical outcome in breast cancer. J. Clin. Pathol. 2011, 65, 159–163. [Google Scholar] [CrossRef]
- Wang, J.; Browne, L.; Slapetova, I.; Shang, F.; Lee, K.; Lynch, J.; Beretov, J.; Whan, R.; Graham, P.H.; Millar, E.K.A. Multiplexed immunofluorescence identifies high stromal CD68+PD-L1+ macrophages as a predictor of improved survival in triple negative breast cancer. Sci. Rep. 2021, 11, 1–12. [Google Scholar] [CrossRef]
- Ahmed, F.S.; Gaule, P.; McGuire, J.; Patel, K.; Blenman, K.; Pusztai, L.; Rimm, D.L. PD-L1 Protein Expression on Both Tumor Cells and Macrophages are Associated with Response to Neoadjuvant Durvalumab with Chemotherapy in Triple-negative Breast Cancer. Clin. Cancer Res. 2020, 26, 5456–5461. [Google Scholar] [CrossRef]
- Reddy, S.M.; Reuben, A.; Barua, S.; Jiang, H.; Zhang, S.; Wang, L.; Gopalakrishnan, V.; Hudgens, C.W.; Tetzlaff, M.T.; Reuben, J.M.; et al. Poor Response to Neoadjuvant Chemotherapy Correlates with Mast Cell Infiltration in Inflammatory Breast Cancer. Cancer Immunol. Res. 2019, 7, 1025–1035. [Google Scholar] [CrossRef]
- Hobbs, E.; Yang, F.; Kumar, T.; Contreras, A.; Cuentas, E.R.P.; Garber, H.; Scoggins, M.; E Adrada, B.; Whitman, G.J.; Arun, B.; et al. Tumor immune microenvironment (TiME) changes by multiplex IF staining in a pilot study of neoadjuvant talazoparib for early-stage breast cancer patients with a BRCA mutation. J. Clin. Oncol. 2019, 37, 585. [Google Scholar] [CrossRef]
- Griguolo, G.; Tosi, A.; Dieci, M.V.; Fineberg, S.; Rossi, V.; Ventura, A.; Bottosso, M.; Bauchet, L.; Miglietta, F.; Jacob, J.; et al. A Comprehensive Profiling of the Immune Microenvironment of Breast Cancer Brain Metastases. Neuro Oncol. 2022. [Google Scholar] [CrossRef]
- Zheng, G.; Guo, Z.; Li, W.; Xi, W.; Zuo, B.; Zhang, R.; Wen, W.; Yang, A.-G.; Jia, L. Interaction between HLA-G and NK cell receptor KIR2DL4 orchestrates HER2-positive breast cancer resistance to trastuzumab. Signal Transduct. Target. Ther. 2021, 6, 236. [Google Scholar] [CrossRef]
- Bassez, A.; Vos, H.; Van Dyck, L.; Floris, G.; Arijs, I.; Desmedt, C.; Boeckx, B.; Bempt, M.V.; Nevelsteen, I.; Lambein, K.; et al. A single-cell map of intratumoral changes during anti-PD1 treatment of patients with breast cancer. Nat. Med. 2021, 27, 820–832. [Google Scholar] [CrossRef]
- Pittet, M.J.; Michielin, O.; Migliorini, D. Clinical relevance of tumour-associated macrophages. Nat. Rev. Clin. Oncol. 2022, 19, 402–421. [Google Scholar] [CrossRef]
- Ma, R.-Y.; Black, A.; Qian, B.-Z. Macrophage diversity in cancer revisited in the era of single-cell omics. Trends Immunol. 2022, 43, 546–563. [Google Scholar] [CrossRef]
- Laviron, M.; Petit, M.; Weber-Delacroix, E.; Combes, A.J.; Arkal, A.R.; Barthélémy, S.; Courau, T.; Hume, D.A.; Combadière, C.; Krummel, M.F.; et al. Tumor-associated macrophage heterogeneity is driven by tissue territories in breast cancer. Cell Rep. 2022, 39. [Google Scholar] [CrossRef]
- Ramos, R.N.; Missolo-Koussou, Y.; Gerber-Ferder, Y.; Bromley, C.P.; Bugatti, M.; Núñez, N.G.; Boari, J.T.; Richer, W.; Menger, L.; Denizeau, J.; et al. Tissue-resident FOLR2+ macrophages associate with CD8+ T cell infiltration in human breast cancer. Cell 2022, 185, 1189–1207.e25. [Google Scholar] [CrossRef]
- Yu, J.; Green, M.D.; Li, S.; Sun, Y.; Journey, S.N.; Choi, J.E.; Rizvi, S.M.; Qin, A.; Waninger, J.J.; Lang, X.; et al. Liver metastasis restrains immunotherapy efficacy via macrophage-mediated T cell elimination. Nat. Med. 2021, 27, 152–164. [Google Scholar] [CrossRef]
- Tumeh, P.C.; Hellmann, M.D.; Hamid, O.; Tsai, K.K.; Loo, K.L.; Gubens, M.A.; Rosenblum, M.; Harview, C.L.; Taube, J.M.; Handley, N.; et al. Liver Metastasis and Treatment Outcome with Anti-PD-1 Monoclonal Antibody in Patients with Melanoma and NSCLC. Cancer Immunol. Res. 2017, 5, 417–424. [Google Scholar] [CrossRef]
- Hu, G.; Wang, S. Prognostic role of tumor-infiltrating CD57-positive lymphocytes in solid tumors: A meta-analysis. Oncotarget 2017, 9, 8111–8119. [Google Scholar] [CrossRef]
- Carter, J.M.; Chumsri, S.; Hillman, D.W.; Zahrieh, D.M.; Ma, Y.; Wang, X.; Kachergus, J.M.; Boughey, J.C.; Liu, M.C.; Kalari, K.R.; et al. Abstract PS16-01: Intra-epithelial tumor immune landscapes are associated with clinical outcomes in early-stage triple-negative breast cancer. Cancer Res. 2021, 81, PS16-01. [Google Scholar] [CrossRef]
- Fuertes, M.B.; Domaica, C.I.; Zwirner, N.W. Leveraging NKG2D Ligands in Immuno-Oncology. Front. Immunol. 2021, 12, 713158. [Google Scholar] [CrossRef]
- Subedi, N.; Verhagen, L.P.; Bosman, E.M.; van Roessel, I.; Tel, J. Understanding natural killer cell biology from a single cell perspective. Cell. Immunol. 2022, 373, 104497. [Google Scholar] [CrossRef]
- Long, E.O.; Sik Kim, H.; Liu, D.; Peterson, M.E.; Rajagopalan, S. Controlling Natural Killer Cell Responses: Integration of Signals for Activation and Inhibition. Annu. Rev. Immunol. 2013, 31, 227–258. [Google Scholar] [CrossRef]
- Muñoz, D.P.; Yannone, S.M.; Daemen, A.; Sun, Y.; Vakar-Lopez, F.; Kawahara, M.; Freund, A.M.; Rodier, F.; Wu, J.D.; Desprez, P.-Y.; et al. Targetable mechanisms driving immunoevasion of persistent senescent cells link chemotherapy-resistant cancer to aging. JCI Insight 2019, 4, e124716. [Google Scholar] [CrossRef]
- Zheng, G.; Jia, L.; Yang, A.-G. Roles of HLA-G/KIR2DL4 in Breast Cancer Immune Microenvironment. Front. Immunol. 2022, 13, 791975. [Google Scholar] [CrossRef]
- López-Soto, A.; Gonzalez, S.; Smyth, M.J.; Galluzzi, L. Control of Metastasis by NK Cells. Cancer Cell 2017, 32, 135–154. [Google Scholar] [CrossRef]
- Chan, I.S.; Ewald, A.J. The changing role of natural killer cells in cancer metastasis. J. Clin. Investig. 2022, 132, e143762. [Google Scholar] [CrossRef]
- Correia, A.L.; Guimaraes, J.C.; der Maur, P.A.; De Silva, D.; Trefny, M.P.; Okamoto, R.; Bruno, S.; Schmidt, A.; Mertz, K.; Volkmann, K.; et al. Hepatic stellate cells suppress NK cell-sustained breast cancer dormancy. Nature 2021, 594, 566–571. [Google Scholar] [CrossRef]
- Chen, W.; Shen, L.; Jiang, J.; Zhang, L.; Zhang, Z.; Pan, J.; Ni, C.; Chen, Z. Antiangiogenic therapy reverses the immunosuppressive breast cancer microenvironment. Biomark. Res. 2021, 9, 1–16. [Google Scholar] [CrossRef]
- Vella, G.; Guelfi, S.; Bergers, G. High Endothelial Venules: A Vascular Perspective on Tertiary Lymphoid Structures in Cancer. Front. Immunol. 2021, 12, 736670. [Google Scholar] [CrossRef]
- Buckanovich, R.J.; Facciabene, A.; Kim, S.; Benencia, F.; Sasaroli, D.; Balint, K.; Katsaros, D.; O’Brien-Jenkins, A.; A Gimotty, P.; Coukos, G. Endothelin B receptor mediates the endothelial barrier to T cell homing to tumors and disables immune therapy. Nat. Med. 2008, 14, 28–36. [Google Scholar] [CrossRef]
- Motz, G.T.; Santoro, S.P.; Wang, L.-P.; Garrabrant, T.; Lastra, R.R.; Hagemann, I.S.; Lal, P.; Feldman, M.D.; Benencia, F.; Coukos, G. Tumor endothelium FasL establishes a selective immune barrier promoting tolerance in tumors. Nat. Med. 2014, 20, 607–615. [Google Scholar] [CrossRef]
- Amersfoort, J.; Eelen, G.; Carmeliet, P. Immunomodulation by endothelial cells—Partnering up with the immune system? Nat. Rev. Immunol. 2022, 22, 576–588. [Google Scholar] [CrossRef]
- Blanchard, L.; Girard, J.-P. High endothelial venules (HEVs) in immunity, inflammation and cancer. Angiogenesis 2021, 24, 719–753. [Google Scholar] [CrossRef] [PubMed]
- Milutinovic, S.; Abe, J.; Godkin, A.; Stein, J.V.; Gallimore, A. The Dual Role of High Endothelial Venules in Cancer Progression versus Immunity. Trends Cancer 2020, 7, 214–225. [Google Scholar] [CrossRef] [PubMed]
- Song, I.H.; Heo, S.-H.; Bang, W.S.; Park, H.S.; Park, I.A.; Kim, Y.-A.; Park, S.Y.; Roh, J.; Gong, G.; Lee, H.J. Predictive Value of Tertiary Lymphoid Structures Assessed by High Endothelial Venule Counts in the Neoadjuvant Setting of Triple-Negative Breast Cancer. Cancer Res. Treat. 2017, 49, 399–407. [Google Scholar] [CrossRef]
- Asrir, A.; Tardiveau, C.; Coudert, J.; Laffont, R.; Blanchard, L.; Bellard, E.; Veerman, K.; Bettini, S.; Lafouresse, F.; Vina, E.; et al. Tumor-associated high endothelial venules mediate lymphocyte entry into tumors and predict response to PD-1 plus CTLA-4 combination immunotherapy. Cancer Cell 2022, 40, 318–334.e9. [Google Scholar] [CrossRef]
- Simon, T.; Salhia, B. Cancer-Associated Fibroblast Subpopulations With Diverse and Dynamic Roles in the Tumor Microenvironment. Mol. Cancer Res. 2022, 20, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.; Kieffer, Y.; Scholer-Dahirel, A.; Pelon, F.; Bourachot, B.; Cardon, M.; Sirven, P.; Magagna, I.; Fuhrmann, L.; Bernard, C.; et al. Fibroblast heterogeneity and immunosuppressive environment in human breast cancer. Cancer Cell 2018, 33, 463–479.e10. [Google Scholar] [CrossRef]
- Pelon, F.; Bourachot, B.; Kieffer, Y.; Magagna, I.; Mermet-Meillon, F.; Bonnet, I.; Costa, A.; Givel, A.-M.; Attieh, Y.; Barbazan, J.; et al. Cancer-associated fibroblast heterogeneity in axillary lymph nodes drives metastases in breast cancer through complementary mechanisms. Nat. Commun. 2020, 11, 404. [Google Scholar] [CrossRef] [PubMed]
- Magagna, I.; Gourdin, N.; Kieffer, Y.; Licaj, M.; Mhaidly, R.; Andre, P.; Morel, A.; Vincent-Salomon, A.; Paturel, C.; Mechta-Grigoriou, F. CD73-Mediated Immunosuppression Is Linked to a Specific Fibroblast Population That Paves the Way for New Therapy in Breast Cancer. Cancers 2021, 13, 5878. [Google Scholar] [CrossRef]
- Fowell, D.J.; Kim, M. The spatio-temporal control of effector T cell migration. Nat. Rev. Immunol. 2021, 21, 582–596. [Google Scholar] [CrossRef]
- Kuczek, D.E.; Larsen, A.M.H.; Thorseth, M.-L.; Carretta, M.; Kalvisa, A.; Siersbæk, M.S.; Simões, A.M.C.; Roslind, A.; Engelholm, L.H.; Noessner, E.; et al. Collagen density regulates the activity of tumor-infiltrating T cells. J. Immunother. Cancer 2019, 7, 68. [Google Scholar] [CrossRef]
- Baker, A.T.; Abuwarwar, M.H.; Poly, L.; Wilkins, S.; Fletcher, A.L. Cancer-Associated Fibroblasts and T Cells: From Mechanisms to Outcomes. J. Immunol. 2021, 206, 310–320. [Google Scholar] [CrossRef]
- Chen, I.X.; Chauhan, V.P.; Posada, J.; Ng, M.R.; Wu, M.W.; Adstamongkonkul, P.; Huang, P.; Lindeman, N.; Langer, R.; Jain, R.K. Blocking CXCR4 Alleviates Desmoplasia, Increases T-Lymphocyte Infiltration, and Im-proves Immunotherapy in Metastatic Breast Cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 4558–4566. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Wu, B.; Chiang, H.-C.; Deng, H.; Zhang, X.; Xiong, W.; Liu, J.; Rozeboom, A.M.; Harris, B.T.; Blommaert, E.; et al. Tumour DDR1 promotes collagen fibre alignment to instigate immune exclusion. Nature 2021, 599, 673–678. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.E.; Creason, A.L.; Stommel, J.M.; Keck, J.M.; Parmar, S.; Betts, C.B.; Blucher, A.; Boniface, C.; Bucher, E.; Burlingame, E.; et al. An omic and multidimensional spatial atlas from serial biopsies of an evolving metastatic breast cancer. Cell Rep. Med. 2022, 3, 100525. [Google Scholar] [CrossRef] [PubMed]
- Baldominos, P.; Barbera-Mourelle, A.; Barreiro, O.; Huang, Y.; Wight, A.; Cho, J.-W.; Zhao, X.; Estivill, G.; Adam, I.; Sanchez, X.; et al. Quiescent cancer cells resist T cell attack by forming an immunosuppressive niche. Cell 2022, 185, 1694–1708.e19. [Google Scholar] [CrossRef] [PubMed]
- Lakins, M.A.; Ghorani, E.; Munir, H.; Martins, C.P.; Shields, J.D. Cancer-associated fibroblasts induce antigen-specific deletion of CD8+ T Cells to protect tumour cells. Nat. Commun. 2018, 9, 948. [Google Scholar] [CrossRef]
- Nederlof, I.; Hajizadeh, S.; Sobhani, F.; Raza, S.E.A.; AbdulJabbar, K.; Harkes, R.; van de Vijver, M.J.; Salgado, R.; Desmedt, C.; Kok, M.; et al. Spatial Interplay of Lymphocytes and Fibroblasts in Estrogen Receptor-Positive HER2-Negative Breast Cancer. NPJ Breast Cancer 2022, 8, 56. [Google Scholar] [CrossRef]
- Nia, H.T.; Munn, L.L.; Jain, R.K. Physical traits of cancer. Science 2020, 370, eaaz0868. [Google Scholar] [CrossRef]
- Ozga, A.J.; Chow, M.T.; Luster, A.D. Chemokines and the immune response to cancer. Immunity 2021, 54, 859–874. [Google Scholar] [CrossRef]
- Hoch, T.; Schulz, D.; Eling, N.; Gómez, J.M.; Levesque, M.P.; Bodenmiller, B. Multiplexed imaging mass cytometry of the chemokine milieus in melanoma characterizes features of the response to immunotherapy. Sci. Immunol. 2022, 7, eabk1692. [Google Scholar] [CrossRef]
- Schulz, D.; Zanotelli, V.R.T.; Fischer, J.R.; Schapiro, D.; Engler, S.; Lun, X.-K.; Jackson, H.W.; Bodenmiller, B. Simultaneous Multiplexed Imaging of mRNA and Proteins with Subcellular Resolution in Breast Cancer Tissue Samples by Mass Cytometry. Cell Syst. 2017, 6, 25–36.e5. [Google Scholar] [CrossRef]
- Duckworth, B.C.; Lafouresse, F.; Wimmer, V.C.; Broomfield, B.J.; Dalit, L.; Alexandre, Y.O.; Sheikh, A.A.; Qin, R.Z.; Alvarado, C.; Mielke, L.A.; et al. Effector and stem-like memory cell fates are imprinted in distinct lymph node niches directed by CXCR3 ligands. Nat. Immunol. 2021, 22, 434–448. [Google Scholar] [CrossRef] [PubMed]
- Dieci, M.V.; Miglietta, F.; Guarneri, V. Immune Infiltrates in Breast Cancer: Recent Updates and Clinical Implications. Cells 2021, 10, 233. [Google Scholar] [CrossRef]
- Denkert, C.; Von Minckwitz, G.; Darb-Esfahani, S.; Lederer, B.; Heppner, B.I.; Weber, K.E.; Budczies, J.; Huober, J.; Klauschen, F.; Furlanetto, J.; et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: A pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018, 19, 40–50. [Google Scholar] [CrossRef]
- Loi, S.; Drubay, D.; Adams, S.; Pruneri, G.; Francis, P.A.; Lacroix-Triki, M.; Joensuu, H.; Dieci, M.V.; Badve, S.; Demaria, S.; et al. Tumor-Infiltrating Lymphocytes and Prognosis: A Pooled Individual Patient Analysis of Early-Stage Triple-Negative Breast Cancers. J. Clin. Oncol. 2019, 37, 559–569. [Google Scholar] [CrossRef]
- Loi, S.; Sirtaine, N.; Piette, F.; Salgado, R.; Viale, G.; Van Eenoo, F.; Rouas, G.; Francis, P.; Crown, J.P.; Hitre, E.; et al. Prognostic and Predictive Value of Tumor-Infiltrating Lymphocytes in a Phase III Randomized Adjuvant Breast Cancer Trial in Node-Positive Breast Cancer Comparing the Addition of Docetaxel to Doxorubicin With Doxorubicin-Based Chemotherapy: BIG 02-98. J. Clin. Oncol. 2013, 31, 860–867. [Google Scholar] [CrossRef]
- Loi, S.; Michiels, S.; Salgado, R.; Sirtaine, N.; Jose, V.; Fumagalli, D.; Kellokumpu-Lehtinen, P.-L.; Bono, P.; Kataja, V.; Desmedt, C.; et al. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: Results from the FinHER trial. Ann. Oncol. 2014, 25, 1544–1550. [Google Scholar] [CrossRef]
- Loi, S.; Salgado, R.; Adams, S.; Pruneri, G.; Francis, P.A.; Lacroix-Triki, M.; Joensuu, H.; Dieci, M.V.; Badve, S.; Demaria, S.; et al. Tumor infiltrating lymphocyte stratification of prognostic staging of early-stage triple negative breast cancer. npj Breast Cancer 2022, 8, 1–4. [Google Scholar] [CrossRef]
- Dieci, M.V.; Mathieu, M.C.; Guarneri, V.; Conte, P.; Delaloge, S.; Andre, F.; Goubar, A. Prognostic and predictive value of tumor-infiltrating lymphocytes in two phase III randomized adjuvant breast cancer trials. Ann. Oncol. 2015, 26, 1698–1704. [Google Scholar] [CrossRef]
- Park, J.H.; Jonas, S.F.; Bataillon, G.; Criscitiello, C.; Salgado, R.; Loi, S.; Viale, G.; Lee, H.J.; Dieci, M.V.; Kim, S.-B.; et al. Prognostic value of tumor-infiltrating lymphocytes in patients with early-stage triple-negative breast cancers (TNBC) who did not receive adjuvant chemotherapy. Ann. Oncol. 2019, 30, 1941–1949. [Google Scholar] [CrossRef]
- Dieci, M.V.; Criscitiello, C.; Goubar, A.; Viale, G.; Conte, P.; Guarneri, V.; Ficarra, G.; Mathieu, M.C.; Delaloge, S.; Curigliano, G.; et al. Prognostic value of tumor-infiltrating lymphocytes on residual disease after primary chemotherapy for triple-negative breast cancer: A retrospective multicenter study. Ann. Oncol. 2014, 25, 611–618. [Google Scholar] [CrossRef]
- Loibl, S.; Untch, M.; Burchardi, N.; Huober, J.; Sinn, B.V.; Blohmer, J.-U.; Grischke, E.-M.; Furlanetto, J.; Tesch, H.; Hanusch, C.; et al. A randomised phase II study investigating durvalumab in addition to an anthracycline taxane-based neoadjuvant therapy in early triple-negative breast cancer: Clinical results and biomarker analysis of GeparNuevo study. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2019, 30, 1279–1288. [Google Scholar] [CrossRef]
- Sharma, P.; Stecklein, S.R.; Yoder, R.; Staley, J.M.; Schwensen, K.; O’Dea, A.; Nye, L.E.; Elia, M.; Satelli, D.; Crane, G.; et al. Clinical and biomarker results of neoadjuvant phase II study of pembrolizumab and carboplatin plus docetaxel in triple-negative breast cancer (TNBC) (NeoPACT). J. Clin. Oncol. 2022, 40, 513. [Google Scholar] [CrossRef]
- Loi, S.; Schmid, P.; Aktan, G.; Karantza, V.; Salgado, R. Relationship between tumor infiltrating lymphocytes (TILs) and response to pembrolizumab (pembro)+chemotherapy (CT) as neoadjuvant treatment (NAT) for triple-negative breast cancer (TNBC): Phase Ib KEYNOTE-173 trial. Ann. Oncol. 2019, 30, iii2. [Google Scholar] [CrossRef]
- Bianchini, G.; Huang, C.-S.; Egle, D.; Bermejo, B.; Zamagni, C.; Thill, M.; Anton, A.; Zambelli, S.; Russo, S.; Ciruelos, E.; et al. LBA13 Tumour infiltrating lymphocytes (TILs), PD-L1 expression and their dynamics in the NeoTRIPaPDL1 trial. Ann. Oncol. 2020, 31, S1145–S1146. [Google Scholar] [CrossRef]
- Molinero, L.L.; Mittendorf, E.A.; Zhang, H.; Barrios, C.H.; Saji, S.; Jung, K.H.; Patel, S.; Chang, C.-W.; Liste-Hermoso, M.; Chui, S.Y.; et al. Abstract LB162: Evaluation of tumor immune biomarkers with pathologic complete response (pCR) in patients receiving atezolizumab + chemotherapy in early triple negative breast cancer (eTNBC): Exploratory analyses from the IMpassion031 study. Cancer Res. 2021, 81, LB162. [Google Scholar] [CrossRef]
- Dieci, M.V.; Tsvetkova, V.; Orvieto, E.; Piacentini, F.; Ficarra, G.; Griguolo, G.; Miglietta, F.; Giarratano, T.; Omarini, C.; Bonaguro, S.; et al. Immune characterization of breast cancer metastases: Prognostic implications. Breast Cancer Res. 2018, 20, 62. [Google Scholar] [CrossRef]
- Adams, S.; Loi, S.; Toppmeyer, D.; Cescon, D.W.; De Laurentiis, M.; Nanda, R.; Winer, E.P.; Mukai, H.; Tamura, K.; Armstrong, A.; et al. Pembrolizumab monotherapy for previously untreated, PD-L1-positive, metastatic triple-negative breast cancer: Cohort B of the phase II KEYNOTE-086 study. Ann. Oncol. 2019, 30, 405–411. [Google Scholar] [CrossRef]
- Adams, S.; Schmid, P.; Rugo, H.S.; Winer, E.P.; Loirat, D.; Awada, A.; Cescon, D.W.; Iwata, H.; Campone, M.; Nanda, R.; et al. Pembrolizumab monotherapy for previously treated metastatic triple-negative breast cancer: Cohort A of the phase II KEYNOTE-086 study. Ann. Oncol. 2019, 30, 397–404. [Google Scholar] [CrossRef]
- Loi, S.; Schmid, P.; Cortes, J.; Cescon, D.W.; Winer, E.P.; Toppmeyer, D.L.; Rugo, H.S.; De Laurentiis, M.; Nanda, R.; Iwata, H.; et al. Abstract PD14-07: Association between biomarkers and response to pembrolizumab in patients with metastatic triple-negative breast cancer (mTNBC): Exploratory analysis from KEYNOTE-086. Cancer Res. 2021, 81, PD14-07. [Google Scholar] [CrossRef]
- Emens, L.; Molinero, L.; Loi, S.; Rugo, H.S.; Schneeweiss, A.; Diéras, V.; Iwata, H.; Barrios, C.H.; Nechaeva, M.; Duc, A.N.; et al. Atezolizumab and nab-Paclitaxel in Advanced Triple-Negative Breast Cancer: Biomarker Evaluation of the IMpassion130 Study. J. Natl. Cancer Inst. 2021, 113, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Dieci, M.; Conte, P.; Bisagni, G.; Brandes, A.; Frassoldati, A.; Cavanna, L.; Musolino, A.; Giotta, F.; Rimanti, A.; Garrone, O.; et al. Association of tumor-infiltrating lymphocytes with distant disease-free survival in the ShortHER randomized adjuvant trial for patients with early HER2+ breast cancer. Ann. Oncol. 2019, 30, 418–423. [Google Scholar] [CrossRef] [PubMed]
- Gelber, R.D.; Wang, X.V.; Cole, B.F.; Cameron, D.; Cardoso, F.; Tjan-Heijnen, V.; Krop, I.; Loi, S.; Salgado, R.; Kiermaier, A.; et al. Six-year absolute invasive disease-free survival benefit of adding adjuvant pertuzumab to trastuzumab and chemotherapy for patients with early HER2-positive breast cancer: A Subpopulation Treatment Effect Pattern Plot (STEPP) analysis of the APHINITY (BIG 4-11) trial. Eur. J. Cancer 2022, 166, 219–228. [Google Scholar] [CrossRef] [PubMed]
- Nuciforo, P.; Pascual, T.; Cortés, J.; Llombart-Cussac, A.; Fasani, R.; Pare, L.; Oliveira, M.; Galvan, P.; Martínez, N.; Bermejo, B.; et al. A predictive model of pathologic response based on tumor cellularity and tumor-infiltrating lymphocytes (CelTIL) in HER2-positive breast cancer treated with chemo-free dual HER2 blockade. Ann. Oncol. 2017, 29, 170–177. [Google Scholar] [CrossRef]
- Griguolo, G.; Serna, G.; Pascual, T.; Fasani, R.; Guardia, X.; Chic, N.; Paré, L.; Pernas, S.; Muñoz, M.; Oliveira, M.; et al. Immune microenvironment characterisation and dynamics during anti-HER2-based neoadjuvant treatment in HER2-positive breast cancer. Npj Precis. Oncol. 2021, 5, 23. [Google Scholar] [CrossRef]
- Salgado, R.; Denkert, C.; Campbell, C.C.; Savas, P.P.; Nuciforo, P.; Aura, C.M.; de Azambuja, E.; Eidtmann, H.; Ellis, C.E.; Baselga, J.; et al. Tumor-Infiltrating Lymphocytes and Associations With Pathological Complete Response and Event-Free Survival in HER2-Positive Early-Stage Breast Cancer Treated With Lapatinib and Trastuzumab. JAMA Oncol. 2015, 1, 448–454. [Google Scholar] [CrossRef]
- Hamy, A.-S.; Bonsang-Kitzis, H.; De Croze, D.; Laas, E.; Darrigues, L.; Topciu, L.; Menet, E.; Vincent-Salomon, A.; Lerebours, F.; Pierga, J.-Y.; et al. Interaction between Molecular Subtypes and Stromal Immune Infiltration before and after Treatment in Breast Cancer Patients Treated with Neoadjuvant Chemotherapy. Clin. Cancer Res. 2019, 25, 6731–6741. [Google Scholar] [CrossRef]
- Luen, S.J.; Salgado, R.; Fox, S.; Savas, P.; Eng-Wong, J.; Clark, E.; Kiermaier, A.; Swain, S.; Baselga, J.; Michiels, S.; et al. Tumour-infiltrating lymphocytes in advanced HER2-positive breast cancer treated with pertuzumab or placebo in addition to trastuzumab and docetaxel: A retrospective analysis of the CLEOPATRA study. Lancet Oncol. 2016, 18, 52–62. [Google Scholar] [CrossRef]
- Emens, L.A.; Esteva, F.J.; Beresford, M.; Saura, C.; De Laurentiis, M.; Kim, S.-B.; Im, S.-A.; Wang, Y.; Salgado, R.; Mani, A.; et al. Trastuzumab emtansine plus atezolizumab versus trastuzumab emtansine plus placebo in previously treated, HER2-positive advanced breast cancer (KATE2): A phase 2, multicentre, randomised, double-blind trial. Lancet Oncol. 2020, 21, 1283–1295. [Google Scholar] [CrossRef]
- Criscitiello, C.; Vingiani, A.; Maisonneuve, P.; Viale, G.; Curigliano, G. Tumor-infiltrating lymphocytes (TILs) in ER+/HER2− breast cancer. Breast Cancer Res. Treat. 2020, 183, 347–354. [Google Scholar] [CrossRef]
- Galon, J.; Bruni, D. Approaches to Treat Immune Hot, Altered and Cold Tumours with Combination Immuno-therapies. Nat. Rev. Drug Discov. 2019, 18, 197–218. [Google Scholar] [CrossRef] [PubMed]
- Nederlof, I.; Horlings, H.; Curtis, C.; Kok, M. A High-Dimensional Window into the Micro-Environment of Triple Negative Breast Cancer. Cancers 2021, 13, 316. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Cole, K.; Martinez-Morilla, S.; Ahmed, F.S.; Zugazagoitia, J.; Staaf, J.; Bosch, A.; Ehinger, A.; Nimeus, E.; Hartman, J.; et al. An Open-Source, Automated Tumor-Infiltrating Lymphocyte Algorithm for Prognosis in Triple-Negative Breast Cancer. Clin. Cancer Res. 2021, 27, 5557–5565. [Google Scholar] [CrossRef] [PubMed]
- Saltz, J.; Gupta, R.; Hou, L.; Kurc, T.; Singh, P.; Nguyen, V.; Samaras, D.; Shroyer, K.R.; Zhao, T.; Batiste, R.; et al. Spatial Organization and Molecular Correlation of Tumor-Infiltrating Lymphocytes Using Deep Learning on Pathology Images. Cell Rep. 2018, 23, 181–193.e7. [Google Scholar] [CrossRef]
- Hendry, S.; Salgado, R.; Gevaert, T.; Russell, P.A.; John, T.; Thapa, B.; Christie, M.; van de Vijver, K.; Estrada, M.; Gonzalez-Ericsson, P.I.; et al. Assessing Tumor-infiltrating Lymphocytes in Solid Tumors: A Practical Review for Pathologists and Proposal for a Standardized Method From the International Immunooncology Biomarkers Working Group: Part 1: Assessing the Host Immune Response, TILs in Invasive Breast Carcinoma and Ductal Carcinoma In Situ, Metastatic Tumor Deposits and Areas for Further Research. Adv. Anat. Pathol. 2017, 24, 235–251. [Google Scholar] [CrossRef]
- Gunderson, A.J.; Yamazaki, T.; Mccarty, K.; Fox, N.; Phillips, M.; Alice, A.; Blair, T.; Whiteford, M.; O’Brien, D.; Ahmad, R.; et al. TGFβ suppresses CD8+ T cell expression of CXCR3 and tumor trafficking. Nat. Commun. 2020, 11, 1749. [Google Scholar] [CrossRef]
- Altan, M.; Kidwell, K.M.; Pelekanou, V.; Carvajal-Hausdorf, D.E.; Schalper, K.A.; Toki, M.; Thomas, D.G.; Sabel, M.S.; Hayes, D.F.; Rimm, D.L. Association of B7-H4, PD-L1, and tumor infiltrating lymphocytes with outcomes in breast cancer. NPJ Breast Cancer 2018, 4, 40. [Google Scholar] [CrossRef]
- Lu, M.Y.; Sater, H.A.; Mahmood, F. Multiplex computational pathology for treatment response prediction. Cancer Cell 2021, 39, 1053–1055. [Google Scholar] [CrossRef]
- Sanchez, K.; Kim, I.; Chun, B.; Pucilowska, J.; Redmond, W.L.; Urba, W.J.; Martel, M.; Wu, Y.; Campbell, M.; Sun, Z.; et al. Multiplex immunofluorescence to measure dynamic changes in tumor-infiltrating lymphocytes and PD-L1 in early-stage breast cancer. Breast Cancer Res. 2021, 23, 1–15. [Google Scholar] [CrossRef]
- Blenman, K.R.; Marczyk, M.; Karn, T.; Qing, T.; Li, X.; Gunasekharan, V.; Yaghoobi, V.; Bai, Y.; Ibrahim, E.Y.; Park, T.; et al. Predictive Markers of Response to Neoadjuvant Durvalumab with Nab-Paclitaxel and Dose-Dense Doxorubicin/Cyclophosphamide in Basal-Like Triple-Negative Breast Cancer. Clin. Cancer Res. 2022, 28, 2587–2597. [Google Scholar] [CrossRef]
- Loibl, S.; Schneeweiss, A.; Huober, J.; Braun, M.; Rey, J.; Blohmer, J.-U.; Furlanetto, J.; Zahm, D.-M.; Hanusch, C.; Thomalla, J.; et al. Neoadjuvant durvalumab improves survival in early triple-negative breast cancer independent of pathological complete response. Ann. Oncol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; Schneeweiss, A.; Rey, J.; Karn, T.; Braun, M.; Huober, J.B.; Sinn, H.-P.; Zahm, D.M.; Hanusch, C.; Marmé, F.; et al. Biomarkers for response to immunotherapy in triple-negative breast cancer: Differences between survival and pCR biomarkers. J. Clin. Oncol. 2022, 40, 583. [Google Scholar] [CrossRef]
- Karn, T.; Denkert, C.; Weber, K.; Holtrich, U.; Hanusch, C.; Sinn, B.; Higgs, B.; Jank, P.; Sinn, H.; Huober, J.; et al. Tumor mutational burden and immune infiltration as independent predictors of response to neoadjuvant immune checkpoint inhibition in early TNBC in GeparNuevo. Ann. Oncol. 2020, 31, 1216–1222. [Google Scholar] [CrossRef]
- Tolaney, S.M.; Kalinsky, K.; Kaklamani, V.G.; D’Adamo, D.R.; Aktan, G.; Tsai, M.L.; O’Regan, R.M.; Kaufman, P.A.; Wilks, S.T.; Andreopoulou, E.; et al. Eribulin plus Pembrolizumab in Patients with Metastatic Tri-ple-Negative Breast Cancer (ENHANCE 1): A Phase Ib/II Study. Clin. Cancer Res. 2021, 27, 3061–3068. [Google Scholar] [CrossRef]
- Miles, D.; Gligorov, J.; André, F.; Cameron, D.; Schneeweiss, A.; Barrios, C.; Xu, B.; Wardley, A.; Kaen, D.; Andrade, L.; et al. Primary results from IMpassion131, a double-blind, placebo-controlled, randomised phase III trial of first-line paclitaxel with or without atezolizumab for unresectable locally advanced/metastatic triple-negative breast cancer. Ann. Oncol. 2021, 32, 994–1004. [Google Scholar] [CrossRef]
- Huober, J.; Barrios, C.H.; Niikura, N.; Jarząb, M.; Chang, Y.-C.; Huggins-Puhalla, S.L.; Pedrini, J.; Zhukova, L.; Graupner, V.; Eiger, D.; et al. Atezolizumab With Neoadjuvant Anti–Human Epidermal Growth Factor Receptor 2 Therapy and Chemotherapy in Human Epidermal Growth Factor Receptor 2–Positive Early Breast Cancer: Primary Results of the Randomized Phase III IMpassion050 Trial. J. Clin. Oncol. 2022, 40, 2946–2956. [Google Scholar] [CrossRef]
- Hamilton, E.P.; Kaklamani, V.; Falkson, C.; Vidal, G.A.; Ward, P.J.; Patre, M.; Chui, S.Y.; Rotmensch, J.; Gupta, K.; Molinero, L.; et al. Impact of Anti-HER2 Treatments Combined With Atezolizumab on the Tumor Immune Microenvironment in Early or Metastatic Breast Cancer: Results From a Phase Ib Study. Clin. Breast Cancer 2021, 21, 539–551. [Google Scholar] [CrossRef]
- Loi, S.; Giobbie-Hurder, A.; Gombos, A.; Bachelot, T.; Hui, R.; Curigliano, G.; Campone, M.; Biganzoli, L.; Bonnefoi, H.; Jerusalem, G.; et al. Pembrolizumab plus trastuzumab in trastuzumab-resistant, advanced, HER2-positive breast cancer (PANACEA): A single-arm, multicentre, phase 1b–2 trial. Lancet Oncol. 2019, 20, 371–382. [Google Scholar] [CrossRef]
- Rugo, H.S.; Delord, J.-P.; Im, S.-A.; Ott, P.A.; Piha-Paul, S.A.; Bedard, P.L.; Sachdev, J.; Le Tourneau, C.; van Brummelen, E.M.; Varga, A.; et al. Safety and Antitumor Activity of Pembrolizumab in Patients with Estrogen Receptor–Positive/Human Epidermal Growth Factor Receptor 2–Negative Advanced Breast Cancer. Clin. Cancer Res. 2018, 24, 2804–2811. [Google Scholar] [CrossRef]
- Pérez-García, J.M.; Llombart-Cussac, A.; Gion, M.; Curigliano, G.; López-Miranda, E.; Alonso, J.L.; Bermejo, B.; Calvo, L.; Carañana, V.; Sánchez, S.D.l.C.; et al. Pembrolizumab plus eribulin in hormone-receptor–positive, HER2-negative, locally recurrent or metastatic breast cancer (KELLY): An open-label, multicentre, single-arm, phase Ⅱ trial. Eur. J. Cancer 2021, 148, 382–394. [Google Scholar] [CrossRef]
- Tolaney, S.M.; Barroso-Sousa, R.; Keenan, T.; Li, T.; Trippa, L.; Vaz-Luis, I.; Wulf, G.; Spring, L.; Sinclair, N.F.; Andrews, C.; et al. Effect of Eribulin With or Without Pembrolizumab on Progression-Free Survival for Patients With Hormone Receptor–Positive, ERBB2-Negative Metastatic Breast Cancer. JAMA Oncol. 2020, 6, 1598–1605. [Google Scholar] [CrossRef]
- Shah, A.N.; Flaum, L.; Helenowski, I.; A Santa-Maria, C.; Jain, S.; Rademaker, A.; Nelson, V.; Tsarwhas, D.; Cristofanilli, M.; Gradishar, W. Phase II study of pembrolizumab and capecitabine for triple negative and hormone receptor-positive, HER2−negative endocrine-refractory metastatic breast cancer. J. Immunother. Cancer 2019, 8, e000173. [Google Scholar] [CrossRef]
- Ali, H.R.; Provenzano, E.; Dawson, S.-J.; Blows, F.M.; Liu, B.; Shah, M.; Earl, H.M.; Poole, C.J.; Hiller, L.; Dunn, J.A.; et al. Association between CD8+ T-cell infiltration and breast cancer survival in 12 439 patients. Ann. Oncol. 2014, 25, 1536–1543. [Google Scholar] [CrossRef]
- Sobral-Leite, M.; Salomon, I.; Opdam, M.; Kruger, D.T.; Beelen, K.J.; Van Der Noort, V.; Van Vlierberghe, R.L.P.; Blok, E.J.; Giardiello, D.; Sanders, J.; et al. Cancer-immune interactions in ER-positive breast cancers: PI3K pathway alterations and tumor-infiltrating lymphocytes. Breast Cancer Res. 2019, 21, 1–12. [Google Scholar] [CrossRef]
- Andreatta, M.; Corria-Osorio, J.; Müller, S.; Cubas, R.; Coukos, G.; Carmona, S.J. Interpretation of T cell states from single-cell transcriptomics data using reference atlases. Nat. Commun. 2021, 12, 1–19. [Google Scholar] [CrossRef]
- van der Leun, A.M.; Schumacher, T.N. An atlas of intratumoral T cells. Science 2021, 374, 1446–1447. [Google Scholar] [CrossRef]
- Ghoneim, H.E.; Fan, Y.; Moustaki, A.; Abdelsamed, H.A.; Dash, P.; Dogra, P.; Carter, R.; Awad, W.; Neale, G.; Thomas, P.G.; et al. De Novo Epigenetic Programs Inhibit PD-1 Blockade-Mediated T Cell Rejuvenation. Cell 2017, 170, 142–157.e19. [Google Scholar] [CrossRef]
- Wu, T.; Ji, Y.; Moseman, E.A.; Xu, H.C.; Manglani, M.; Kirby, M.; Anderson, S.M.; Handon, R.; Kenyon, E.; Elkahloun, A.; et al. The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci. Immunol. 2016, 1, eaai8593. [Google Scholar] [CrossRef]
- Utzschneider, D.T.; Charmoy, M.; Chennupati, V.; Pousse, L.; Ferreira, D.P.; Calderon-Copete, S.; Danilo, M.; Alfei, F.; Hofmann, M.; Wieland, D.; et al. T Cell Factor 1-Expressing Memory-like CD8+ T Cells Sustain the Immune Response to Chronic Viral Infections. Immunity 2016, 45, 415–427. [Google Scholar] [CrossRef] [PubMed]
- Sade-Feldman, M.; Yizhak, K.; Bjorgaard, S.L.; Ray, J.P.; de Boer, C.G.; Jenkins, R.W.; Lieb, D.J.; Chen, J.H.; Frederick, D.T.; Barzily-Rokni, M.; et al. Defining T Cell States Associated with Response to Checkpoint Immunotherapy in Melanoma. Cell 2018, 175, 998–1013.e1020. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, I.; Schaeuble, K.; Chennupati, V.; Marraco, S.A.F.; Calderon-Copete, S.; Ferreira, D.P.; Carmona, S.J.; Scarpellino, L.; Gfeller, D.; Pradervand, S.; et al. Intratumoral Tcf1+PD-1+CD8+ T Cells with Stem-like Properties Promote Tumor Control in Response to Vaccination and Checkpoint Blockade Immunotherapy. Immunity 2019, 50, 195–211.e10. [Google Scholar] [CrossRef]
- Tooley, K.A.; Escobar, G.; Anderson, A.C. Spatial determinants of CD8+ T cell differentiation in cancer. Trends Cancer 2022, 8, 642–654. [Google Scholar] [CrossRef]
- Stoltzfus, C.R.; Sivakumar, R.; Kunz, L.; Pope, B.E.O.; Menietti, E.; Speziale, D.; Adelfio, R.; Bacac, M.; Colombetti, S.; Perro, M.; et al. Multi-Parameter Quantitative Imaging of Tumor Microenvironments Reveals Perivascular Immune Niches Associated With Anti-Tumor Immunity. Front. Immunol. 2021, 12, 726492. [Google Scholar] [CrossRef]
- Jansen, C.S.; Prokhnevska, N.; Master, V.A.; Sanda, M.G.; Carlisle, J.W.; Bilen, M.A.; Cardenas, M.; Wilkinson, S.; Lake, R.; Sowalsky, A.G.; et al. An intra-tumoral niche maintains and differentiates stem-like CD8 T cells. Nature 2019, 576, 465–470. [Google Scholar] [CrossRef]
- Egelston, C.; Avalos, C.; Huang, Y.; Wang, R.; Solomon, S.; Tu, T.Y.; Simons, D.L.; Lee, P.P. Complex Pheno-typing of PD-1+ CD39+ Exhausted CD8+ T Cells in Human Carcinomas. J. Immunol. 2018, 200, 57.23. [Google Scholar]
- Lee, Y.; Park, J.; Park, S.-H.; Shin, E.-C. CD39+CD8+ T Cells Exhibit a Distinct Phenotype among Tumor-Infiltrating Tumor-Antigenspecific CD8+ T Cells. J. Immunol. 2019, 202, 195.2. [Google Scholar]
- Canale, F.P.; Ramello, M.C.; Núñez, N.; Furlan, C.L.A.; Bossio, S.N.; Serrán, M.G.; Boari, J.T.; del Castillo, A.; Ledesma, M.; Sedlik, C.; et al. CD39 Expression Defines Cell Exhaustion in Tumor-Infiltrating CD8+ T Cells. Cancer Res. 2018, 78, 115–128. [Google Scholar] [CrossRef]
- Simoni, Y.; Becht, E.; Fehlings, M.; Loh, C.Y.; Koo, S.-L.; Teng, K.W.W.; Yeong, J.P.S.; Nahar, R.; Zhang, T.; Kared, H.; et al. Bystander CD8+ T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 2018, 557, 575–579. [Google Scholar] [CrossRef] [PubMed]
- Duhen, T.; Duhen, R.; Montler, R.; Moses, J.; Moudgil, T.; de Miranda, N.F.; Goodall, C.P.; Blair, T.C.; Fox, B.A.; McDermott, J.E.; et al. Co-expression of CD39 and CD103 identifies tumor-reactive CD8 T cells in human solid tumors. Nat. Commun. 2018, 9, 2724. [Google Scholar] [CrossRef] [PubMed]
- Luoma, A.M.; Suo, S.; Wang, Y.; Gunasti, L.; Porter, C.B.; Nabilsi, N.; Tadros, J.; Ferretti, A.P.; Liao, S.; Gurer, C.; et al. Tissue-resident memory and circulating T cells are early responders to pre-surgical cancer immunotherapy. Cell 2022, 185, 2918–2935.e29. [Google Scholar] [CrossRef]
- Banchereau, R.; Chitre, A.S.; Scherl, A.; Wu, T.D.; Patil, N.S.; de Almeida, P.; Kadel, I.E.E.; Madireddi, S.; Au-Yeung, A.; Takahashi, C.; et al. Intratumoral CD103+ CD8+ T cells predict response to PD-L1 blockade. J. Immunother. Cancer 2021, 9, e002231. [Google Scholar] [CrossRef]
- Byrne, A.; Savas, P.; Sant, S.; Li, R.; Virassamy, B.; Luen, S.J.; Beavis, P.; Mackay, L.K.; Neeson, P.J.; Loi, S. Tissue-resident memory T cells in breast cancer control and immunotherapy responses. Nat. Rev. Clin. Oncol. 2020, 17, 341–348. [Google Scholar] [CrossRef]
- Park, S.; Gebhardt, T.; Mackay, L.K. Tissue-Resident Memory T Cells in Cancer Immunosurveillance. Trends Immunol. 2019, 40, 735–747. [Google Scholar] [CrossRef]
- Yost, K.E.; Satpathy, A.T.; Wells, D.K.; Qi, Y.; Wang, C.; Kageyama, R.; McNamara, K.L.; Granja, J.M.; Sarin, K.Y.; Brown, R.A.; et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat. Med. 2019, 25, 1251–1259. [Google Scholar] [CrossRef]
- Egelston, C.A.; Avalos, C.; Tu, T.Y.; Rosario, A.; Wang, R.; Solomon, S.; Srinivasan, G.; Nelson, M.S.; Huang, Y.; Lim, M.H.; et al. Resident memory CD8+ T cells within cancer islands mediate survival in breast cancer patients. JCI Insight 2019, 4, e130000. [Google Scholar] [CrossRef]
- Wang, Z.-Q.; Milne, K.; DeRocher, H.; Webb, J.R.; Nelson, B.H.; Watson, P.H. CD103 and Intratumoral Immune Response in Breast Cancer. Clin. Cancer Res. 2016, 22, 6290–6297. [Google Scholar] [CrossRef]
- de Lara, P.T.; Castañón, H.; Vermeer, M.; Núñez, N.; Silina, K.; Sobottka, B.; Urdinez, J.; Cecconi, V.; Yagita, H.; Attar, F.M.; et al. CD39+PD-1+CD8+ T cells mediate metastatic dormancy in breast cancer. Nat. Commun. 2021, 12, 1–14. [Google Scholar] [CrossRef]
- Meier, S.L.; Satpathy, A.T.; Wells, D.K. Bystander T cells in cancer immunology and therapy. Nat. Cancer 2022, 3, 143–155. [Google Scholar] [CrossRef]
- Yeong, J.; Suteja, L.; Simoni, Y.; Lau, K.W.; Tan, A.C.; Li, H.H.; Lim, S.; Loh, J.H.; Wee, F.Y.; Nerurkar, S.N.; et al. Intratumoral CD39+CD8+ T Cells Predict Response to Programmed Cell Death Protein-1 or Programmed Death Ligand-1 Blockade in Patients With NSCLC. J. Thorac. Oncol. 2021, 16, 1349–1358. [Google Scholar] [CrossRef]
- Takeuchi, A.; Saito, T. CD4 CTL, a Cytotoxic Subset of CD4+ T Cells, Their Differentiation and Function. Front. Immunol. 2017, 8, 194. [Google Scholar] [CrossRef]
- Li, S.; Zhuang, S.; Heit, A.; Koo, S.-L.; Tan, A.C.; Chow, I.-T.; Kwok, W.W.; Tan, I.B.; Tan, D.S.; Simoni, Y.; et al. Bystander CD4+ T cells infiltrate human tumors and are phenotypically distinct. OncoImmunology 2022, 11, 2012961. [Google Scholar] [CrossRef]
- Hori, S.; Nomura, T.; Sakaguchi, S. Control of Regulatory T Cell Development by the Transcription Factor Foxp3. Science 2003, 299, 1057–1061. [Google Scholar] [CrossRef]
- Lu, Y.; Wang, X.; Gu, J.; Lu, H.; Zhang, F.; Li, X.; Qian, X.; Wang, X.; Lu, L. iTreg induced from CD39+ naive T cells demonstrate enhanced proliferate and suppressive ability. Int. Immunopharmacol. 2015, 28, 925–930. [Google Scholar] [CrossRef]
- Plitas, G.; Konopacki, C.; Wu, K.; Bos, P.D.; Morrow, M.; Putintseva, E.V.; Chudakov, D.M.; Rudensky, A.Y. Regulatory T Cells Exhibit Distinct Features in Human Breast Cancer. Immunity 2016, 45, 1122–1134. [Google Scholar] [CrossRef]
- Plitas, G.; Rudensky, A.Y. Regulatory T Cells in Cancer. Annu. Rev. Cancer Biol. 2020, 4, 459–477. [Google Scholar] [CrossRef]
- Kos, K.; de Visser, K.E. The Multifaceted Role of Regulatory T Cells in Breast Cancer. Annu. Rev. Cancer Biol. 2021, 5, 291–310. [Google Scholar] [CrossRef]
- Liu, S.; Foulkes, W.D.; Leung, S.; Gao, D.; Lau, S.; Kos, Z.; Nielsen, T.O. Prognostic significance of FOXP3+ tumor-infiltrating lymphocytes in breast cancer depends on estrogen receptor and human epidermal growth factor receptor-2 expression status and concurrent cytotoxic T-cell infiltration. Breast Cancer Res. 2014, 16, 1–12. [Google Scholar] [CrossRef]
- Mahmoud, S.; Paish, E.C.; Powe, D.G.; Macmillan, R.D.; Lee, A.H.S.; Ellis, I.; Green, A. An evaluation of the clinical significance of FOXP3+ infiltrating cells in human breast cancer. Breast Cancer Res. Treat. 2010, 127, 99–108. [Google Scholar] [CrossRef]
- West, N.; Kost, S.E.; Martin, S.; Milne, K.; DeLeeuw, R.J.; Nelson, B.; Watson, P.H. Tumour-infiltrating FOXP3+ lymphocytes are associated with cytotoxic immune responses and good clinical outcome in oestrogen receptor-negative breast cancer. Br. J. Cancer 2012, 108, 155–162. [Google Scholar] [CrossRef]
- Brown, C.C.; Gottschalk, R.A. Volume control: Turning the dial on regulatory T cells. Cell 2021, 184, 3847–3849. [Google Scholar] [CrossRef]
- Marangoni, F.; Zhakyp, A.; Corsini, M.; Geels, S.N.; Carrizosa, E.; Thelen, M.; Mani, V.; Prüßmann, J.N.; Warner, R.D.; Ozga, A.J.; et al. Expansion of tumor-associated Treg cells upon disruption of a CTLA-4-dependent feedback loop. Cell 2021, 184, 3998–4015.e19. [Google Scholar] [CrossRef] [PubMed]
- Wong, H.S.; Park, K.; Gola, A.; Baptista, A.P.; Miller, C.H.; Deep, D.; Lou, M.; Boyd, L.F.; Rudensky, A.Y.; Savage, P.A.; et al. A local regulatory T cell feedback circuit maintains immune homeostasis by pruning self-activated T cells. Cell 2021, 184, 3981–3997.e22. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, S.; Togashi, Y.; Kamada, T.; Sugiyama, E.; Nishinakamura, H.; Takeuchi, Y.; Vitaly, K.; Itahashi, K.; Maeda, Y.; Matsui, S.; et al. The PD-1 expression balance between effector and regulatory T cells predicts the clinical efficacy of PD-1 blockade therapies. Nat. Immunol. 2020, 21, 1346–1358. [Google Scholar] [CrossRef] [PubMed]
- Kamada, T.; Togashi, Y.; Tay, C.; Ha, D.; Sasaki, A.; Nakamura, Y.; Sato, E.; Fukuoka, S.; Tada, Y.; Tanaka, A.; et al. PD-1+ regulatory T cells amplified by PD-1 blockade promote hyperprogression of cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 9999–10008. [Google Scholar] [CrossRef] [PubMed]
- Garaud, S.; Dieu-Nosjean, M.-C.; Willard-Gallo, K. T follicular helper and B cell crosstalk in tertiary lymphoid structures and cancer immunotherapy. Nat. Commun. 2022, 13, 2259. [Google Scholar] [CrossRef]
- Laumont, C.M.; Banville, A.C.; Gilardi, M.; Hollern, D.P.; Nelson, B.H. Tumour-infiltrating B cells: Immunological mechanisms, clinical impact and therapeutic opportunities. Nat. Cancer 2022, 22, 414–430. [Google Scholar] [CrossRef]
- Knutson, K.L.; Clynes, R.; Shreeder, B.; Yeramian, P.; Kemp, K.P.; Ballman, K.; Tenner, K.S.; Erskine, C.L.; Norton, N.; Northfelt, D.; et al. Improved Survival of HER2+ Breast Cancer Patients Treated with Trastuzumab and Chemotherapy Is Associated with Host Antibody Immunity against the HER2 Intracellular Domain. Cancer Res. 2016, 76, 3702–3710. [Google Scholar] [CrossRef]
- Garaud, S.; Buisseret, L.; Solinas, C.; Gu-Trantien, C.; De Wind, A.; Eynden, G.V.D.; Naveaux, C.; Lodewyckx, J.-N.; Boisson, A.; Duvillier, H.; et al. Tumor-infiltrating B cells signal functional humoral immune responses in breast cancer. JCI Insight 2019, 4, e129641. [Google Scholar] [CrossRef]
- Harris, R.J.; Cheung, A.; Ng, J.C.F.; Laddach, R.; Chenoweth, A.M.; Crescioli, S.; Fittall, M.; Dominguez-Rodriguez, D.; Roberts, J.; Levi, D.; et al. Tumor-Infiltrating B Lymphocyte Profiling Identifies IgG-Biased, Clonally Expanded Prognostic Phenotypes in Triple-Negative Breast Cancer. Cancer Res. 2021, 81, 4290–4304. [Google Scholar] [CrossRef]
- Liu, X.; Tsang, J.Y.; Hlaing, T.; Hu, J.; Ni, Y.-B.; Chan, S.K.; Cheung, S.Y.; Tse, G.M. Distinct Tertiary Lymphoid Structure Associations and Their Prognostic Relevance in HER2 Positive and Negative Breast Cancers. Oncologist 2017, 22, 1316–1324. [Google Scholar] [CrossRef]
- Millar, E.; Browne, L.; Slapetova, I.; Shang, F.; Ren, Y.; Bradshaw, R.; Brauer, H.A.; O’Toole, S.; Beretov, J.; Whan, R.; et al. TILs Immunophenotype in Breast Cancer Predicts Local Failure and Overall Survival: Analysis in a Large Radiotherapy Trial with Long-Term Follow-Up. Cancers 2020, 12, 2365. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, S.; Lee, A.H.S.; Paish, E.C.; Macmillan, R.D.; Ellis, I.; Green, A.R. The prognostic significance of B lymphocytes in invasive carcinoma of the breast. Breast Cancer Res. Treat. 2011, 132, 545–553. [Google Scholar] [CrossRef] [PubMed]
- Wortman, J.C.; He, T.-F.; Solomon, S.; Zhang, R.Z.; Rosario, A.; Wang, R.; Tu, T.Y.; Schmolze, D.; Yuan, Y.; Yost, S.E.; et al. Spatial distribution of B cells and lymphocyte clusters as a predictor of triple-negative breast cancer outcome. NPJ Breast Cancer 2021, 7, 84. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; Loibl, S.; Noske, A.; Roller, M.; Müller, B.M.; Komor, M.; Budczies, J.; Darb-Esfahani, S.; Kronenwett, R.; Hanusch, C.; et al. Tumor-Associated Lymphocytes As an Independent Predictor of Response to Neoadjuvant Chemotherapy in Breast Cancer. J. Clin. Oncol. 2010, 28, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.R.; Wimberly, H.; Lannin, D.R.; Nixon, C.; Rimm, D.L.; Bossuyt, V. Multiplexed Quantitative Analysis of CD3, CD8, and CD20 Predicts Response to Neoadjuvant Chemotherapy in Breast Cancer. Clin. Cancer Res. 2014, 20, 5995–6005. [Google Scholar] [CrossRef]
- García-Martínez, E.; Gil, G.L.; Benito, A.C.; González-Billalabeitia, E.; Conesa, M.A.V.; García, T.G.; García-Garre, E.; Vicente, V.; Ayala De La Peña, F. Tumor-infiltrating immune cell profiles and their change after neoadjuvant chemotherapy predict response and prognosis of breast cancer. Breast Cancer Res. 2014, 16, 488. [Google Scholar] [CrossRef]
- De Angelis, C.; Nagi, C.; Hoyt, C.C.; Liu, L.; Roman, K.; Wang, C.; Zheng, Y.; Veeraraghavan, J.; Sethunath, V.; Nuciforo, P.; et al. Evaluation of the Predictive Role of Tumor Immune Infiltrate in Patients with HER2-Positive Breast Cancer Treated with Neoadjuvant Anti-HER2 Therapy without Chemotherapy. Clin. Cancer Res. 2020, 26, 738–745. [Google Scholar] [CrossRef]
- Ishigami, E.; Sakakibara, M.; Sakakibara, J.; Masuda, T.; Fujimoto, H.; Hayama, S.; Nagashima, T.; Sangai, T.; Nakagawa, A.; Nakatani, Y.; et al. Coexistence of Regulatory B Cells and Regulatory T Cells in Tu-mor-Infiltrating Lymphocyte Aggregates Is a Prognostic Factor in Patients with Breast Cancer. Breast Cancer 2019, 26, 180–189. [Google Scholar] [CrossRef]
- Helmink, B.A.; Reddy, S.M.; Gao, J.; Zhang, S.; Basar, R.; Thakur, R.; Yizhak, K.; Sade-Feldman, M.; Blando, J.; Han, G.; et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature 2020, 577, 549–555. [Google Scholar] [CrossRef]
- Petitprez, F.; de Reyniès, A.; Keung, E.Z.; Chen, T.W.-W.; Sun, C.-M.; Calderaro, J.; Jeng, Y.-M.; Hsiao, L.-P.; Lacroix, L.; Bougoüin, A.; et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature 2020, 577, 556–560. [Google Scholar] [CrossRef]
- Gao, Z.-H.; Li, C.-X.; Liu, M.; Jiang, J.-Y. Predictive and prognostic role of tumour-infiltrating lymphocytes in breast cancer patients with different molecular subtypes: A meta-analysis. BMC Cancer 2020, 20, 1150. [Google Scholar] [CrossRef] [PubMed]
- Cabrita, R.; Lauss, M.; Sanna, A.; Donia, M.; Larsen, M.S.; Mitra, S.; Johansson, I.; Phung, B.; Harbst, K.; Vallon-Christersson, J.; et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature 2020, 577, 561–565. [Google Scholar] [CrossRef] [PubMed]
- Gu-Trantien, C.; Loi, S.; Garaud, S.; Equeter, C.; Libin, M.; De Wind, A.; Ravoet, M.; Le Buanec, H.; Sibille, C.; Manfouo-Foutsop, G.; et al. CD4+ follicular helper T cell infiltration predicts breast cancer survival. J. Clin. Investig. 2013, 123, 2873–2892. [Google Scholar] [CrossRef]
- Gu-Trantien, C.; Migliori, E.; Buisseret, L.; De Wind, A.; Brohée, S.; Garaud, S.; Noël, G.; Chi, V.L.; Lodewyckx, J.N.J.; Naveaux, C.C.; et al. CXCL13-producing TFH cells link immune suppression and adaptive memory in human breast cancer. JCI Insight 2017, 2, e91487. [Google Scholar] [CrossRef]
- Meylan, M.; Petitprez, F.; Becht, E.; Bougoüin, A.; Pupier, G.; Calvez, A.; Giglioli, I.; Verkarre, V.; Lacroix, G.; Verneau, J.; et al. Tertiary lymphoid structures generate and propagate anti-tumor antibody-producing plasma cells in renal cell cancer. Immunity 2022, 55, 527–541.e5. [Google Scholar] [CrossRef] [PubMed]
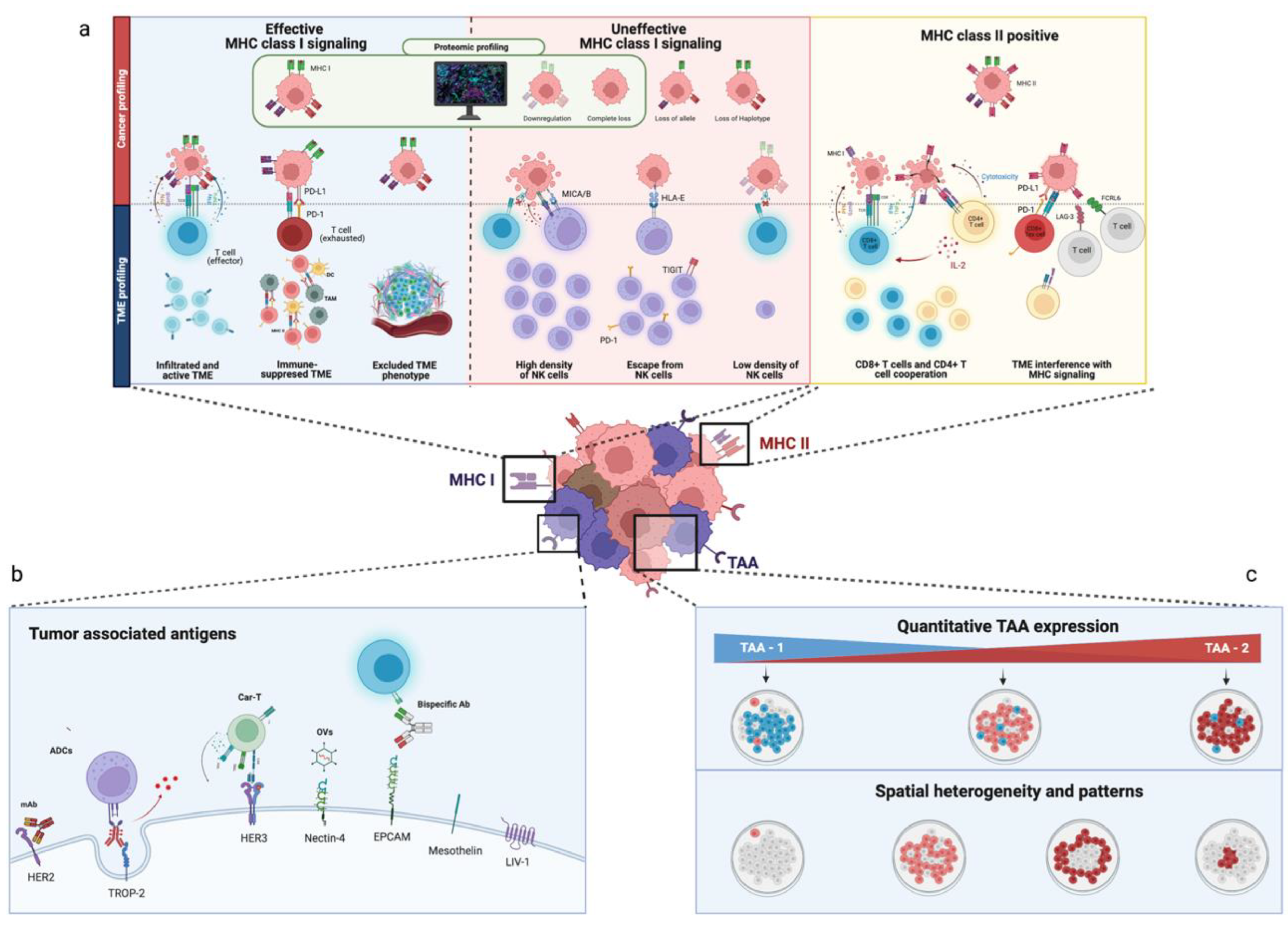
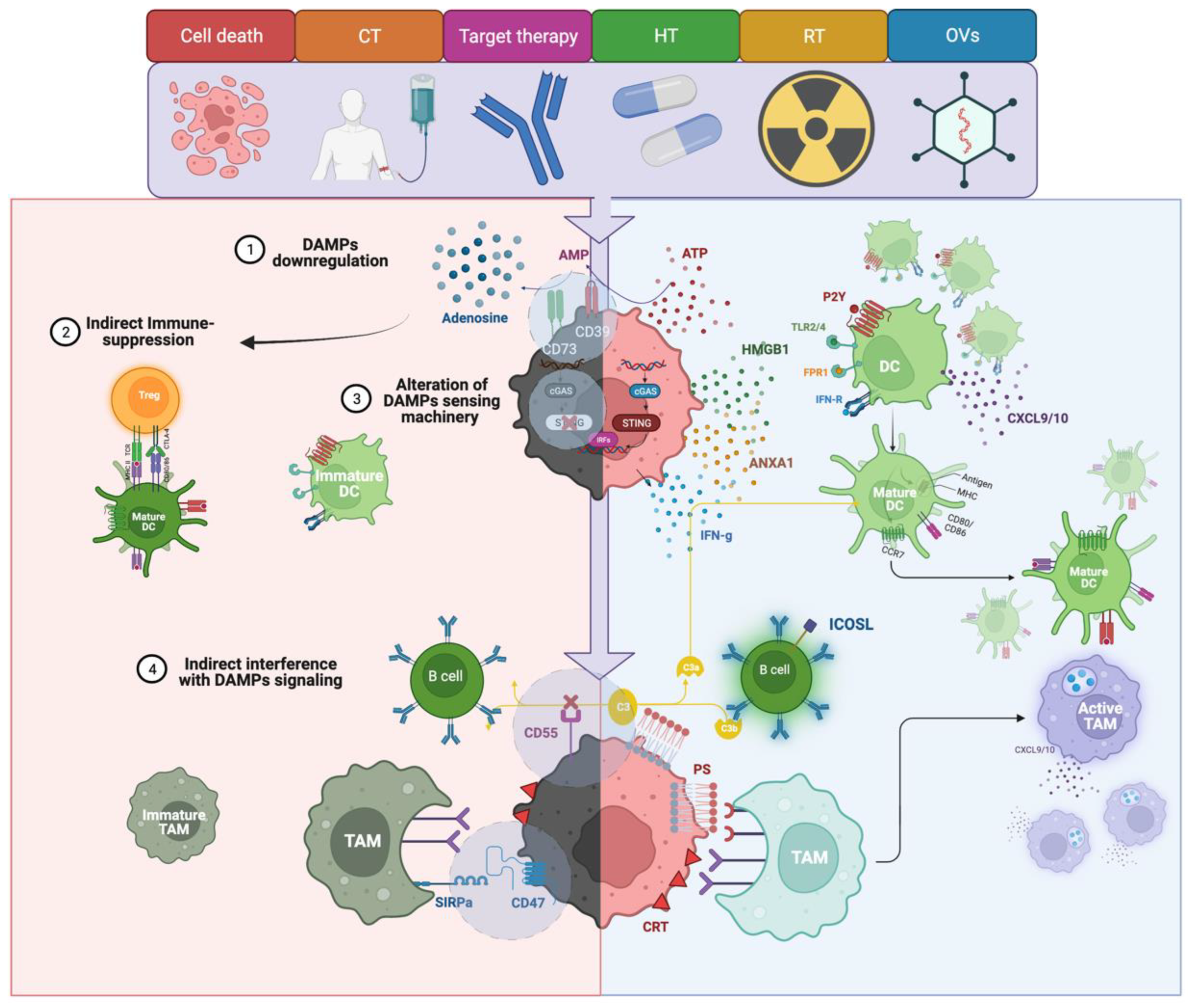
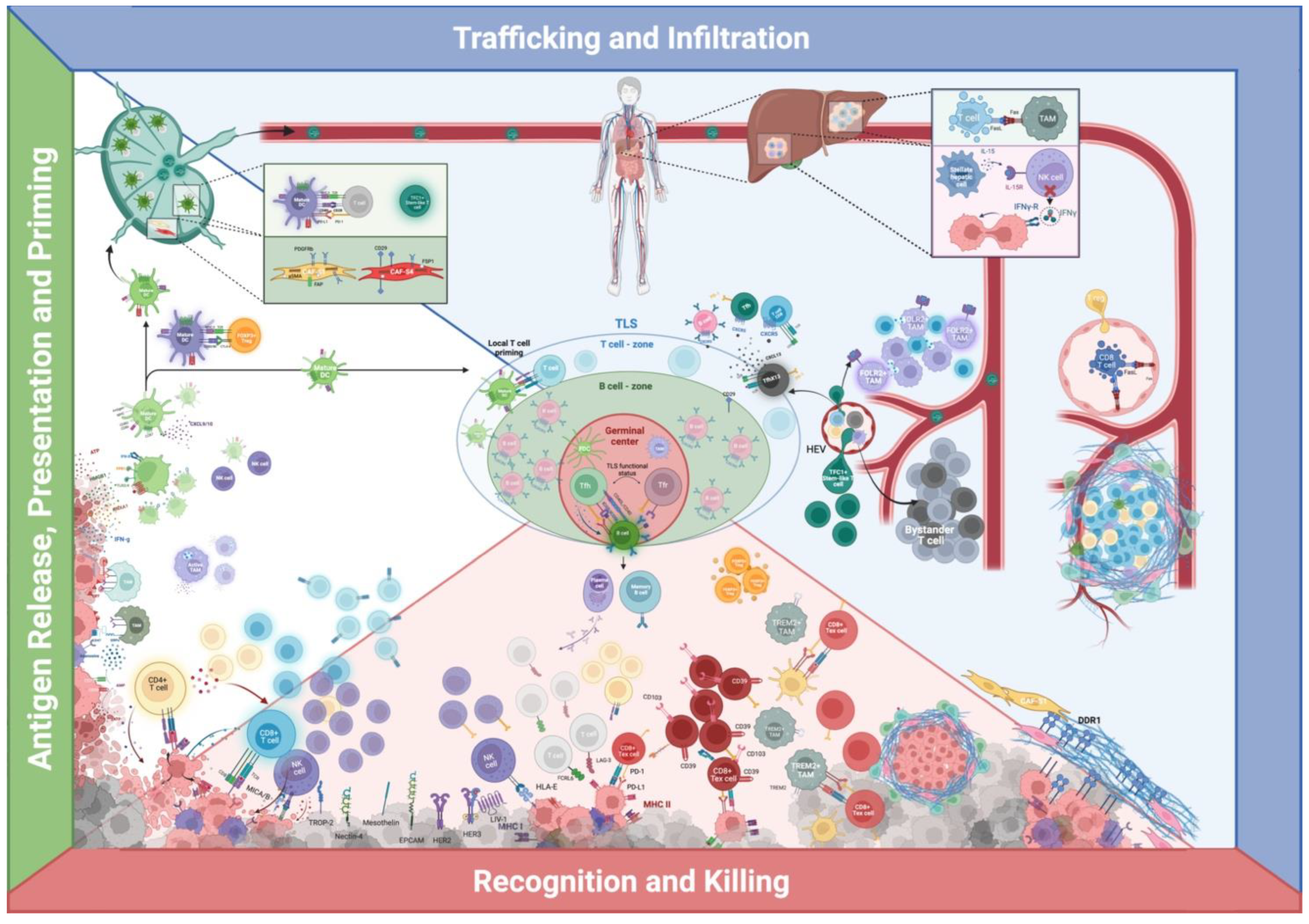
| BC Subtype | Trial | Phase | Treatment Arms | Primary Efficacy Endpoints | Biomarker Analysis | N. Patients (Trial Status) | References | |
|---|---|---|---|---|---|---|---|---|
| TNBC | Early TNBC | Keynote-173 | I/II | Pembro + T +/− Cb > AC− > S− > pembro adj x 1yr | pCR | PT: sTILs and PD-L1 associated with pCR and ORR; and OT: sTILs associated with pCR and ORR | 60 (completed) | [205] |
| I-SPY 2 | II | Pembro/placebo + T > AC | pCR | MHC II expression predictive of response to ICI | 64 (completed) | [82] | ||
| NeoPACT | II | Pembro + CbD | pCR | High sTILs are associated with higher pCR | 117 (active, not recruiting) | [204] | ||
| Keynote-522 | III | Pembro/Placebo + CbT > AC > CH > pembro adj x 1yr | pCR + EFS | PD-L1 CPS not predictive of response to ICI | 602 (completed | [12] | ||
| NeoTRIPaPDL1 | III | Atezo/placebo + NabP + Cb > S > anthracycline-based CT | EFS | pCR rate + 10% to atezo in immune-rich TME (PDL1 IC+, high/intermediate sTILs/iTILs) High expression of GATA3 and CD20, epithelial of HLA-DR and Ki67 in both epithelial and TME favors atezo arm; and PD-L1 + IDO+ APC and CD56 NE cells were associated with a higher response rate to atezo. | 278 (active, not recruiting) | [83,206] | ||
| Impassion031 | III | Atezo/placebo + NabT > AC | pCR | PT: PD-L1 IC+ and TC+, sTILs, iTILs, and TLS linked to improved pCR in placebo arm; and OT: numerical increase in iTILs and PD-L1 in immune cells in patients with pCR in ICI arm; further, ICI can promote close contact of TILs to tumor nests | 455 (active, not recruiting) | [207] | ||
| NCT02489448 | I/II | Durva + nab-paclitaxel > AC | pCR | IHC: sTILs associated with higher pCR, sTILs, and PD-L1 do not predict benefit in multivariate analysis; MHC II expression predicts response to ICI; and mIF: PD-L1 TC+, IC+ in the stroma and PD-L1+ CD68+ TAM compartment each associated with higher rates of pCR | 69 (completed) | [82,231] | ||
| GeparNuevo | II | Durva/placebo + nab-paclitaxel | pCR | PT: High sTILs associated with higher pCR in both arms; OT: iTILs increase = higher pCR in ICI arm; and RD: high TILs associated low rates of relapse in both arms | 174 (completed) | [203,232,233,234] | ||
| Metastatic TNBC | Keynote-119 | III | Pembro/CT | OS | sTILs associated with ICI benefit, in particular in previously untreated mTNBC; and PD-L1 TC+ adds predictive power to pembro arm | 622 (completed) | [26,209,210] | |
| Keynote-086 | II | Pembro | DCR, ORR, DoR, PFS, OS | sTILs, PD-L1 CPS+, and CD8 IHC evaluation correlate with the response rate to pembro | 254 (completed) | [211] | ||
| Keynote-355 | III | Pembro/placebo + NabP/T/Gem + Cb | PFS, OS | PD-L1 CPS ≥ 10 correlates with improved PFS and OS | 847 (active, not recruiting) | [11] | ||
| ENHANCE-I | Ib/II | Pembro + eribulin | ORR | PD-L1 numerically higher ORR | 167 (completed) | [235] | ||
| Impassion130 | III | Atezo/placebo + NabT | PFS, OS | PD-L1 IC+ predictive of ICI benefit; and PD-L1 IC+ and either PD-L1 TC+ or 10% or more sTILs had the highest clinical activity with A + nP | 902 (completed) | [10,212] | ||
| Impassion131 | III | Atezo/placebo + T | PFS | PD-L1 IC+ does not predict benefit | 651 (active, not recruiting) | [28,236] | ||
| TONIC | II | Nivo/nivo after induction with CT or RT | PFS | PD-L1 IC+, sTILs, and CD8+ higher in responders | 67 (active, not recruiting) | [109] | ||
| HER2 | Early HER2+ BC | Impassion-050 | III | Atezo/placebo + THP + AC > S > atezo/placebo + HP | pCR | PD-L1 IC+ does not predict pCR | 454 (active, not recruiting) | [237] |
| LABC/Metastatic HER2+ BC | NCT02605915 | Ib | In LABC: Atezo + HP or atezo/T-DM1 > THP + Cb In mBC: Atezo with trastuzumab/pertuzumab, atezo with T-DM1, or atezo with THP | ORR + DoR | PT: No correlation between response and PD-L1 IC+, TC+, sTILs, and CD8+ T-cell density in central tumor area and immune phenotypes (ID, IE, or IN); OT: increase in PD-L1 IC+ in both cohorts, no association with response; and LABC: significant increase in CD8+ T cells density in the central tumoral area, but not correlated with pCR, no increase in mBC | 76 (completed) | [238] | |
| Metastatic HER2+ BC | PANACEA | I/II | Pembro + trastuzumab | ORR | sTILs correlate with ORR and disease control, as well as higher clinical benefit in PD-L1 + CPS | 58 (completed) | [239] | |
| KATE-2 | II | Atezo/placebo + T-DM1 | PFS | High CD8 T cells at invasive margins favor atezo arm in subgroup analysis | 1486 (completed) | [220] | ||
| HR+ | Early HR+ BC | GIADA | II | Nivo + exemestane + triptorelin + EC | pCR | PT: in pCR patients higher in sTILs, iTILs (iCD4, I CD8, and iCD4+ FOXP3+), and TAMs (intratumoral); TAMs: stromal CD68+ CD163+ TAMs) immune-checkpoints co-expression: PD-1+ on T cells, and PD-L1 on TAMs (CD68+ PD-L1+ and CD68+ CD163+ PD-L1) higher in pCR; OT after CT: sTILs increase, increase in CD8+ T cells, decrease in FOXP3+ CD4+ T cells, and CD68+ CD163+ TAMs; and OT after Nivo: increase in intratumoral and stromal CD8+, CD8+ Granzyme+ T cells and stromal CD4+ Granzyme B+ T cells. | 43 (completed) | [23] |
| ISPY-2 | II | Pembro/placebo + T > AC | pCR | MHC II expression predicts response to ICI | 89 (completed) | [82] | ||
| Metastatic HR+ BC | Keynote-028 | Ib | Pembro | ORR | sTILs do not predict PFS | 83 (completed) | [240] | |
| KELLY | II | Pembro + eribulin | CBR | PD-L1 does not predict benefit | 44 (completed) | [241] | ||
| NCT03051659 | II | Pembro + eribulin | PFS | sTILs and PD-L1 do not predict benefit | 88 (active, not recruiting) | [242] | ||
| NCT03044730 | II | Pembro + capecitabine | PFS | sTILs and PD-L1 do not predict benefit | 14 (completed) | [243] | ||
| NIMBUS | II | Pembro + nivo in TMB-H | ORR | sTILs and PD-L1 do not predict benefit | 20 (active, not recruiting) | [28] | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Massa, D.; Tosi, A.; Rosato, A.; Guarneri, V.; Dieci, M.V. Multiplexed In Situ Spatial Protein Profiling in the Pursuit of Precision Immuno-Oncology for Patients with Breast Cancer. Cancers 2022, 14, 4885. https://doi.org/10.3390/cancers14194885
Massa D, Tosi A, Rosato A, Guarneri V, Dieci MV. Multiplexed In Situ Spatial Protein Profiling in the Pursuit of Precision Immuno-Oncology for Patients with Breast Cancer. Cancers. 2022; 14(19):4885. https://doi.org/10.3390/cancers14194885
Chicago/Turabian StyleMassa, Davide, Anna Tosi, Antonio Rosato, Valentina Guarneri, and Maria Vittoria Dieci. 2022. "Multiplexed In Situ Spatial Protein Profiling in the Pursuit of Precision Immuno-Oncology for Patients with Breast Cancer" Cancers 14, no. 19: 4885. https://doi.org/10.3390/cancers14194885
APA StyleMassa, D., Tosi, A., Rosato, A., Guarneri, V., & Dieci, M. V. (2022). Multiplexed In Situ Spatial Protein Profiling in the Pursuit of Precision Immuno-Oncology for Patients with Breast Cancer. Cancers, 14(19), 4885. https://doi.org/10.3390/cancers14194885







