Molecular Characterization Reveals Subclasses of 1q Gain in Intermediate Risk Wilms Tumors
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cohort Selection and Sequencing
2.2. Variant Calling
2.3. Mutation Burden
2.4. Mutational Signatures
2.5. Copy Number Clustering
2.6. Expression Profile Extraction and Expression Clustering
2.7. Gene Set Enrichment of Expression Profiles
2.8. Effects of Copy Number Alterations on Gene Expression
2.9. Cancer Genes
2.10. Wnt Signaling Pathway Analysis
2.11. Joint Analysis of Recurrent CNs/SVs and Recurrent Expression Changes
2.12. Gene-Level Integration of SNVs, CNs and SVs
2.13. Visualization
3. Results
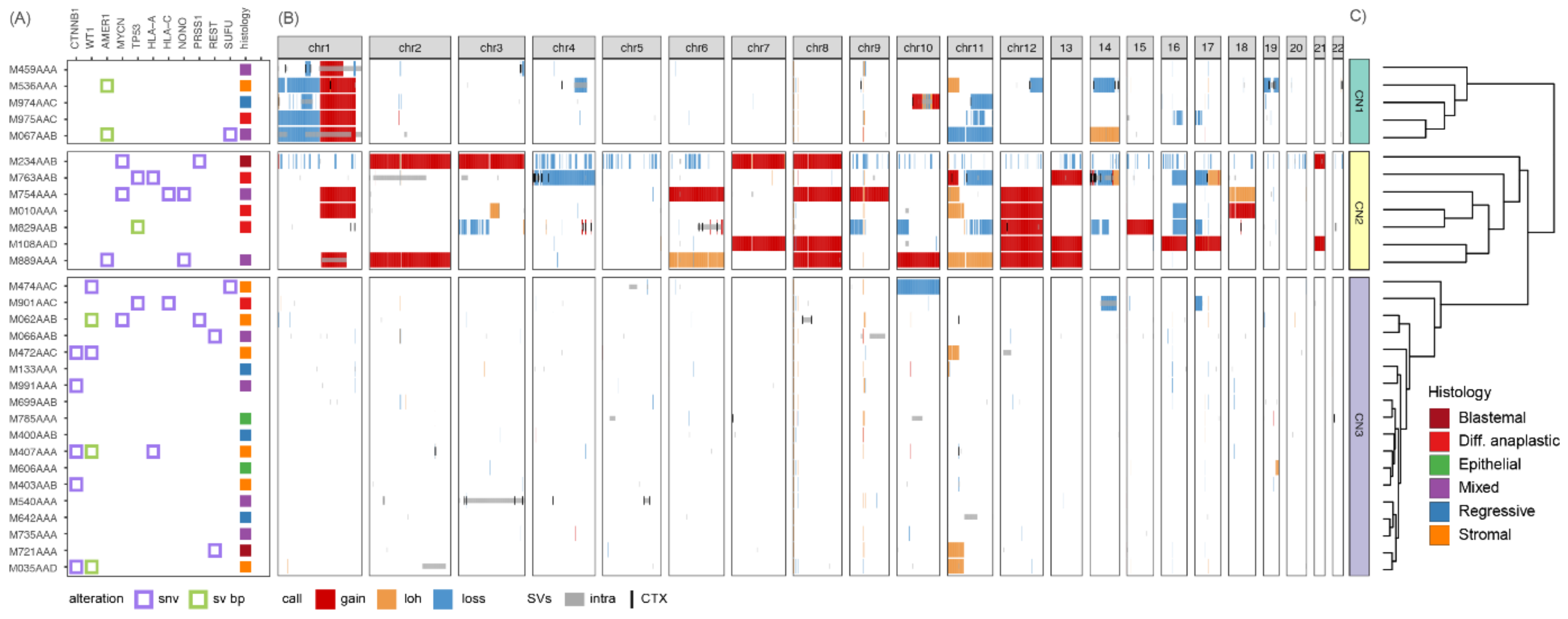
3.1. Copy Number Profiles Cluster Tumors into Three Groups Reflecting Different Degrees of Genomic Instability
3.2. Different Somatic Alterations Converge on Four Gene Expression Clusters
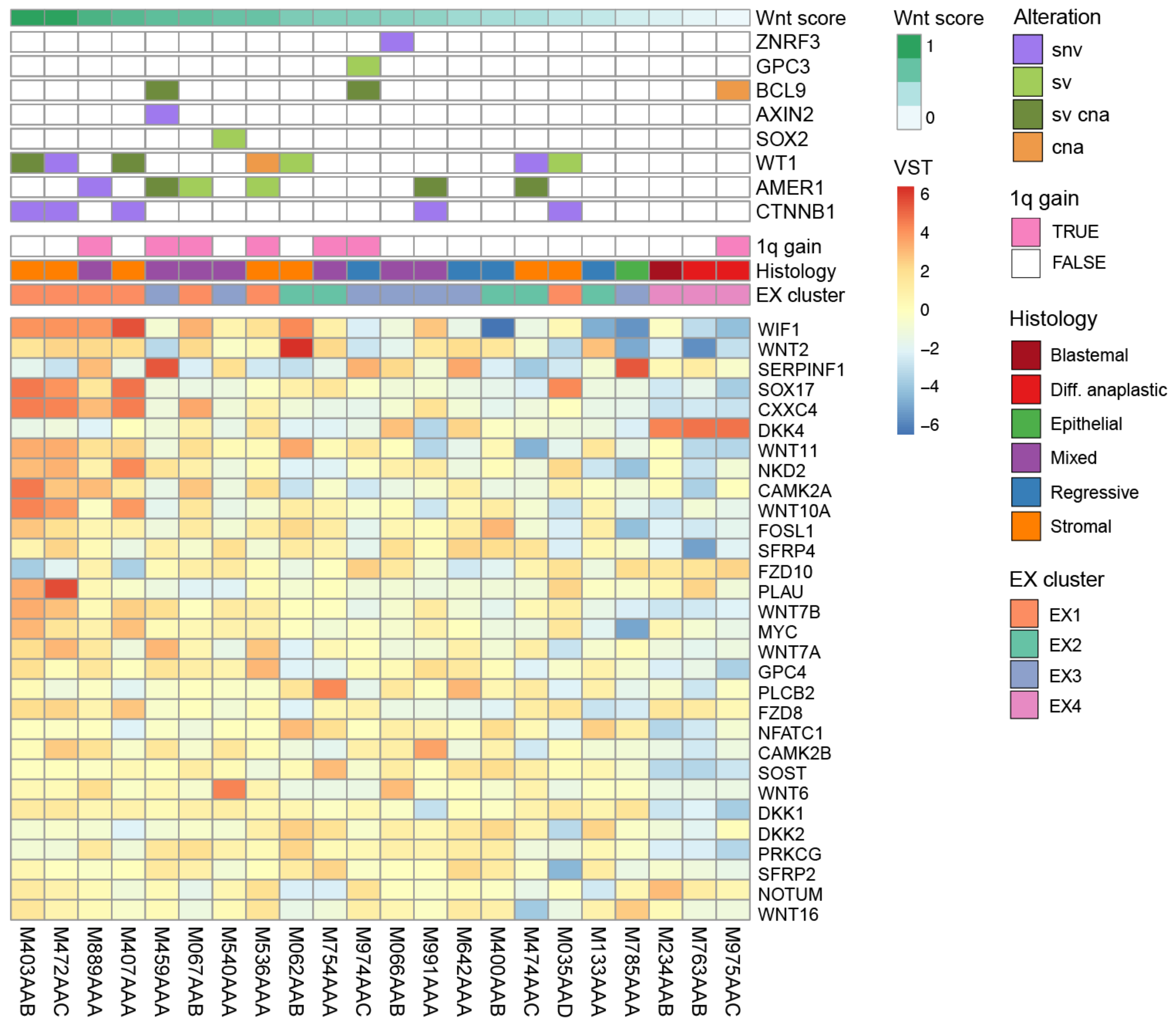
3.3. CNs/SVs Result in Wnt Pathway Activation
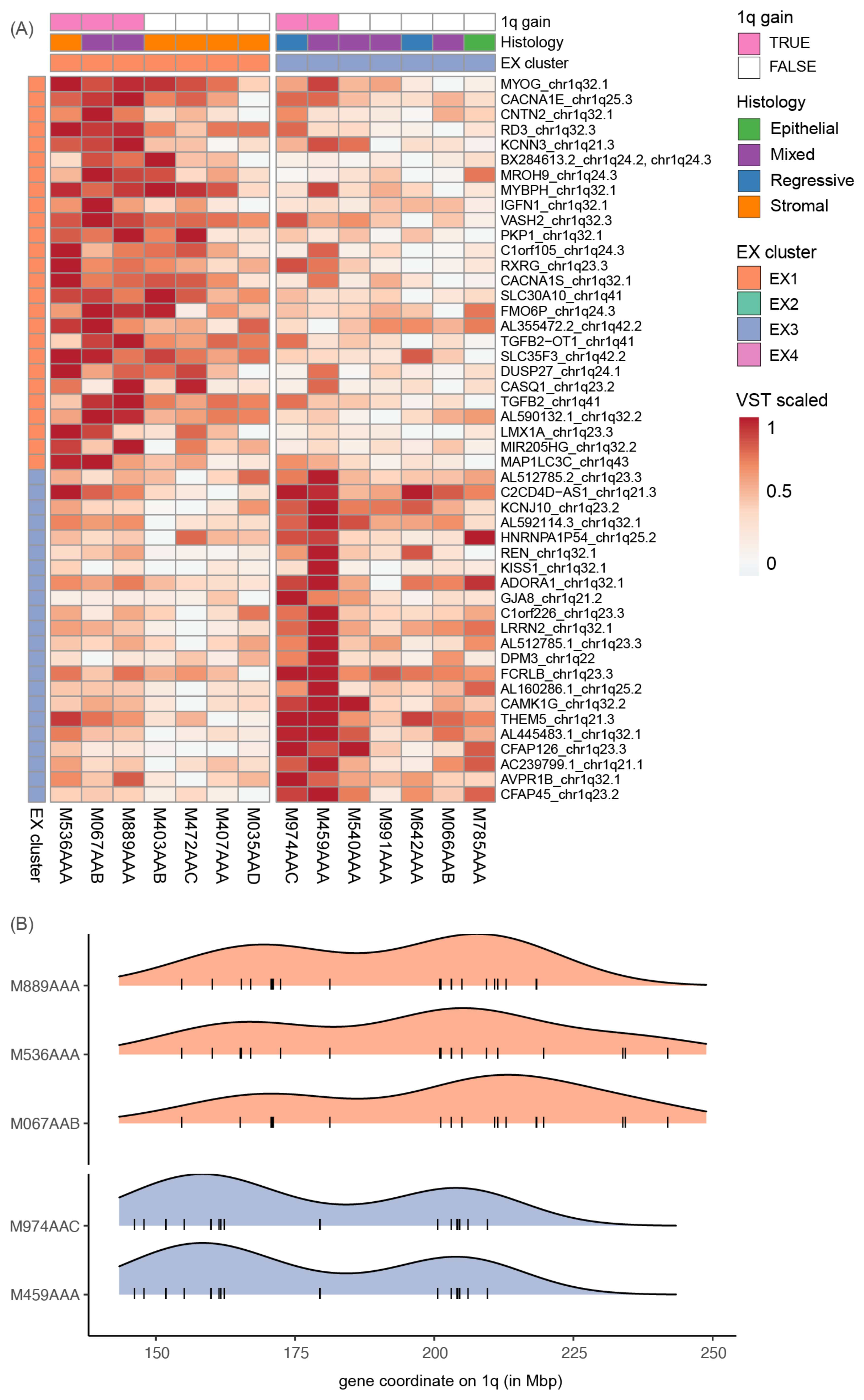
3.4. 1q Gain Is Associated with Overexpression of Different Gene Sets
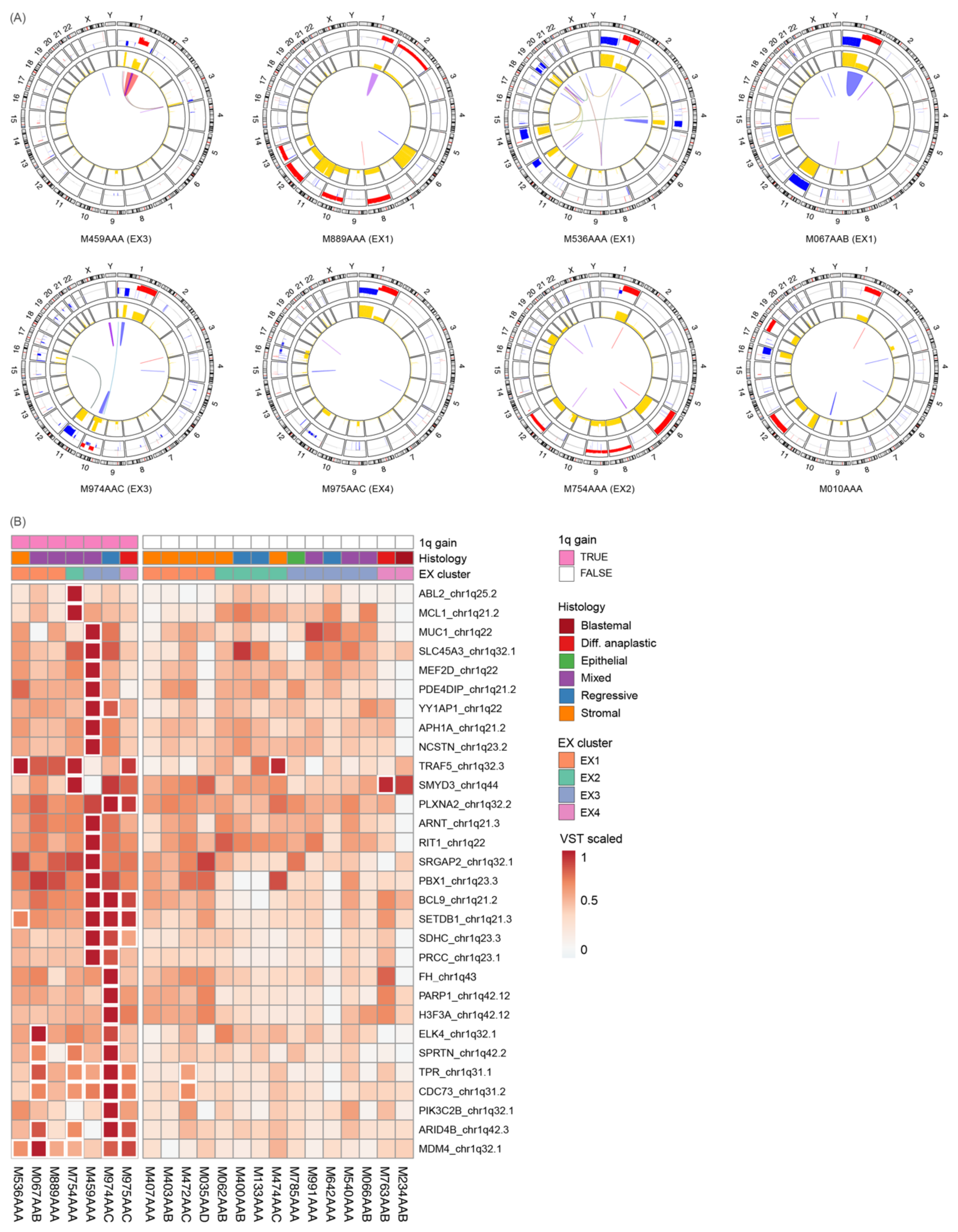
3.5. 1q Gain Arises through Different Mutational Mechanisms
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Byron, S.A.; Hendricks, W.P.D.; Nagulapally, A.B.; Kraveka, J.M.; Ferguson, W.S.; Brown, V.I.; Eslin, D.E.; Mitchell, D.; Cornelius, A.; Roberts, W.; et al. Genomic and Transcriptomic Analysis of Relapsed and Refractory Childhood Solid Tumors Reveals a Diverse Molecular Landscape and Mechanisms of Immune Evasion. Cancer Res. 2021, 81, 5818–5832. [Google Scholar] [CrossRef] [PubMed]
- Gratias, E.J.; Dome, J.S.; Jennings, L.J.; Chi, Y.Y.; Tian, J.; Anderson, J.; Grundy, P.; Mullen, E.A.; Geller, J.I.; Fernandez, C.V.; et al. Association of Chromosome 1q Gain With Inferior Survival in Favorable-Histology Wilms Tumor: A Report From the Children’s Oncology Group. J. Clin. Oncol. 2016, 34, 3189–3194. [Google Scholar] [CrossRef] [PubMed]
- Gratias, E.J.; Jennings, L.J.; Anderson, J.R.; Dome, J.S.; Grundy, P.; Perlman, E.J. Gain of 1q Is Associated with Inferior Event-Free and Overall Survival in Patients with Favorable Histology Wilms Tumor: A Report from the Children’s Oncology Group. Cancer 2013, 119, 3887–3894. [Google Scholar] [CrossRef] [PubMed]
- Chagtai, T.; Zill, C.; Dainese, L.; Wegert, J.; Savola, S.; Popov, S.; Mifsud, W.; Vujanić, G.; Sebire, N.; Le Bouc, Y.; et al. Gain of 1q As a Prognostic Biomarker in Wilms Tumors (WTs) Treated With Preoperative Chemotherapy in the International Society of Paediatric Oncology (SIOP) WT 2001 Trial: A SIOP Renal Tumours Biology Consortium Study. J. Clin. Oncol. 2016, 34. [Google Scholar] [CrossRef] [PubMed]
- Van den Heuvel-Eibrink, M.M.; Hol, J.A.; Pritchard-Jones, K.; van Tinteren, H.; Furtwängler, R.; Verschuur, A.C.; Vujanic, G.M.; Leuschner, I.; Brok, J.; Rübe, C.; et al. Position Paper: Rationale for the Treatment of Wilms Tumour in the UMBRELLA SIOP-RTSG 2016 Protocol. Nat. Rev. Urol. 2017, 14, 743–752. [Google Scholar] [CrossRef]
- Vujanić, G.M.; Gessler, M.; Ooms, A.H.; Collini, P.; Coulomb-l’Hermine, A.; D’Hooghe, E.; de Krijger, R.R.; Perotti, D.; Pritchard-Jones, K.; Vokuhl, C.; et al. The UMBRELLA SIOP–RTSG 2016 Wilms Tumour Pathology and Molecular Biology Protocol. Nat. Rev. Urol. 2018, 15, 693–701. [Google Scholar] [CrossRef]
- Treger, T.D.; Chowdhury, T.; Pritchard-Jones, K.; Behjati, S. The Genetic Changes of Wilms Tumour. Nat. Rev. Nephrol. 2019, 15, 240–251. [Google Scholar] [CrossRef]
- Gadd, S.; Huff, V.; Walz, A.L.; Ooms, A.H.A.G.; Armstrong, A.E.; Gerhard, D.S.; Smith, M.A.; Auvil, J.M.G.; Meerzaman, D.; Chen, Q.-R.; et al. A Children’s Oncology Group and TARGET Initiative Exploring the Genetic Landscape of Wilms Tumor. Nat. Genet. 2017, 49, 1487–1494. [Google Scholar] [CrossRef]
- Deng, C.; Dai, R.; Li, X.; Liu, F. Genetic Variation Frequencies in Wilms’ Tumor: A Meta-Analysis and Systematic Review. Cancer Sci. 2016, 107, 690–699. [Google Scholar] [CrossRef]
- Grundy, P.E.; Breslow, N.E.; Li, S.; Perlman, E.; Bruce Beckwith, J.; Ritchey, M.L.; Shamberger, R.C.; Haase, G.M.; D’Angio, G.J.; Donaldson, M.; et al. Loss of Heterozygosity for Chromosomes 1p and 16q Is an Adverse Prognostic Factor in Favorable-Histology Wilms Tumor: A Report From the National Wilms Tumor Study Group. J. Clin. Oncol. 2005, 23, 7312–7321. [Google Scholar] [CrossRef]
- Gadd, S.; Huff, V.; Skol, A.D.; Renfro, L.A.; Fernandez, C.V.; Mullen, E.A.; Jones, C.D.; Hoadley, K.A.; Yap, K.L.; Ramirez, N.C.; et al. Genetic Changes Associated with Relapse in Favorable Histology Wilms Tumor: A Children’s Oncology Group AREN03B2 Study. Cell Rep. Med. 2022, 3, 100644. [Google Scholar] [CrossRef] [PubMed]
- Cresswell, G.D.; Apps, J.R.; Chagtai, T.; Mifsud, B.; Bentley, C.C.; Maschietto, M.; Popov, S.D.; Weeks, M.E.; Olsen, Ø.E.; Sebire, N.J.; et al. Intra-Tumor Genetic Heterogeneity in Wilms Tumor: Clonal Evolution and Clinical Implications. EBioMedicine 2016, 9, 120–129. [Google Scholar] [CrossRef] [PubMed]
- Haruta, M.; Arai, Y.; Okita, H.; Tanaka, Y.; Takimoto, T.; Sugino, R.P.; Yamada, Y.; Kamijo, T.; Oue, T.; Fukuzawa, M.; et al. Combined Genetic and Chromosomal Characterization of Wilms Tumors Identifies Chromosome 12 Gain as a Potential New Marker Predicting a Favorable Outcome. Neoplasia 2019, 21, 117. [Google Scholar] [CrossRef] [PubMed]
- Van Belzen, I.A.; Cai, C.; van Tuil, M.; Badloe, S.; Strengman, E.; Janse, A.; Verwiel, E.T.; van der Leest, D.F.; Kester, L.; Molenaar, J.J.; et al. Systematic Discovery of Gene Fusions in Pediatric Cancer by Integrating RNA-Seq and WGS. bioRxiv 2021, 8, 458342. [Google Scholar]
- Van der Auwera, G.A.; Carneiro, M.O.; Hartl, C.; Poplin, R.; Del Angel, G.; Levy-Moonshine, A.; Jordan, T.; Shakir, K.; Roazen, D.; Thibault, J.; et al. From FastQ Data to High Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Curr. Protoc. Bioinform. 2013, 43, 11.10.1–11.10.33. [Google Scholar]
- Kerstens, H.H.D.; Hehir-Kwa, J.Y.; van de Geer, E.; van Run, C.; Verwiel, E.T.P.; van der Leest, D.; Tops, B.B.J.; Kemmeren, P. Trecode: A FAIR Eco-System for the Analysis and Archiving of Omics Data in a Combined Diagnostic and Research Setting. bioRxiv 2020, 11, 363689. [Google Scholar]
- Benjamin, D.; Sato, T.; Cibulskis, K.; Getz, G.; Stewart, C.; Lichtenstein, L. Calling Somatic SNVs and Indels with Mutect2. bioRxiv 2019, 861054. [Google Scholar] [CrossRef]
- Amemiya, H.M.; Kundaje, A.; Boyle, A.P. The ENCODE Blacklist: Identification of Problematic Regions of the Genome. Sci. Rep. 2019, 9, 9354. [Google Scholar] [CrossRef]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef]
- Chen, X.; Schulz-Trieglaff, O.; Shaw, R.; Barnes, B.; Schlesinger, F.; Källberg, M.; Cox, A.J.; Kruglyak, S.; Saunders, C.T. Manta: Rapid Detection of Structural Variants and Indels for Germline and Cancer Sequencing Applications. Bioinformatics 2016, 32, 1220–1222. [Google Scholar] [CrossRef] [PubMed]
- Rausch, T.; Zichner, T.; Schlattl, A.; Stutz, A.M.; Benes, V.; Korbel, J.O. DELLY: Structural Variant Discovery by Integrated Paired-End and Split-Read Analysis. Bioinformatics 2012, 28, i333–i339. [Google Scholar] [CrossRef] [PubMed]
- Cameron, D.L.; Schröder, J.; Penington, J.S.; Do, H.; Molania, R.; Dobrovic, A.; Speed, T.P.; Papenfuss, A.T. GRIDSS: Sensitive and Specific Genomic Rearrangement Detection Using Positional de Bruijn Graph Assembly. Genome Res. 2017, 27, 2050–2060. [Google Scholar] [CrossRef]
- Collins, R.L.; Brand, H.; Karczewski, K.J.; Zhao, X.; Alföldi, J.; Francioli, L.C.; Khera, A.V.; Lowther, C.; Gauthier, L.D.; Wang, H.; et al. A Structural Variation Reference for Medical and Population Genetics. Nature 2020, 581, 444–451. [Google Scholar] [CrossRef]
- dbVar. Available online: https://www.ncbi.nlm.nih.gov/dbvar/studies/nstd186/ (accessed on 1 July 2021).
- MacDonald, J.R.; Ziman, R.; Yuen, R.K.C.; Feuk, L.; Scherer, S.W. The Database of Genomic Variants: A Curated Collection of Structural Variation in the Human Genome. Nucleic Acids Res. 2014, 42, D986–D992. [Google Scholar] [CrossRef] [PubMed]
- Manders, F.; Brandsma, A.M.; de Kanter, J.; Verheul, M.; Oka, R.; van Roosmalen, M.J.; van der Roest, B.; van Hoeck, A.; Cuppen, E.; van Boxtel, R. MutationalPatterns: The One Stop Shop for the Analysis of Mutational Processes. BMC Genom. 2022, 23, 134. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Kim, J.; Haradhvala, N.J.; Huang, M.N.; Tian Ng, A.W.; Wu, Y.; Boot, A.; Covington, K.R.; Gordenin, D.A.; Bergstrom, E.N.; et al. The Repertoire of Mutational Signatures in Human Cancer. Nature 2020, 578, 94–101. [Google Scholar] [CrossRef]
- Chaves-Urbano, B.; Hernando, B.; Garcia, M.J.; Macintyre, G. CNpare: Matching DNA Copy Number Profiles. Bioinformatics 2022, 38, 3638–3641. [Google Scholar] [CrossRef]
- Brunet, J.P.; Tamayo, P.; Golub, T.R.; Mesirov, J.P. Metagenes and Molecular Pattern Discovery Using Matrix Factorization. Proc. Natl. Acad. Sci. USA 2004, 101, 4164–4169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Chakravarty, D.; Gao, J.; Phillips, S.M.; Kundra, R.; Zhang, H.; Wang, J.; Rudolph, J.E.; Yaeger, R.; Soumerai, T.; Nissan, M.H.; et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol 2017, 2017, PO.17.00011. [Google Scholar] [CrossRef] [PubMed]
- Gröbner, S.N.; Worst, B.C.; Weischenfeldt, J.; Buchhalter, I.; Kleinheinz, K.; Rudneva, V.A.; Johann, P.D.; Balasubramanian, G.P.; Segura-Wang, M.; Brabetz, S.; et al. The Landscape of Genomic Alterations across Childhood Cancers. Nature 2018, 555, 321–327. [Google Scholar] [CrossRef] [PubMed]
- Martens, M.; Ammar, A.; Riutta, A.; Waagmeester, A.; Slenter, D.N.; Hanspers, K.; A Miller, R.; Digles, D.; Lopes, E.N.; Ehrhart, F.; et al. WikiPathways: Connecting Communities. Nucleic Acids Res. 2021, 49, D613–D621. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: Berlin/Heidelberg, Germany, 2016; ISBN 9783319242750. [Google Scholar]
- Kolde, R. Pheatmap: Pretty Heatmaps. Available online: https://rdrr.io/cran/pheatmap/ (accessed on 11 August 2022).
- Ggpubr: “ggplot2” Based Publication Ready Plots. Available online: https://rdrr.io/cran/ggpubr/ (accessed on 11 August 2022).
- Rstatix: Pipe-Friendly Framework for Basic Statistical Tests. Available online: https://rdrr.io/cran/rstatix/ (accessed on 11 August 2022).
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. Circlize Implements and Enhances Circular Visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef]
- Wegert, J.; Ishaque, N.; Vardapour, R.; Geörg, C.; Gu, Z.; Bieg, M.; Ziegler, B.; Bausenwein, S.; Nourkami, N.; Ludwig, N.; et al. Mutations in the SIX1/2 Pathway and the DROSHA/DGCR8 miRNA Microprocessor Complex Underlie High-Risk Blastemal Type Wilms Tumors. Cancer Cell 2015, 27, 298–311. [Google Scholar] [CrossRef]
- Mohanty, V.; Wang, F.; Mills, G.B.; CTD2 Research Network; Chen, K. Uncoupling of Gene Expression from Copy Number Presents Therapeutic Opportunities in Aneuploid Cancers. Cell Rep. Med. 2021, 2, 100349. [Google Scholar] [CrossRef]
- Systematic Review of the Immunological Landscape of Wilms Tumors. Mol. Ther. Oncolytics 2021, 22, 454–467. [CrossRef]
- Austinat, M.; Dunsch, R.; Wittekind, C.; Tannapfel, A.; Gebhardt, R.; Gaunitz, F. Correlation between Beta-Catenin Mutations and Expression of Wnt-Signaling Target Genes in Hepatocellular Carcinoma. Mol. Cancer 2008, 7, 21. [Google Scholar] [CrossRef]
- Gadd, S.; Huff, V.; Huang, C.-C.; Ruteshouser, E.C.; Dome, J.S.; Grundy, P.E.; Breslow, N.; Jennings, L.; Green, D.M.; Beckwith, J.B.; et al. Clinically Relevant Subsets Identified by Gene Expression Patterns Support a Revised Ontogenic Model of Wilms Tumor: A Children’s Oncology Group Study. Neoplasia 2012, 14, 742–756. [Google Scholar] [CrossRef] [PubMed]
- Corbin, M.; de Reyniès, A.; Rickman, D.S.; Berrebi, D.; Boccon-Gibod, L.; Cohen-Gogo, S.; Fabre, M.; Jaubert, F.; Faussillon, M.; Yilmaz, F.; et al. WNT/β-Catenin Pathway Activation in Wilms Tumors: A Unifying Mechanism with Multiple Entries? Genes Chromosomes Cancer 2009, 48, 816–827. [Google Scholar] [CrossRef] [PubMed]
- Royer-Pokora, B.; Busch, M.; Beier, M.; Duhme, C.; de Torres, C.; Mora, J.; Brandt, A.; Royer, H.-D. Wilms Tumor Cells with WT1 Mutations Have Characteristic Features of Mesenchymal Stem Cells and Express Molecular Markers of Paraxial Mesoderm. Hum. Mol. Genet. 2010, 19, 1651–1668. [Google Scholar] [CrossRef] [PubMed]
- Royer-Pokora, B.; Beier, M.; Brandt, A.; Duhme, C.; Busch, M.; de Torres, C.; Royer, H.; Mora, J. Chemotherapy and Terminal Skeletal Muscle Differentiation in WT1-mutant Wilms Tumors. Cancer Med. 2018, 7, 1359. [Google Scholar] [CrossRef] [PubMed]
- Nayak, P.; Colas, A.; Mercola, M.; Varghese, S.; Subramaniam, S. Temporal Mechanisms of Myogenic Specification in Human Induced Pluripotent Stem Cells. Sci. Adv. 2021, 7, eabf7412. [Google Scholar] [CrossRef] [PubMed]
- Gisselsson, D.; Jin, Y.; Lindgren, D.; Persson, J.; Gisselsson, L.; Hanks, S.; Sehic, D.; Mengelbier, L.H.; Øra, I.; Rahman, N.; et al. Generation of Trisomies in Cancer Cells by Multipolar Mitosis and Incomplete Cytokinesis. Proc. Natl. Acad. Sci. USA 2010, 107, 20489–20493. [Google Scholar] [CrossRef]
- Krepischi, A.C.V.; Maschietto, M.; Ferreira, E.N.; Silva, A.G.; Costa, S.S.; da Cunha, I.W.; Barros, B.D.F.; Grundy, P.E.; Rosenberg, C.; Carraro, D.M. Genomic Imbalances Pinpoint Potential Oncogenes and Tumor Suppressors in Wilms Tumors. Mol. Cytogenet. 2016, 9, 1–10. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
van Belzen, I.A.E.M.; van Tuil, M.; Badloe, S.; Strengman, E.; Janse, A.; Verwiel, E.T.P.; van der Leest, D.F.M.; de Vos, S.; Baker-Hernandez, J.; Groenendijk, A.; et al. Molecular Characterization Reveals Subclasses of 1q Gain in Intermediate Risk Wilms Tumors. Cancers 2022, 14, 4872. https://doi.org/10.3390/cancers14194872
van Belzen IAEM, van Tuil M, Badloe S, Strengman E, Janse A, Verwiel ETP, van der Leest DFM, de Vos S, Baker-Hernandez J, Groenendijk A, et al. Molecular Characterization Reveals Subclasses of 1q Gain in Intermediate Risk Wilms Tumors. Cancers. 2022; 14(19):4872. https://doi.org/10.3390/cancers14194872
Chicago/Turabian Stylevan Belzen, Ianthe A. E. M., Marc van Tuil, Shashi Badloe, Eric Strengman, Alex Janse, Eugène T. P. Verwiel, Douwe F. M. van der Leest, Sam de Vos, John Baker-Hernandez, Alissa Groenendijk, and et al. 2022. "Molecular Characterization Reveals Subclasses of 1q Gain in Intermediate Risk Wilms Tumors" Cancers 14, no. 19: 4872. https://doi.org/10.3390/cancers14194872
APA Stylevan Belzen, I. A. E. M., van Tuil, M., Badloe, S., Strengman, E., Janse, A., Verwiel, E. T. P., van der Leest, D. F. M., de Vos, S., Baker-Hernandez, J., Groenendijk, A., de Krijger, R., Kerstens, H. H. D., Drost, J., van den Heuvel-Eibrink, M. M., Tops, B. B. J., Holstege, F. C. P., Kemmeren, P., & Hehir-Kwa, J. Y. (2022). Molecular Characterization Reveals Subclasses of 1q Gain in Intermediate Risk Wilms Tumors. Cancers, 14(19), 4872. https://doi.org/10.3390/cancers14194872









