Oncologic Imaging and Radiomics: A Walkthrough Review of Methodological Challenges
Abstract
Simple Summary
Abstract
1. Introduction
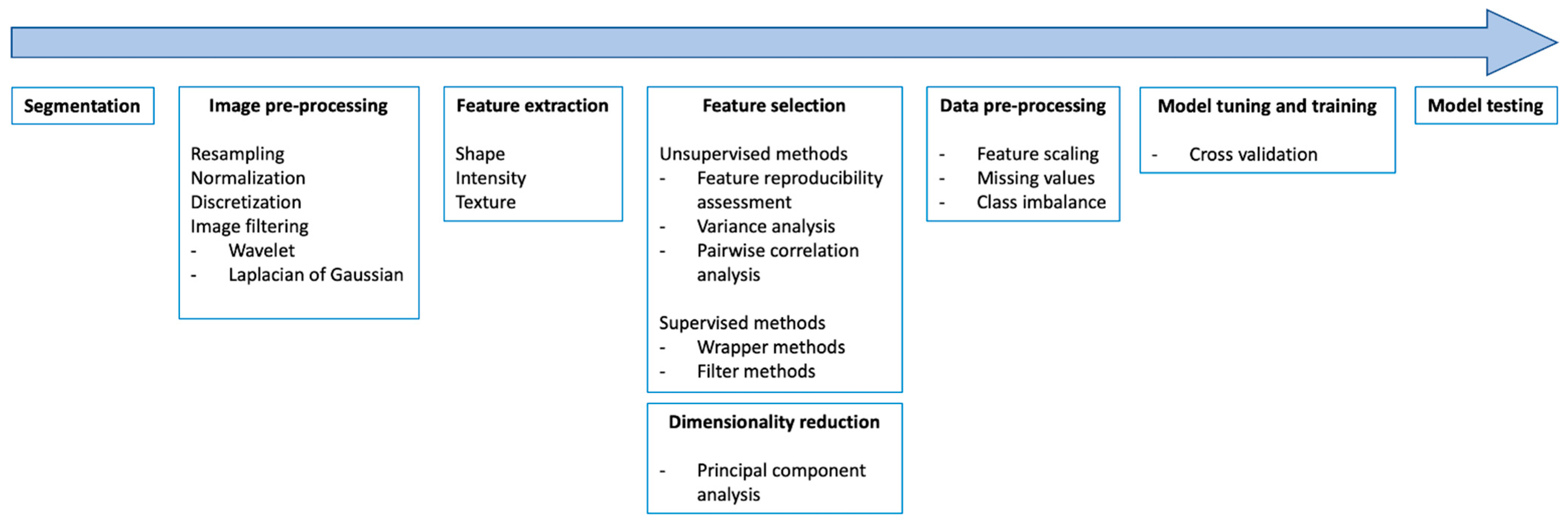
2. Image Preprocessing and Quality Control of Imaging Datasets
3. Lesion Segmentation
4. Feature Extraction and Selection
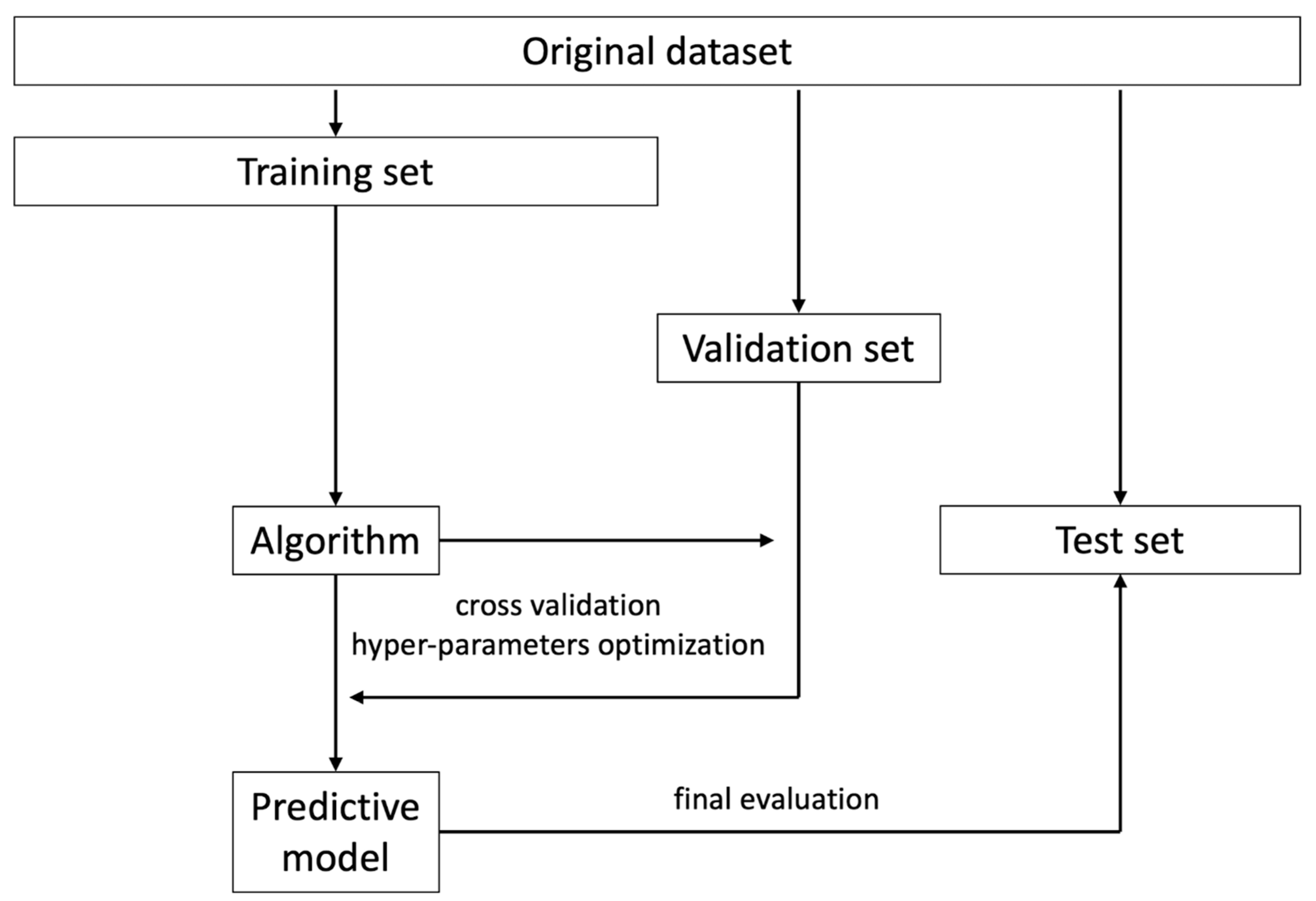
5. Data Processing for Model Training
6. Validation, Calibration, and Reporting Accuracy
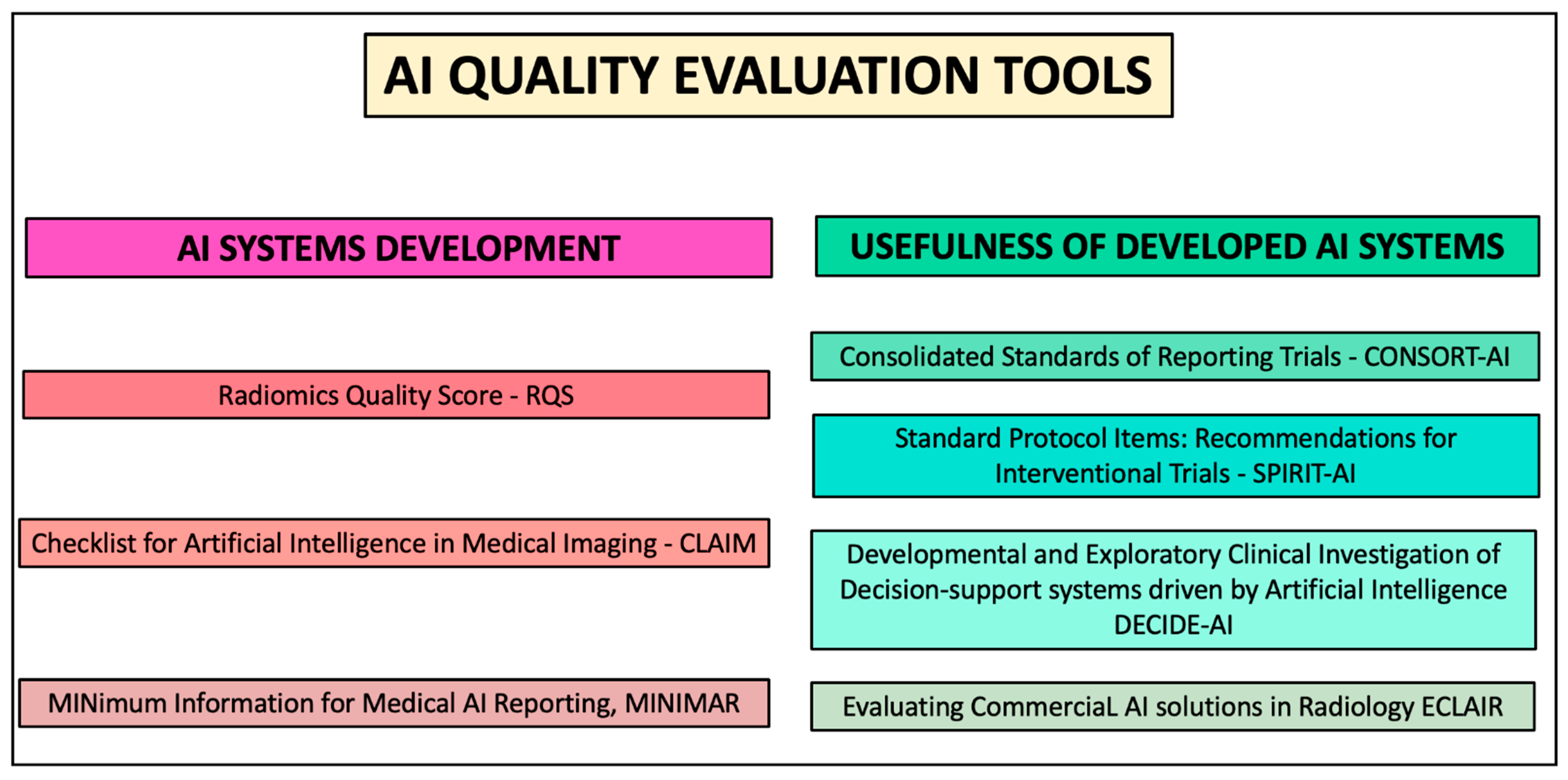
7. Quality Evaluation Tools
8. Further Considerations
9. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mainenti, P.P.; Stanzione, A.; Guarino, S.; Romeo, V.; Ugga, L.; Romano, F.; Storto, G.; Maurea, S.; Brunetti, A. Colorectal Cancer: Parametric Evaluation of Morphological, Functional and Molecular Tomographic Imaging. World J. Gastroenterol. 2019, 25, 5233–5256. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Gu, Y.; Basu, S.; Berglund, A.; Eschrich, S.A.; Schabath, M.B.; Forster, K.; Aerts, H.J.W.L.; Dekker, A.; Fenstermacher, D.; et al. Radiomics: The Process and the Challenges. Magn. Reson. Imaging 2012, 30, 1234–1248. [Google Scholar] [CrossRef]
- Ding, H.; Wu, C.; Liao, N.; Zhan, Q.; Sun, W.; Huang, Y.; Jiang, Z.; Li, Y. Radiomics in Oncology: A 10-Year Bibliometric Analysis. Front. Oncol. 2021, 11, 689802. [Google Scholar] [CrossRef] [PubMed]
- Park, J.E.; Kim, D.; Kim, H.S.; Park, S.Y.; Kim, J.Y.; Cho, S.J.; Shin, J.H.; Kim, J.H. Quality of Science and Reporting of Radiomics in Oncologic Studies: Room for Improvement According to Radiomics Quality Score and TRIPOD Statement. Eur. Radiol. 2020, 30, 523–536. [Google Scholar] [CrossRef] [PubMed]
- Stanzione, A.; Galatola, R.; Cuocolo, R.; Romeo, V.; Verde, F.; Mainenti, P.P.; Brunetti, A.; Maurea, S. Radiomics in Cross-Sectional Adrenal Imaging: A Systematic Review and Quality Assessment Study. Diagnostics 2022, 12, 578. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, C.; Zhang, J.; Hu, X.; Fan, Y.; Ma, K.; Sparrelid, E.; Brismar, T.B. Radiomics Models for Predicting Microvascular Invasion in Hepatocellular Carcinoma: A Systematic Review and Radiomics Quality Score Assessment. Cancers 2021, 13, 5864. [Google Scholar] [CrossRef]
- Park, J.E.; Park, S.Y.; Kim, H.J.; Kim, H.S. Reproducibility and Generalizability in Radiomics Modeling: Possible Strategies in Radiologic and Statistical Perspectives. Korean J. Radiol. 2019, 20, 1124. [Google Scholar] [CrossRef]
- Tiwari, P.; Verma, R. The Pursuit of Generalizability to Enable Clinical Translation of Radiomics. Radiol. Artif. Intell. 2021, 3, e200227. [Google Scholar] [CrossRef]
- Li, Y.; Ammari, S.; Balleyguier, C.; Lassau, N.; Chouzenoux, E. Impact of Preprocessing and Harmonization Methods on the Removal of Scanner Effects in Brain MRI Radiomic Features. Cancers 2021, 13, 3000. [Google Scholar] [CrossRef]
- Esses, S.J.; Taneja, S.S.; Rosenkrantz, A.B. Imaging Facilities’ Adherence to PI-RADS v2 Minimum Technical Standards for the Performance of Prostate MRI. Acad. Radiol. 2018, 25, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Cuocolo, R.; Stanzione, A.; Ponsiglione, A.; Verde, F.; Ventimiglia, A.; Romeo, V.; Petretta, M.; Imbriaco, M. Prostate MRI Technical Parameters Standardization: A Systematic Review on Adherence to PI-Radsv2 Acquisition Protocol. Eur. J. Radiol. 2019, 120, 108662. [Google Scholar] [CrossRef]
- Rizzo, S.; Botta, F.; Raimondi, S.; Origgi, D.; Fanciullo, C.; Morganti, A.G.; Bellomi, M. Radiomics: The Facts and the Challenges of Image Analysis. Eur. Radiol. Exp. 2018, 2, 36. [Google Scholar] [CrossRef] [PubMed]
- Lennartz, S.; O’Shea, A.; Parakh, A.; Persigehl, T.; Baessler, B.; Kambadakone, A. Robustness of Dual-Energy CT-Derived Radiomic Features across Three Different Scanner Types. Eur. Radiol. 2022, 32, 1959–1970. [Google Scholar] [CrossRef] [PubMed]
- Euler, A.; Laqua, F.C.; Cester, D.; Lohaus, N.; Sartoretti, T.; Pinto dos Santos, D.; Alkadhi, H.; Baessler, B. Virtual Monoenergetic Images of Dual-Energy CT—Impact on Repeatability, Reproducibility, and Classification in Radiomics. Cancers 2021, 13, 4710. [Google Scholar] [CrossRef]
- Van Timmeren, J.E.; Leijenaar, R.T.H.; van Elmpt, W.; Wang, J.; Zhang, Z.; Dekker, A.; Lambin, P. Test–Retest Data for Radiomics Feature Stability Analysis: Generalizable or Study-Specific? Tomography 2016, 2, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Jha, A.K.; Mithun, S.; Jaiswar, V.; Sherkhane, U.B.; Purandare, N.C.; Prabhash, K.; Rangarajan, V.; Dekker, A.; Wee, L.; Traverso, A. Repeatability and Reproducibility Study of Radiomic Features on a Phantom and Human Cohort. Sci. Rep. 2021, 11, 2055. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, J.P.B.; Aboagye, E.O.; Adams, J.E.; Aerts, H.J.W.L.; Barrington, S.F.; Beer, A.J.; Boellaard, R.; Bohndiek, S.E.; Brady, M.; Brown, G.; et al. Imaging Biomarker Roadmap for Cancer Studies. Nat. Rev. Clin. Oncol. 2017, 14, 169–186. [Google Scholar] [CrossRef]
- Altazi, B.A.; Zhang, G.G.; Fernandez, D.C.; Montejo, M.E.; Hunt, D.; Werner, J.; Biagioli, M.C.; Moros, E.G. Reproducibility of F18-FDG PET Radiomic Features for Different Cervical Tumor Segmentation Methods, Gray-Level Discretization, and Reconstruction Algorithms. J. Appl. Clin. Med. Phys. 2017, 18, 32–48. [Google Scholar] [CrossRef]
- Van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Granzier, R.W.Y.; Ibrahim, A.; Primakov, S.; Keek, S.A.; Halilaj, I.; Zwanenburg, A.; Engelen, S.M.E.; Lobbes, M.B.I.; Lambin, P.; Woodruff, H.C.; et al. Test–Retest Data for the Assessment of Breast MRI Radiomic Feature Repeatability. J. Magn. Reson. Imaging 2022, 56, 592–604. [Google Scholar] [CrossRef]
- Schwier, M.; van Griethuysen, J.; Vangel, M.G.; Pieper, S.; Peled, S.; Tempany, C.; Aerts, H.J.W.L.; Kikinis, R.; Fennessy, F.M.; Fedorov, A. Repeatability of Multiparametric Prostate MRI Radiomics Features. Sci. Rep. 2019, 9, 9441. [Google Scholar] [CrossRef] [PubMed]
- Papanikolaou, N.; Matos, C.; Koh, D.M. How to Develop a Meaningful Radiomic Signature for Clinical Use in Oncologic Patients. Cancer Imaging 2020, 20, 33. [Google Scholar] [CrossRef] [PubMed]
- Cuocolo, R.; Stanzione, A.; Castaldo, A.; De Lucia, D.R.; Imbriaco, M. Quality Control and Whole-Gland, Zonal and Lesion Annotations for the Prostatex Challenge Public Dataset. Eur. J. Radiol. 2021, 138, 109647. [Google Scholar] [CrossRef] [PubMed]
- Oakden-Rayner, L. Exploring Large-scale Public Medical Image Datasets. Acad. Radiol. 2020, 27, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Usman, M.; Latif, S.; Asim, M.; Lee, B.-D.; Qadir, J. Retrospective Motion Correction in Multishot MRI Using Generative Adversarial Network. Sci. Rep. 2020, 10, 4786. [Google Scholar] [CrossRef]
- Zhao, B. Understanding Sources of Variation to Improve the Reproducibility of Radiomics. Front. Oncol. 2021, 11, 633176. [Google Scholar] [CrossRef]
- van Timmeren, J.E.; Cester, D.; Tanadini-Lang, S.; Alkadhi, H.; Baessler, B. Radiomics In Medical Imaging—“How-To” Guide and Critical Reflection. Insights Imaging 2020, 11, 91. [Google Scholar] [CrossRef]
- Parmar, C.; Rios Velazquez, E.; Leijenaar, R.; Jermoumi, M.; Carvalho, S.; Mak, R.H.; Mitra, S.; Shankar, B.U.; Kikinis, R.; Haibe-Kains, B.; et al. Robust Radiomics Feature Quantification Using Semiautomatic Volumetric Segmentation. PLoS ONE 2014, 9, e102107. [Google Scholar] [CrossRef]
- Cheng, P.M.; Montagnon, E.; Yamashita, R.; Pan, I.; Cadrin-Chênevert, A.; Perdigón Romero, F.; Chartrand, G.; Kadoury, S.; Tang, A. Deep Learning: An Update for Radiologists. RadioGraphics 2021, 41, 1427–1445. [Google Scholar] [CrossRef]
- Kuhl, C.K.; Truhn, D. The Long Route to Standardized Radiomics: Unraveling the Knot from the End. Radiology 2020, 295, 339–341. [Google Scholar] [CrossRef]
- Kocak, B.; Kus, E.A.; Yardimci, A.H.; Bektas, C.T.; Kilickesmez, O. Machine Learning in Radiomic Renal Mass Characterization: Fundamentals, Applications, Challenges, and Future Directions. Am. J. Roentgenol. 2020, 215, 920–928. [Google Scholar] [CrossRef]
- Kocak, B.; Ates, E.; Durmaz, E.S.; Ulusan, M.B.; Kilickesmez, O. Influence of Segmentation Margin on Machine Learning–Based High-Dimensional Quantitative CT Texture Analysis: A Reproducibility Study on Renal Clear Cell Carcinomas. Eur. Radiol. 2019, 29, 4765–4775. [Google Scholar] [CrossRef]
- Zhang, X.; Zhong, L.; Zhang, B.; Zhang, L.; Du, H.; Lu, L.; Zhang, S.; Yang, W.; Feng, Q. The Effects of Volume of Interest Delineation on MRI-Based Radiomics Analysis: Evaluation with Two Disease Groups. Cancer Imaging 2019, 19, 89. [Google Scholar] [CrossRef] [PubMed]
- Pavic, M.; Bogowicz, M.; Würms, X.; Glatz, S.; Finazzi, T.; Riesterer, O.; Roesch, J.; Rudofsky, L.; Friess, M.; Veit-Haibach, P.; et al. Influence of Inter-Observer Delineation Variability on Radiomics Stability in Different Tumor Sites. Acta Oncol. 2018, 57, 1070–1074. [Google Scholar] [CrossRef]
- Gitto, S.; Cuocolo, R.; Emili, I.; Tofanelli, L.; Chianca, V.; Albano, D.; Messina, C.; Imbriaco, M.; Sconfienza, L.M. Effects of Interobserver Variability on 2D and 3D CT- and MRI-Based Texture Feature Reproducibility of Cartilaginous Bone Tumors. J. Digit. Imaging 2021, 34, 820–832. [Google Scholar] [CrossRef]
- Qiu, Q.; Duan, J.; Duan, Z.; Meng, X.; Ma, C.; Zhu, J.; Lu, J.; Liu, T.; Yin, Y. Reproducibility and Non-Redundancy of Radiomic Features Extracted from Arterial Phase CT Scans in Hepatocellular Carcinoma Patients: Impact of Tumor Segmentation Variability. Quant. Imaging Med. Surg. 2019, 9, 453–464. [Google Scholar] [CrossRef]
- Haarburger, C.; Müller-Franzes, G.; Weninger, L.; Kuhl, C.; Truhn, D.; Merhof, D. Radiomics Feature Reproducibility Under Inter-Rater Variability in Segmentations of CT Images. Sci. Rep. 2020, 10, 12688. [Google Scholar] [CrossRef]
- Kocak, B.; Kus, E.A.; Kilickesmez, O. How to Read and Review Papers on Machine Learning and Artificial Intelligence in Radiology: A Survival Guide to Key Methodological Concepts. Eur. Radiol. 2021, 31, 1819–1830. [Google Scholar] [CrossRef]
- Stanzione, A.; Gambardella, M.; Cuocolo, R.; Ponsiglione, A.; Romeo, V.; Imbriaco, M. Prostate MRI radiomics: A Systematic Review and Radiomic Quality Score Assessment. Eur. J. Radiol. 2020, 129, 109095. [Google Scholar] [CrossRef]
- Kocak, B.; Durmaz, E.S.; Erdim, C.; Ates, E.; Kaya, O.K.; Kilickesmez, O. Radiomics of Renal Masses: Systematic Review of Reproducibility and Validation Strategies. Am. J. Roentgenol. 2020, 214, 129–136. [Google Scholar] [CrossRef] [PubMed]
- Gitto, S.; Cuocolo, R.; Albano, D.; Morelli, F.; Pescatori, L.C.; Messina, C.; Imbriaco, M.; Sconfienza, L.M. CT And MRI Radiomics of Bone and Soft-Tissue Sarcomas: A Systematic Review of Reproducibility and Validation Strategies. Insights Imaging 2021, 12, 68. [Google Scholar] [CrossRef] [PubMed]
- Shur, J.D.; Doran, S.J.; Kumar, S.; ap Dafydd, D.; Downey, K.; O’Connor, J.P.B.; Papanikolaou, N.; Messiou, C.; Koh, D.-M.; Orton, M.R. Radiomics in Oncology: A Practical Guide. RadioGraphics 2021, 41, 1717–1732. [Google Scholar] [CrossRef] [PubMed]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef]
- Jovic, A.; Brkic, K.; Bogunovic, N. A review of feature selection methods with applications. In Proceedings of the 2015 38th International Convention on Information and Communication Technology, Electronics and Microelectronics (MIPRO), Opatija, Croatia, 25–29 May 2015; pp. 1200–1205. [Google Scholar]
- Baeßler, B.; Weiss, K.; Pinto dos Santos, D. Robustness and Reproducibility of Radiomics in Magnetic Resonance Imaging. Investind Radiol. 2019, 54, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Traverso, A.; Wee, L.; Dekker, A.; Gillies, R. Repeatability and Reproducibility of Radiomic Features: A Systematic Review. Int. J. Radiat. Oncol. 2018, 102, 1143–1158. [Google Scholar] [CrossRef]
- Kocak, B.; Durmaz, E.S.; Ates, E.; Kilickesmez, O. Radiomics with Artificial Intelligence: A Practical Guide for Beginners. Diagn. Interv. Radiol. 2019, 25, 485–495. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Müller, A.; Nothman, J.; Louppe, G.; et al. Scikit-Learn: Machine Learning in Python. 2012. Available online: https://arxiv.org/abs/1201.0490 (accessed on 29 August 2022).
- Demircioğlu, A. Measuring the Bias of Incorrect Application of Feature Selection When Using Cross-Validation in Radiomics. Insights Imaging 2021, 12, 172. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Kapoor, S.; Narayanan, A. Leakage and the Reproducibility Crisis in ML-Based Science. 2022. Available online: https://arxiv.org/abs/2207.07048 (accessed on 29 August 2022).
- Wang, H.; Zheng, H. Model Validation, Machine Learning. In Encyclopedia of Systems Biology; Springer: New York, NY, USA, 2013; pp. 1406–1407. [Google Scholar]
- Parmar, C.; Barry, J.D.; Hosny, A.; Quackenbush, J.; Aerts, H.J.W.L. Data Analysis Strategies in Medical Imaging. Clin. Cancer Res. 2018, 24, 3492–3499. [Google Scholar] [CrossRef]
- Van Calster, B.; McLernon, D.J.; van Smeden, M.; Wynants, L.; Steyerberg, E.W. Calibration: The Achilles Heel of Predictive Analytics. BMC Med. 2019, 17, 230. [Google Scholar] [CrossRef]
- Gaube, S.; Suresh, H.; Raue, M.; Merritt, A.; Berkowitz, S.J.; Lermer, E.; Coughlin, J.F.; Guttag, J.V.; Colak, E.; Ghassemi, M. Do as AI Say: Susceptibility in Deployment ff Clinical Decision-Aids. NPJ Digit. Med. 2021, 4, 31. [Google Scholar] [CrossRef]
- Statistical Thinking–Classification, vs. Prediction. Available online: https://www.fharrell.com/post/classification/ (accessed on 29 August 2022).
- Steyerberg, E.W.; Vickers, A.J.; Cook, N.R.; Gerds, T.; Gonen, M.; Obuchowski, N.; Pencina, M.J.; Kattan, M.W. Assessing the Performance of Prediction Models. Epidemiology 2010, 21, 128–138. [Google Scholar] [CrossRef]
- Pepe, M.S. Limitations of the Odds Ratio in Gauging the Performance of a Diagnostic, Prognostic, or Screening Marker. Am. J. Epidemiol. 2004, 159, 882–890. [Google Scholar] [CrossRef]
- Nattino, G.; Pennell, M.L.; Lemeshow, S. Assessing the Goodness of Fit of Logistic Regression Models in Large Samples: A Modification of the Hosmer-Lemeshow test. Biometrics 2020, 76, 549–560. [Google Scholar] [CrossRef]
- Gitto, S.; Cuocolo, R.; van Langevelde, K.; van de Sande, M.A.J.; Parafioriti, A.; Luzzati, A.; Imbriaco, M.; Sconfienza, L.M.; Bloem, J.L. MRI Radiomics-Based Machine Learning Classification of Atypical Cartilaginous Tumour and Grade II Chondrosarcoma of Long Bones. eBioMedicine 2022, 75, 103757. [Google Scholar] [CrossRef]
- Tong, Y.; Li, J.; Huang, Y.; Zhou, J.; Liu, T.; Guo, Y.; Yu, J.; Zhou, S.; Wang, Y.; Chang, C. Ultrasound-Based Radiomic Nomogram for Predicting Lateral Cervical Lymph Node Metastasis in Papillary Thyroid Carcinoma. Acad. Radiol. 2021, 28, 1675–1684. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Li, T.; Cai, Q.; Wang, X.; Liao, Y.; Cheng, Y.; Zhou, Q. Development and Validation of a Radiomics Nomogram for Differentiating Mycoplasma Pneumonia and Bacterial Pneumonia. Diagnostics 2021, 11, 1330. [Google Scholar] [CrossRef] [PubMed]
- Cuocolo, R.; Stanzione, A.; Faletti, R.; Gatti, M.; Calleris, G.; Fornari, A.; Gentile, F.; Motta, A.; Dell’Aversana, S.; Creta, M.; et al. MRI Index Lesion Radiomics and Machine Learning for Detection of Extraprostatic Extension of Disease: A Multicenter Study. Eur. Radiol. 2021, 31, 7575–7583. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The Bridge Between Medical Imaging and Personalized Medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Montesinos López, O.A.; Montesinos López, A.; Crossa, J. Overfitting, Model Tuning, and Evaluation of Prediction Performance. In Multivariate Statistical Machine Learning Methods for Genomic Prediction; Springer International Publishing: Cham, Switzerland, 2022; pp. 109–139. [Google Scholar]
- Pinto dos Santos, D.; Dietzel, M.; Baessler, B. A Decade of Radiomics Research: Are Images Really Data or Just Patterns in the Noise? Eur. Radiol. 2021, 31, 1–4. [Google Scholar] [CrossRef]
- Futoma, J.; Simons, M.; Panch, T.; Doshi-Velez, F.; Celi, L.A. The Myth of Generalisability in Clinical Research and Machine Learning in Health Care. Lancet Digit. Health 2020, 2, e489–e492. [Google Scholar] [CrossRef]
- Mongan, J.; Moy, L.; Kahn, C.E. Checklist for Artificial Intelligence in Medical Imaging (CLAIM): A Guide for Authors and Reviewers. Radiol. Artif. Intell. 2020, 2, e200029. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Boussard, T.; Bozkurt, S.; Ioannidis, J.P.A.; Shah, N.H. MINIMAR (MINimum Information for Medical AI Reporting): Developing Reporting Standards for Artificial Intelligence in Health Care. J. Am. Med. Inform. Assoc. 2020, 27, 2011–2015. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Cruz Rivera, S.; Moher, D.; Calvert, M.J.; Denniston, A.K.; Ashrafian, H.; Beam, A.L.; Chan, A.-W.; Collins, G.S.; Deeks, A.D.J.; et al. Reporting Guidelines for Clinical Trial Reports for Interventions Involving Artificial Intelligence: The CONSORT-AI Extension. Lancet Digit. Health. 2020, 2, e537–e548. [Google Scholar] [CrossRef]
- Cruz Rivera, S.; Liu, X.; Chan, A.-W.; Denniston, A.K.; Calvert, M.J.; Ashrafian, H.; Beam, A.L.; Collins, G.S.; Darzi, A.; Deeks, J.J.; et al. Guidelines for Clinical Trial Protocols for Interventions Involving Artificial Intelligence: The SPIRIT-AI Extension. Lancet Digit. Health. 2020, 2, e549–e560. [Google Scholar] [CrossRef]
- Vasey, B.; Nagendran, M.; Campbell, B.; Clifton, D.A.; Collins, G.S.; Denaxas, S.; Denniston, A.K.; Faes, L.; Geerts, B.; Ibrahim, M.; et al. Reporting Guideline for the Early-Stage Clinical Evaluation of Decision Support Systems Driven by Artificial Intelligence: DECIDE-AI. Nat. Med. 2022, 28, 924–933. [Google Scholar] [CrossRef] [PubMed]
- Omoumi, P.; Ducarouge, A.; Tournier, A.; Harvey, H.; Kahn, C.E.; Louvet-de Verchère, F.; Pinto Dos Santos, D.; Kober, T.; Richiardi, J. To Buy or Not to Buy—Evaluating Commercial AI Solutions in Radiology (The ECLAIR Guidelines). Eur. Radiol. 2021, 31, 3786–3796. [Google Scholar] [CrossRef]
- Collins, G.S.; Dhiman, P.; Andaur Navarro, C.L.; Ma, J.; Hooft, L.; Reitsma, J.B.; Logullo, P.; Beam, A.L.; Peng, L.; Van Calster, B.; et al. Protocol for Development of a Reporting Guideline (Tripod-Ai) and Risk of Bias Tool (Probast-Ai) for Diagnostic and Prognostic Prediction Model Studies Based on Artificial Intelligence. BMJ Open 2021, 11, e048008. [Google Scholar] [CrossRef] [PubMed]
- van Leeuwen, K.G.; Schalekamp, S.; Rutten, M.J.C.M.; van Ginneken, B.; de Rooij, M. Artificial Intelligence in Radiology: 100 Commercially Available Products and Their Scientific Evidence. Eur. Radiol. 2021, 31, 3797–3804. [Google Scholar] [CrossRef]
- Pinto dos Santos, D. Radiomics in Endometrial Cancer and Beyond—A Perspective from the Editors of the EJR. Eur. J. Radiol. 2022, 150, 110266. [Google Scholar] [CrossRef]
- Gatta, R.; Depeursinge, A.; Ratib, O.; Michielin, O.; Leimgruber, A. Integrating Radiomics Into Holomics for Personalised oncology: From Algorithms to edside. Eur. Radiol. Exp. 2020, 4, 11. [Google Scholar] [CrossRef] [PubMed]
- Bodalal, Z.; Trebeschi, S.; Nguyen-Kim, T.D.L.; Schats, W.; Beets-Tan, R. Radiogenomics: Bridging Imaging and Genomics. Abdom. Radiol. 2019, 44, 1960–1984. [Google Scholar] [CrossRef] [PubMed]
- Vickers, A.J.; Elkin, E.B. Decision Curve Analysis: A Novel Method for Evaluating Prediction Models. Med. Decis. Mak. 2006, 26, 565–574. [Google Scholar] [CrossRef] [PubMed]

| Image Preprocessing Technique | Rationale | Advantage |
|---|---|---|
| Normalization | MRI data contain arbitrary intensity units and grey-level intensity that can be homogenized with intensity outlier filtering (e.g., calculating the mean and standard deviation of grey levels and excluding those outside a definite range such as mean ± 3 times the standard variation). | Reducing the heterogeneity due to varying pixel grey-level value distribution across exams |
| Resampling | Images with different spatial resolutions can be uniformed and either upscaled or downscaled to isotropic voxel spacing. | Increases reproducibility by making texture features rotationally invariant |
| Discretization | Grouping pixels into bins based on intensity ranges, which is conceptually similar to creating a histogram. | A greater number of bins (or a smaller bin width) tend to preserve image details at the cost of noise. Conversely, noise reduction can be achieved by reducing the number of bins (or increasing bin width) but will cause the image to lose detail. |
| Bias field correction | MRI can suffer from spatial signal variation caused by the magnetic field being intrinsically inhomogeneous. | Correct undesired inhomogeneities |
| Image filtering | Application of edge enhancing (e.g., Laplacian of Gaussian) or decomposition (e.g., wavelet transform) filters to obtain additional image volumes from which to extract features. | May emphasize useful image characteristics while reducing noise |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stanzione, A.; Cuocolo, R.; Ugga, L.; Verde, F.; Romeo, V.; Brunetti, A.; Maurea, S. Oncologic Imaging and Radiomics: A Walkthrough Review of Methodological Challenges. Cancers 2022, 14, 4871. https://doi.org/10.3390/cancers14194871
Stanzione A, Cuocolo R, Ugga L, Verde F, Romeo V, Brunetti A, Maurea S. Oncologic Imaging and Radiomics: A Walkthrough Review of Methodological Challenges. Cancers. 2022; 14(19):4871. https://doi.org/10.3390/cancers14194871
Chicago/Turabian StyleStanzione, Arnaldo, Renato Cuocolo, Lorenzo Ugga, Francesco Verde, Valeria Romeo, Arturo Brunetti, and Simone Maurea. 2022. "Oncologic Imaging and Radiomics: A Walkthrough Review of Methodological Challenges" Cancers 14, no. 19: 4871. https://doi.org/10.3390/cancers14194871
APA StyleStanzione, A., Cuocolo, R., Ugga, L., Verde, F., Romeo, V., Brunetti, A., & Maurea, S. (2022). Oncologic Imaging and Radiomics: A Walkthrough Review of Methodological Challenges. Cancers, 14(19), 4871. https://doi.org/10.3390/cancers14194871











