Novel Insights into Redox-Based Mechanisms for Auranofin-Induced Rapid Cancer Cell Death
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Line and Reagents
2.2. Evaluation of Cytotoxicity
2.3. Thioredoxin Reductase Assay
2.4. Oxidative Stress Assessment
2.5. Quantitative Redoxome Analysis
2.6. BrdU Incorporation and Cell Cycle Analysis
2.7. Quantitation of Cellular dTTP and dGTP
2.8. Actin Filament Imaging
2.9. Statistical Analysis
3. Results
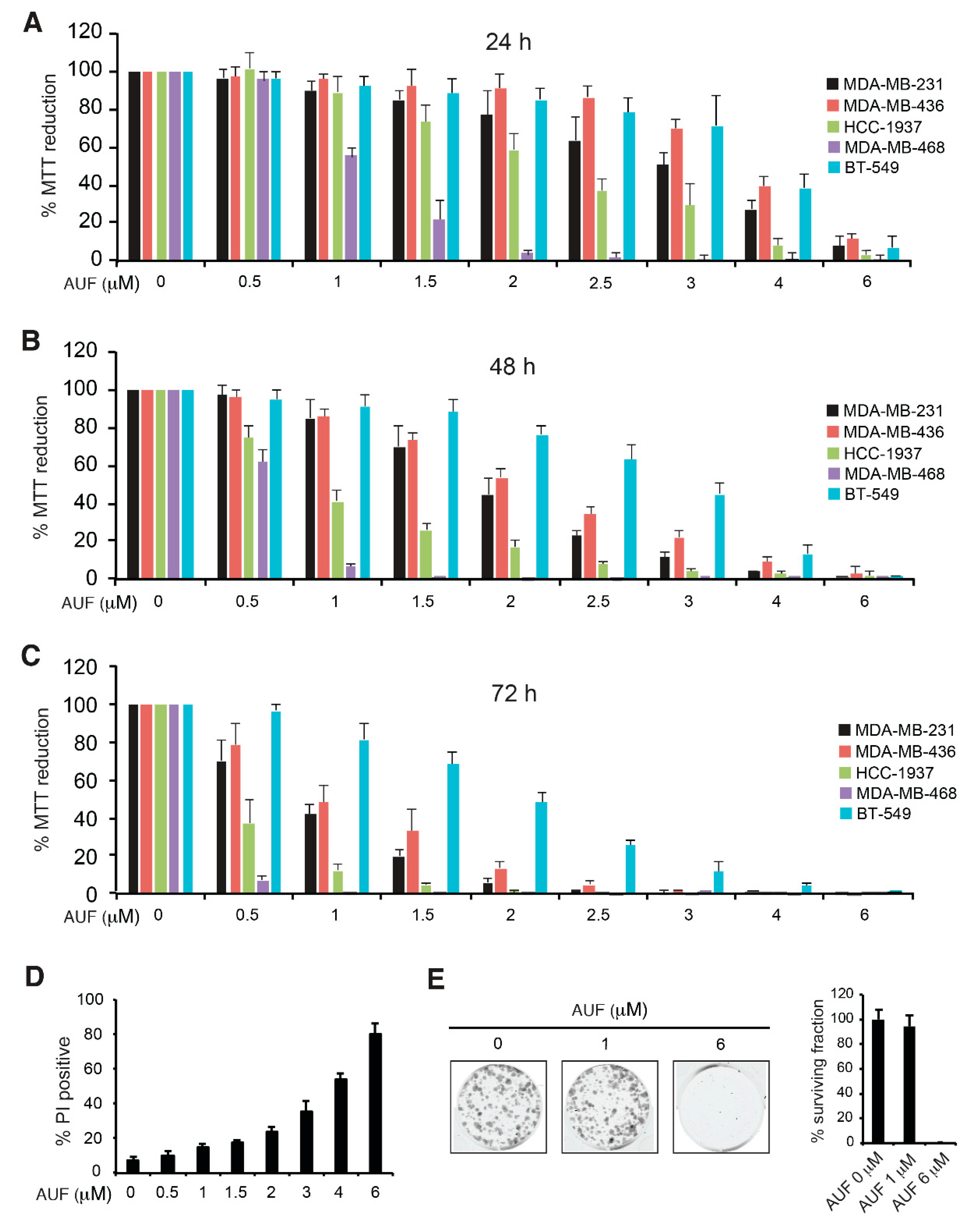
3.1. AUF-Induced Cytotoxicity in TNBC Cells
3.2. AUF-Mediated TrxR Inhibition Alone May Not Be Sufficient to Induce Cell Death
3.3. AUF Treatment Induces Rapid and General Intracellular Oxidation
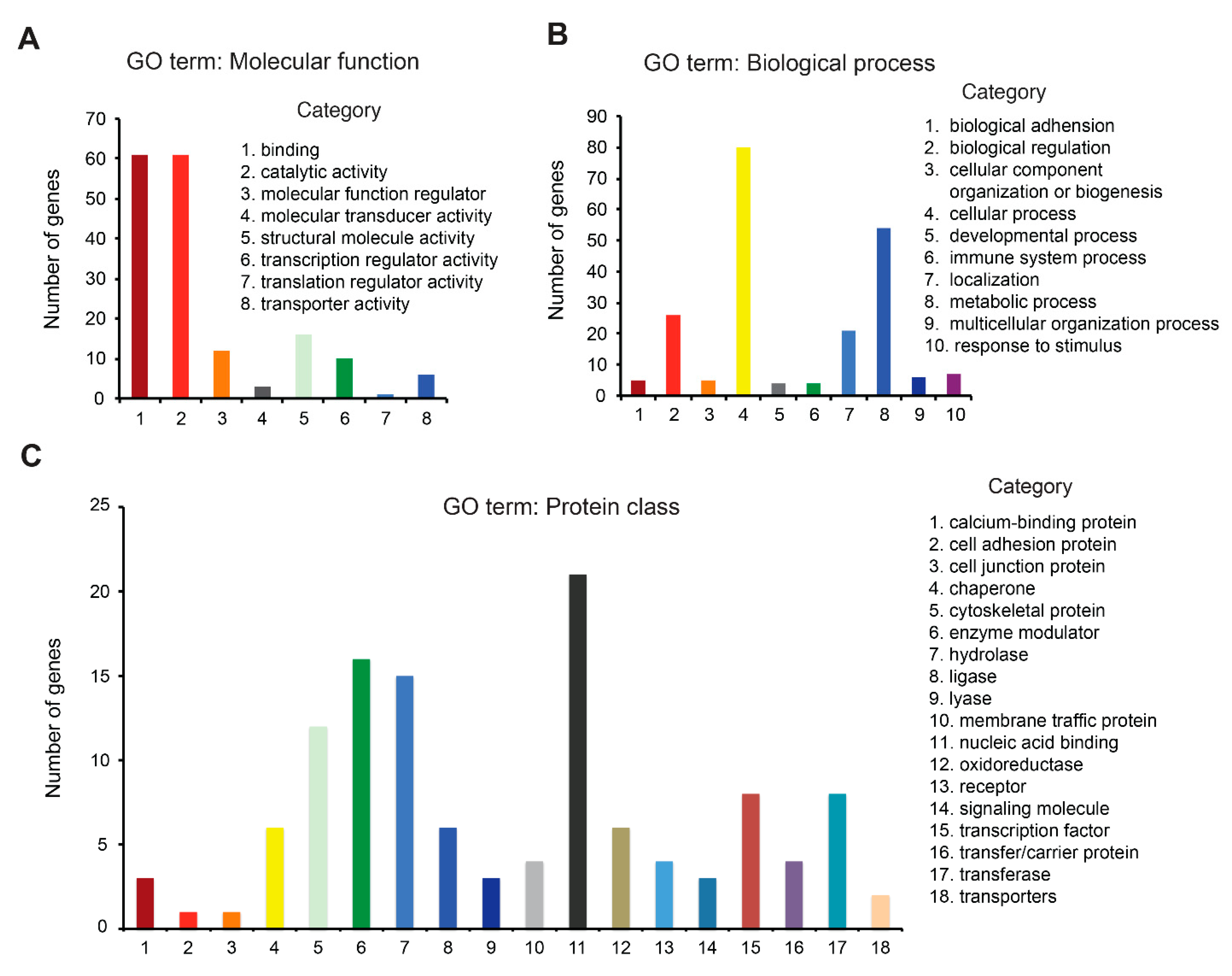
3.4. Redox Proteome Analysis Reveals a Large Spectrum of AUF-Induced Effects
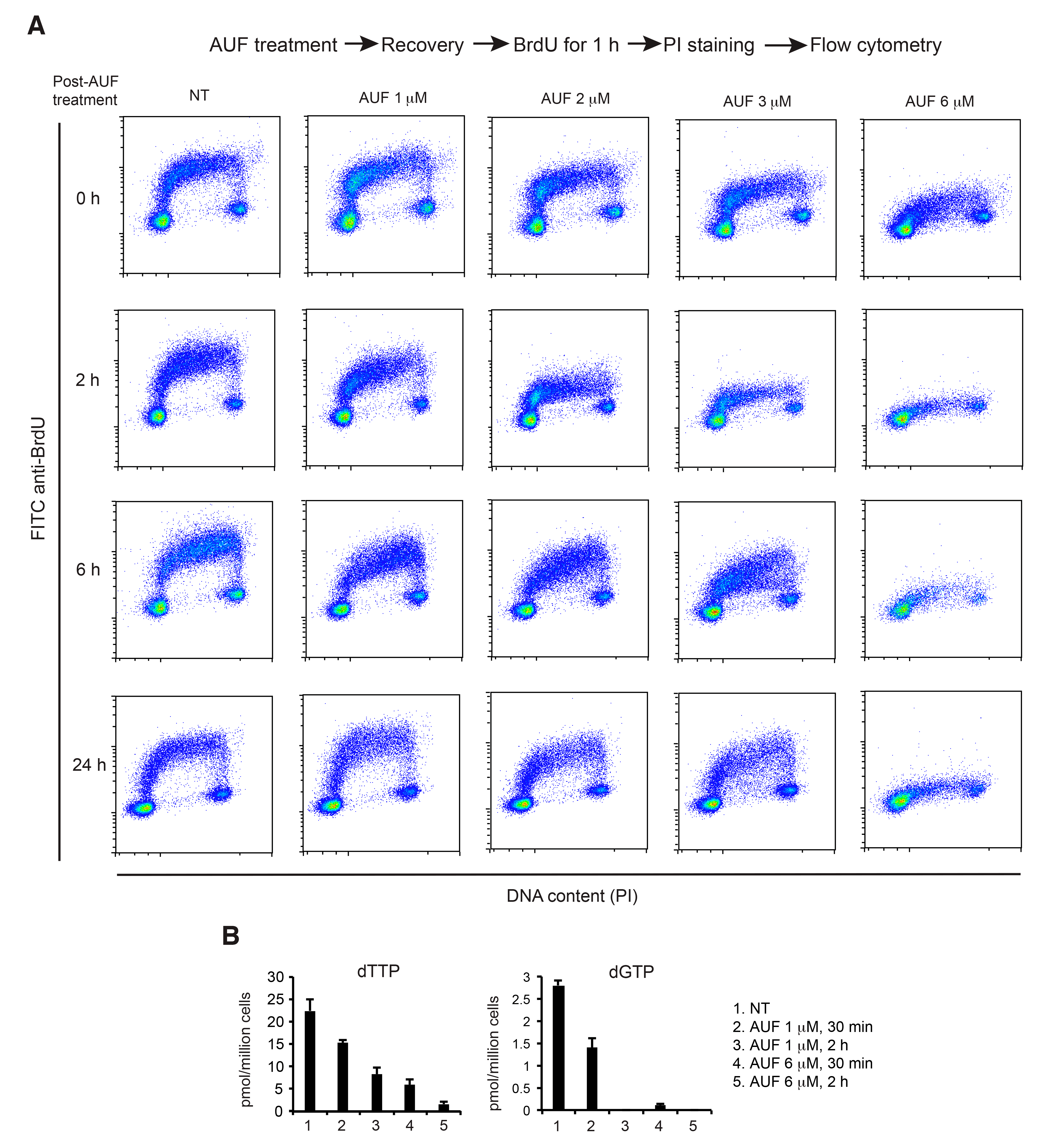
3.5. AUF Treatment Induces a Dose-Dependent Inhibition of DNA Replication
3.6. AUF Treatment Induces a Rapid Disintegration of the Actin Cytoskeleton Structure
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gorrini, C.; Harris, I.S.; Mak, T.W. Modulation of oxidative stress as an anticancer strategy. Nat. Rev. Drug Discov. 2013, 12, 931–947. [Google Scholar] [CrossRef] [PubMed]
- Hatem, E.; El Banna, N.; Huang, M.E. Multifaceted Roles of Glutathione and Glutathione-Based Systems in Carcinogenesis and Anticancer Drug Resistance. Antioxid. Redox Signal. 2017, 27, 1217–1234. [Google Scholar] [CrossRef] [PubMed]
- Kean, W.F.; Kean, I.R. Clinical pharmacology of gold. Inflammopharmacology 2008, 16, 112–125. [Google Scholar] [CrossRef] [PubMed]
- Roder, C.; Thomson, M.J. Auranofin: Repurposing an old drug for a golden new age. Drugs R D 2015, 15, 13–20. [Google Scholar] [CrossRef]
- Madeira, J.M.; Gibson, D.L.; Kean, W.F.; Klegeris, A. The biological activity of auranofin: Implications for novel treatment of diseases. Inflammopharmacology 2012, 20, 297–306. [Google Scholar] [CrossRef]
- Gamberi, T.; Chiappetta, G.; Fiaschi, T.; Modesti, A.; Sorbi, F.; Magherini, F. Upgrade of an old drug: Auranofin in innovative cancer therapies to overcome drug resistance and to increase drug effectiveness. Med. Res. Rev. 2022, 42, 1111–1146. [Google Scholar] [CrossRef]
- Zhang, X.; Selvaraju, K.; Saei, A.A.; D'Arcy, P.; Zubarev, R.A.; Arner, E.S.; Linder, S. Repurposing of auranofin: Thioredoxin reductase remains a primary target of the drug. Biochimie 2019, 162, 46–54. [Google Scholar] [CrossRef]
- Onodera, T.; Momose, I.; Kawada, M. Potential Anticancer Activity of Auranofin. Chem. Pharm. Bull. 2019, 67, 186–191. [Google Scholar] [CrossRef]
- Nobili, S.; Mini, E.; Landini, I.; Gabbiani, C.; Casini, A.; Messori, L. Gold compounds as anticancer agents: Chemistry, cellular pharmacology, and preclinical studies. Med. Res. Rev. 2010, 30, 550–580. [Google Scholar] [CrossRef]
- Liu, N.; Li, X.; Huang, H.; Zhao, C.; Liao, S.; Yang, C.; Liu, S.; Song, W.; Lu, X.; Lan, X.; et al. Clinically used antirheumatic agent auranofin is a proteasomal deubiquitinase inhibitor and inhibits tumor growth. Oncotarget 2014, 5, 5453–5471. [Google Scholar]
- Huang, H.; Liao, Y.; Liu, N.; Hua, X.; Cai, J.; Yang, C.; Long, H.; Zhao, C.; Chen, X.; Lan, X.; et al. Two clinical drugs deubiquitinase inhibitor auranofin and aldehyde dehydrogenase inhibitor disulfiram trigger synergistic anti-tumor effects in vitro and in vivo. Oncotarget 2016, 7, 2796–2808. [Google Scholar] [CrossRef] [PubMed]
- Simon, T.M.; Kunishima, D.H.; Vibert, G.J.; Lorber, A. Inhibitory effects of a new oral gold compound on HeLa cells. Cancer 1979, 44, 1965. [Google Scholar] [CrossRef]
- Mirabelli, C.K.; Johnson, R.K.; Sung, C.M.; Faucette, L.; Muirhead, K.; Crooke, S.T. Evaluation of the in vivo antitumor activity and in vitro cytotoxic properties of auranofin, a coordinated gold compound, in murine tumor models. Cancer Res. 1985, 45, 32–39. [Google Scholar] [PubMed]
- Liu, W.; Gust, R. Metal N-heterocyclic carbene complexes as potential antitumor metallodrugs. Chem. Soc. Rev. 2013, 42, 755–773. [Google Scholar] [PubMed]
- Kim, N.H.; Park, H.J.; Oh, M.K.; Kim, I.S. Antiproliferative effect of gold(I) compound auranofin through inhibition of STAT3 and telomerase activity in MDA-MB 231 human breast cancer cells. BMB Rep. 2013, 46, 59–64. [Google Scholar] [CrossRef] [PubMed]
- Nakaya, A.; Sagawa, M.; Muto, A.; Uchida, H.; Ikeda, Y.; Kizaki, M. The gold compound auranofin induces apoptosis of human multiple myeloma cells through both down-regulation of STAT3 and inhibition of NF-kappaB activity. Leuk. Res. 2011, 35, 243–249. [Google Scholar] [CrossRef]
- Park, S.H.; Lee, J.H.; Berek, J.S.; Hu, M.C. Auranofin displays anticancer activity against ovarian cancer cells through FOXO3 activation independent of p53. Int. J. Oncol. 2014, 45, 1691–1698. [Google Scholar] [CrossRef]
- Wang, Y.; Hill, K.S.; Fields, A.P. PKCiota maintains a tumor-initiating cell phenotype that is required for ovarian tumorigenesis. Mol. Cancer Res. 2013, 11, 1624–1635. [Google Scholar] [CrossRef]
- Li, H.; Hu, J.; Wu, S.; Wang, L.; Cao, X.; Zhang, X.; Dai, B.; Cao, M.; Shao, R.; Zhang, R.; et al. Auranofin-mediated inhibition of PI3K/AKT/mTOR axis and anticancer activity in non-small cell lung cancer cells. Oncotarget 2016, 7, 3548–3558. [Google Scholar] [CrossRef]
- Hatem, E.; Azzi, S.; El Banna, N.; He, T.; Heneman-Masurel, A.; Vernis, L.; Baille, D.; Masson, V.; Dingli, F.; Loew, D.; et al. Auranofin/Vitamin C: A Novel Drug Combination Targeting Triple-Negative Breast Cancer. J. Natl. Cancer Inst. 2019, 111, 597–608. [Google Scholar] [CrossRef]
- He, T.; Hatem, E.; Vernis, L.; Lei, M.; Huang, M.E. PRX1 knockdown potentiates vitamin K3 toxicity in cancer cells: A potential new therapeutic perspective for an old drug. J. Exp. Clin. Cancer Res. 2015, 34, 152. [Google Scholar] [CrossRef] [PubMed]
- Mishina, N.M.; Markvicheva, K.N.; Bilan, D.S.; Matlashov, M.E.; Shirmanova, M.V.; Liebl, D.; Schultz, C.; Lukyanov, S.; Belousov, V.V. Visualization of intracellular hydrogen peroxide with HyPer, a genetically encoded fluorescent probe. Methods Enzymol. 2013, 526, 45–59. [Google Scholar]
- El Banna, N.; Hatem, E.; Heneman-Masurel, A.; Leger, T.; Baille, D.; Vernis, L.; Garcia, C.; Martineau, S.; Dupuy, C.; Vagner, S.; et al. Redox modifications of cysteine-containing proteins, cell cycle arrest and translation inhibition: Involvement in vitamin C-induced breast cancer cell death. Redox Biol. 2019, 26, 101290. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Casagrande, J.T.; Thomas, P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013, 8, 1551–1566. [Google Scholar] [CrossRef] [PubMed]
- Dennis, G., Jr.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, R60. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- Qiu, X.; Guittet, O.; Mingoes, C.; El Banna, N.; Huang, M.E.; Lepoivre, M.; Hildebrandt, N. Quantification of Cellular Deoxyribonucleoside Triphosphates by Rolling Circle Amplification and Forster Resonance Energy Transfer. Anal. Chem. 2019, 91, 14561–14568. [Google Scholar] [CrossRef]
- Bilan, D.S.; Belousov, V.V. HyPer Family Probes: State of the Art. Antioxid. Redox Signal. 2016, 24, 731–751. [Google Scholar] [CrossRef]
- Saei, A.A.; Gullberg, H.; Sabatier, P.; Beusch, C.M.; Johansson, K.; Lundgren, B.; Arvidsson, P.I.; Arner, E.S.J.; Zubarev, R.A. Comprehensive chemical proteomics for target deconvolution of the redox active drug auranofin. Redox Biol. 2020, 32, 101491. [Google Scholar] [CrossRef]
- Hasan, A.A.; Kalinina, E.; Tatarskiy, V.; Shtil, A. The Thioredoxin System of Mammalian Cells and Its Modulators. Biomedicines 2022, 10, 1757. [Google Scholar] [CrossRef]
- Gencheva, R.; Arner, E.S.J. Thioredoxin Reductase Inhibition for Cancer Therapy. Annu. Rev. Pharmacol. Toxicol. 2022, 62, 177–196. [Google Scholar] [CrossRef] [PubMed]
- Mandal, P.K.; Schneider, M.; Kolle, P.; Kuhlencordt, P.; Forster, H.; Beck, H.; Bornkamm, G.W.; Conrad, M. Loss of thioredoxin reductase 1 renders tumors highly susceptible to pharmacologic glutathione deprivation. Cancer Res. 2010, 70, 9505–9514. [Google Scholar] [CrossRef] [PubMed]
- Hellfritsch, J.; Kirsch, J.; Schneider, M.; Fluege, T.; Wortmann, M.; Frijhoff, J.; Dagnell, M.; Fey, T.; Esposito, I.; Kolle, P.; et al. Knockout of mitochondrial thioredoxin reductase stabilizes prolyl hydroxylase 2 and inhibits tumor growth and tumor-derived angiogenesis. Antioxid. Redox Signal. 2015, 22, 938–950. [Google Scholar] [CrossRef] [PubMed]
- Chiappetta, G.; Gamberi, T.; Faienza, F.; Limaj, X.; Rizza, S.; Messori, L.; Filomeni, G.; Modesti, A.; Vinh, J. Redox proteome analysis of auranofin exposed ovarian cancer cells (A2780). Redox Biol. 2022, 52, 102294. [Google Scholar] [CrossRef]
- Fiskus, W.; Saba, N.; Shen, M.; Ghias, M.; Liu, J.; Gupta, S.D.; Chauhan, L.; Rao, R.; Gunewardena, S.; Schorno, K.; et al. Auranofin induces lethal oxidative and endoplasmic reticulum stress and exerts potent preclinical activity against chronic lymphocytic leukemia. Cancer Res. 2014, 74, 2520–2532. [Google Scholar] [CrossRef]
- Go, Y.M.; Roede, J.R.; Walker, D.I.; Duong, D.M.; Seyfried, N.T.; Orr, M.; Liang, Y.; Pennell, K.D.; Jones, D.P. Selective targeting of the cysteine proteome by thioredoxin and glutathione redox systems. Mol. Cell. Proteom. 2013, 12, 3285–3296. [Google Scholar] [CrossRef]
- Lu, J.; Holmgren, A. The thioredoxin antioxidant system. Free Radic. Biol. Med. 2014, 66, 75–87. [Google Scholar] [CrossRef]
- Prigge, J.R.; Coppo, L.; Martin, S.S.; Ogata, F.; Miller, C.G.; Bruschwein, M.D.; Orlicky, D.J.; Shearn, C.T.; Kundert, J.A.; Lytchier, J.; et al. Hepatocyte Hyperproliferation upon Liver-Specific Co-disruption of Thioredoxin-1, Thioredoxin Reductase-1, and Glutathione Reductase. Cell Rep. 2017, 19, 2771–2781. [Google Scholar] [CrossRef]
- Kotsantis, P.; Jones, R.M.; Higgs, M.R.; Petermann, E. Cancer therapy and replication stress: Forks on the road to perdition. Adv. Clin. Chem. 2015, 69, 91–138. [Google Scholar]
- Trenner, A.; Sartori, A.A. Harnessing DNA Double-Strand Break Repair for Cancer Treatment. Front. Oncol. 2019, 9, 1388. [Google Scholar] [CrossRef]
- Ong, M.S.; Deng, S.; Halim, C.E.; Cai, W.; Tan, T.Z.; Huang, R.Y.; Sethi, G.; Hooi, S.C.; Kumar, A.P.; Yap, C.T. Cytoskeletal Proteins in Cancer and Intracellular Stress: A Therapeutic Perspective. Cancers 2020, 12, 238. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Huff, L.P.; Fujii, M.; Griendling, K.K. Redox regulation of the actin cytoskeleton and its role in the vascular system. Free Radic. Biol. Med. 2017, 109, 84–107. [Google Scholar] [CrossRef] [PubMed]
- Chaffman, M.; Brogden, R.N.; Heel, R.C.; Speight, T.M.; Avery, G.S. Auranofin. A preliminary review of its pharmacological properties and therapeutic use in rheumatoid arthritis. Drugs 1984, 27, 378–424. [Google Scholar] [CrossRef] [PubMed]
- Capparelli, E.V.; Bricker-Ford, R.; Rogers, M.J.; McKerrow, J.H.; Reed, S.L. Phase I Clinical Trial Results of Auranofin, a Novel Antiparasitic Agent. Antimicrob. Agents Chemother. 2017, 61, e01947-16. [Google Scholar] [CrossRef]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452.e17. [Google Scholar] [CrossRef]



| GO Term | Biological Process | Count | % | p Value |
|---|---|---|---|---|
| GO:0098609 | cell–cell adhesion | 18 | 11.18 | 6.0825 × 10−10 |
| GO:0055114 | oxidation–reduction process | 13 | 8.07 | 0.0093 |
| GO:0008283 | cell proliferation | 11 | 6.83 | 0.0023 |
| GO:0043066 | negative regulation of apoptotic process | 10 | 6.21 | 0.0264 |
| GO:0007010 | cytoskeleton organization | 9 | 5.59 | 1.3107 × 10−4 |
| GO:0016032 | viral process | 9 | 5.59 | 0.0069 |
| GO:0006457 | protein folding | 8 | 4.97 | 0.0015 |
| GO:0051301 | cell division | 8 | 4.97 | 0.0452 |
| GO:0007049 | cell cycle | 7 | 4.35 | 0.0159 |
| GO:0007067 | mitotic nuclear division | 7 | 4.35 | 0.0284 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hatem, E.; El Banna, N.; Heneman-Masurel, A.; Baïlle, D.; Vernis, L.; Riquier, S.; Golinelli-Cohen, M.-P.; Guittet, O.; Vallières, C.; Camadro, J.-M.; et al. Novel Insights into Redox-Based Mechanisms for Auranofin-Induced Rapid Cancer Cell Death. Cancers 2022, 14, 4864. https://doi.org/10.3390/cancers14194864
Hatem E, El Banna N, Heneman-Masurel A, Baïlle D, Vernis L, Riquier S, Golinelli-Cohen M-P, Guittet O, Vallières C, Camadro J-M, et al. Novel Insights into Redox-Based Mechanisms for Auranofin-Induced Rapid Cancer Cell Death. Cancers. 2022; 14(19):4864. https://doi.org/10.3390/cancers14194864
Chicago/Turabian StyleHatem, Elie, Nadine El Banna, Amélie Heneman-Masurel, Dorothée Baïlle, Laurence Vernis, Sylvie Riquier, Marie-Pierre Golinelli-Cohen, Olivier Guittet, Cindy Vallières, Jean-Michel Camadro, and et al. 2022. "Novel Insights into Redox-Based Mechanisms for Auranofin-Induced Rapid Cancer Cell Death" Cancers 14, no. 19: 4864. https://doi.org/10.3390/cancers14194864
APA StyleHatem, E., El Banna, N., Heneman-Masurel, A., Baïlle, D., Vernis, L., Riquier, S., Golinelli-Cohen, M.-P., Guittet, O., Vallières, C., Camadro, J.-M., Qiu, X., Hildebrandt, N., Lepoivre, M., & Huang, M.-E. (2022). Novel Insights into Redox-Based Mechanisms for Auranofin-Induced Rapid Cancer Cell Death. Cancers, 14(19), 4864. https://doi.org/10.3390/cancers14194864







