A Novel Bayesian Framework Infers Driver Activation States and Reveals Pathway-Oriented Molecular Subtypes in Head and Neck Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection and Preprocessing
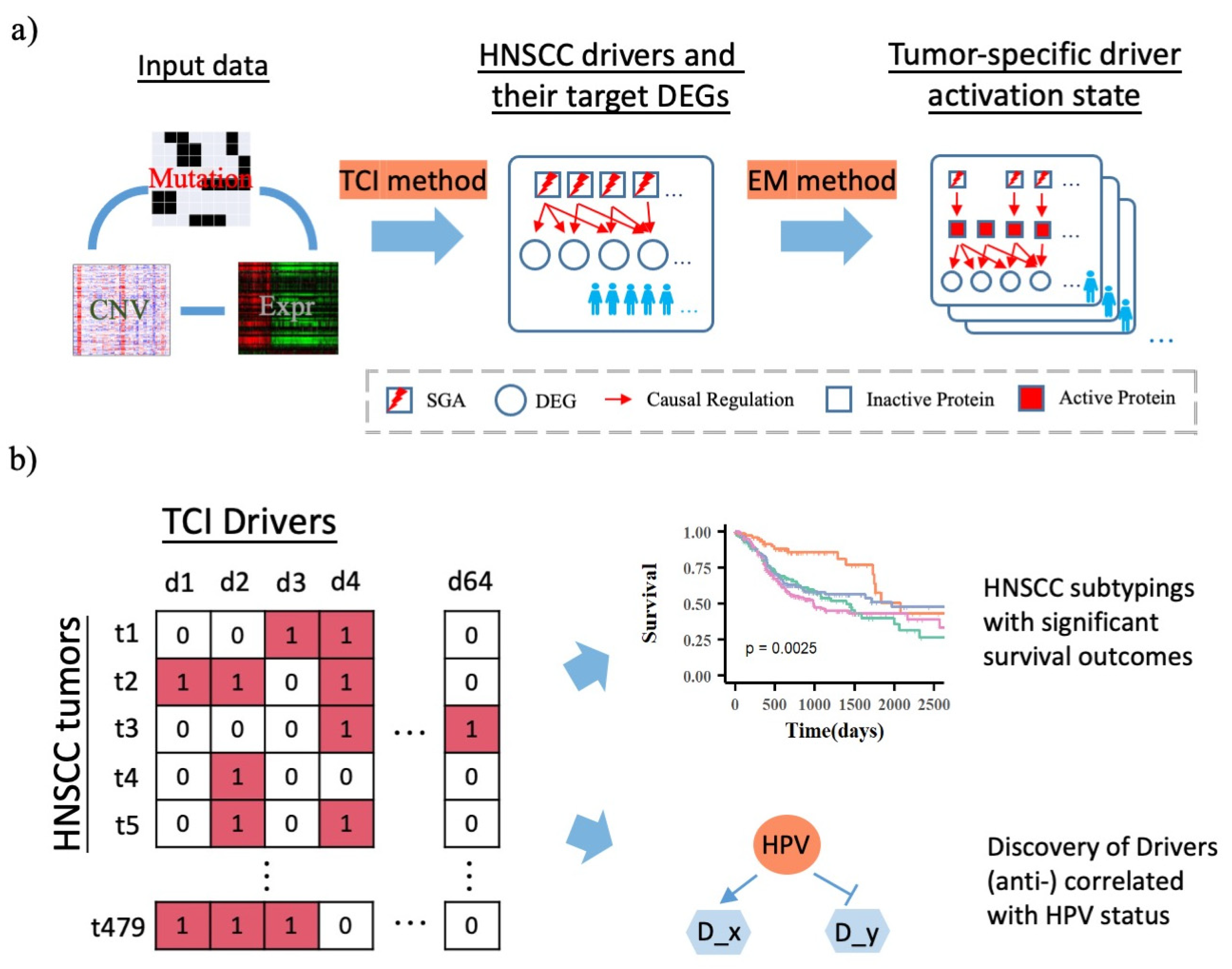
2.2. Bayesian Framework: 1. Tumor-Specific Causal Inference (TCI) Method
2.3. Discovery of TCI Drivers and Target DEGs in HNSCC Patients
2.4. Bayesian Framework: 2. Inference of Tumor Specific Driver Activation State Using Expectation Maximization (EM) Algorithm
2.5. Patient Subtyping Using Consensus Clustering
2.6. Reconstruction of Driver → DEG Causal Network
3. Results
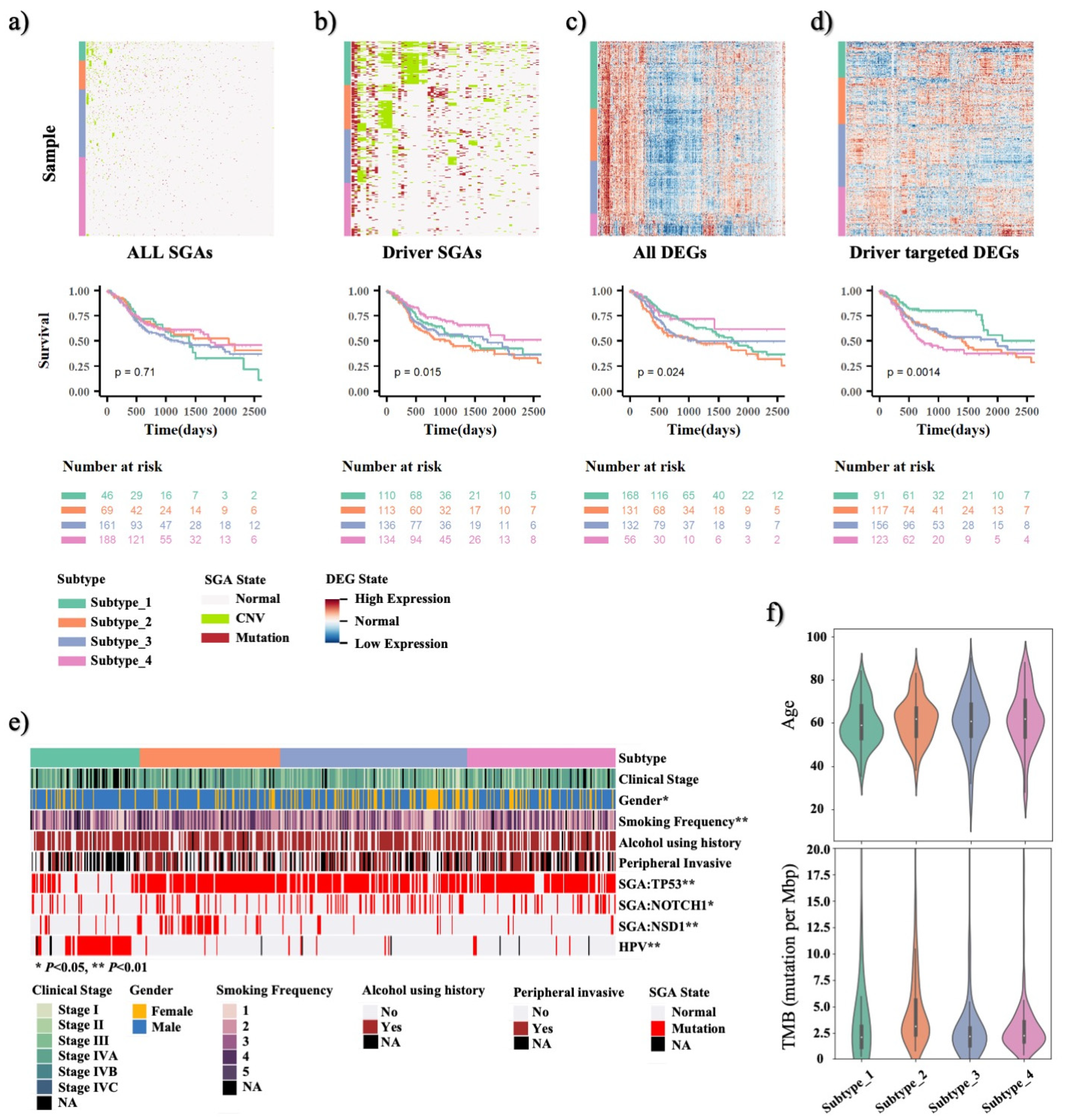
3.1. TCI Method Can Identify Major Cancer Driver Genes and Their Causative DEGs Targets for HNSCC Patients
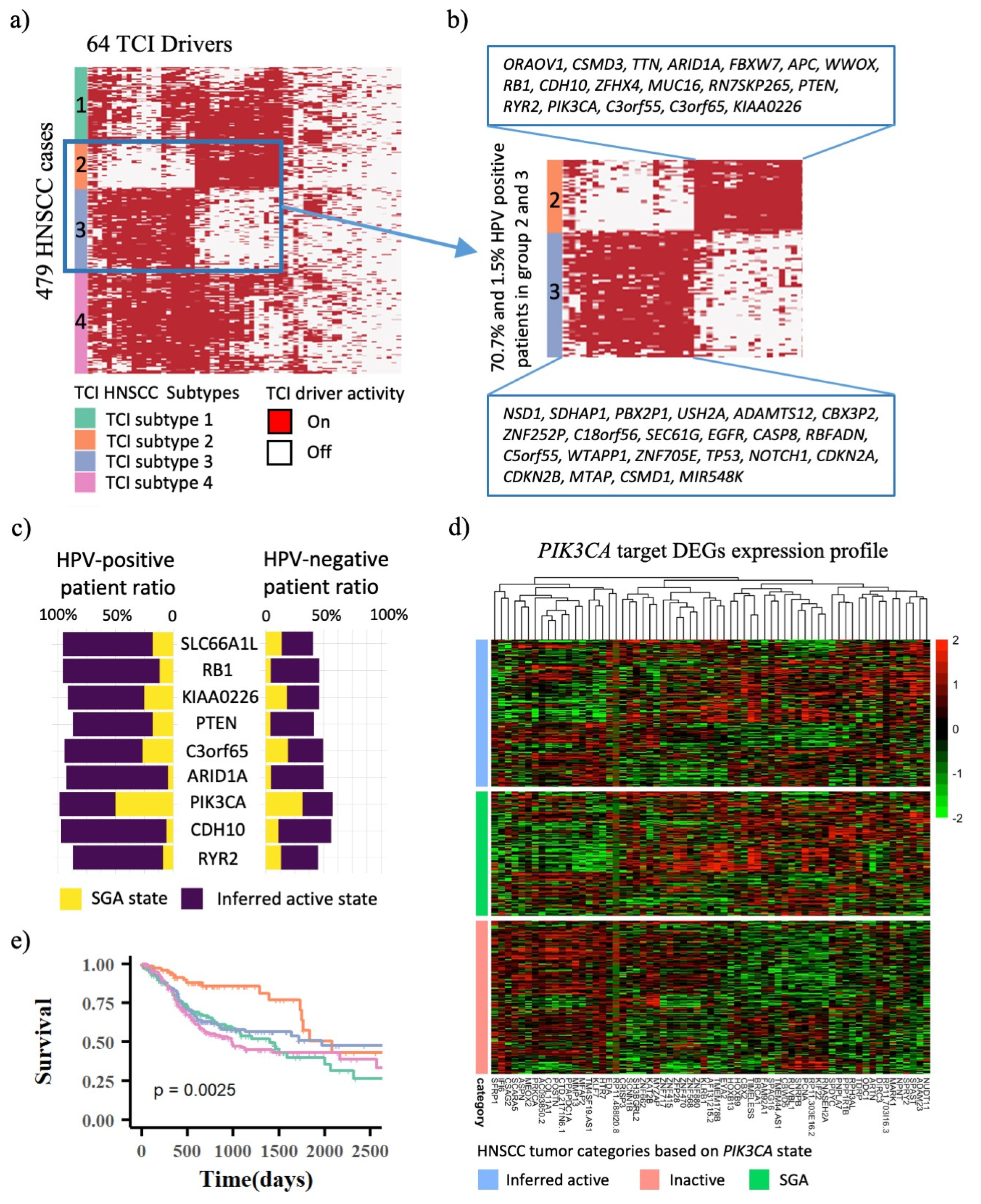
3.2. TCI-Derived Molecular Profiles Predict Significant Prognostic Outcome Differences among HNSCC Patient Subtypes
3.3. Infer Driver Activation States and Represent HNSCC Tumors in the Space of Cellular Signaling Systems
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kandoth, C.; McLellan, M.D.; Vandin, F.; Ye, K.; Niu, B.; Lu, C.; Xie, M.; Zhang, Q.; McMichael, J.F.; Wyczalkowski, M.A.; et al. Mutational landscape and significance across 12 major cancer types. Nature 2013, 502, 333–339. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.S.; Stojanov, P.; Mermel, C.H.; Robinson, J.T.; Garraway, L.A.; Golub, T.R.; Meyerson, M.; Gabriel, S.B.; Lander, E.S.; Getz, G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014, 505, 495–501. [Google Scholar] [CrossRef] [PubMed]
- Zack, T.I.; Schumacher, S.E.; Carter, S.L.; Cherniack, A.D.; Saksena, G.; Tabak, B.; Lawrence, M.S.; Zhsng, C.Z.; Wala, J.; Mermel, C.H.; et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 2013, 45, 1134–1140. [Google Scholar] [CrossRef] [PubMed]
- Ciriello, G.; Miller, M.L.; Aksoy, B.A.; Senbabaoglu, Y.; Schultz, N.; Sander, C. Emerging landscape of oncogenic signatures across human cancers. Nat. Genet. 2013, 45, 1127–1133. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Perez, C.; Tamborero, D.; Schroeder, M.P.; Antolin, A.A.; Deu-Pons, J.; Perez-Llamas, C.; Mestres, J.; Gonzalez-Perez, A.; Lopez-Bigas, N. In silico prescription of anticancer drugs to cohorts of 28 tumor types reveals targeting opportunities. Cancer Cell 2015, 27, 382–396. [Google Scholar] [CrossRef]
- Garraway, L.A.; Verweij, J.; Ballman, K.V. Precision oncology: An overview. J. Clin. Oncol. 2013, 31, 1803–1805. [Google Scholar] [CrossRef]
- Cai, C.; Cooper, G.F.; Lu, K.N.; Ma, X.; Xu, S.; Zhao, Z.; Chen, X.; Xue, Y.; Lee, A.V.; Clark, N.; et al. Systematic discovery of the functional impact of somatic genome alterations in individual tumors through tumor-specific causal inference. PLoS Comput. Biol. 2019, 15, e1007088. [Google Scholar] [CrossRef]
- Cai, C.; Chen, L.; Jiang, X.; Lu, X. Modeling signal transduction from protein phosphorylation to gene expression. Cancer Inform. 2014, 13, 59–67. [Google Scholar] [CrossRef]
- Liu, J.; Ma, X.; Cooper, G.F.; Lu, X. Explicit representation of protein activity states significantly improves causal discovery of protein phosphorylation networks. BMC Bioinform. 2020, 21, 379. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.H.; Parker, J.S.; Karaca, G.; Wu, J.; Funkhouser, W.K.; Moore, D.; Butterfoss, D.; Xiang, D.; Zanation, A.; Yin, X.; et al. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell 2004, 5, 489–500. [Google Scholar] [CrossRef]
- Walter, V.; Yin, X.; Wilkerson, M.D.; Cabanski, C.R.; Zhao, N.; Du, Y.; Ang, M.K.; Hayward, M.C.; Salazar, A.H.; Hoadley, K.A.; et al. Molecular subtypes in head and neck cancer exhibit distinct patterns of chromosomal gain and loss of canonical cancer genes. PLoS ONE 2013, 8, e56823. [Google Scholar] [CrossRef]
- Cancer Genome Atlas, N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Bishop, C.M. Pattern Recognition and Machine Learning (Information Science and Statistics); Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repecka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Basukala, O.; Mittal, S.; Massimi, P.; Bestagno, M.; Banks, L. The HPV-18 E7 CKII phospho acceptor site is required for maintaining the transformed phenotype of cervical tumour-derived cells. PLoS Pathog. 2019, 15, e1007769. [Google Scholar] [CrossRef]
- Gao, C.; McDowell, I.C.; Zhao, S.; Brown, C.D.; Engelhardt, B.E. Context Specific and Differential Gene Co-expression Networks via Bayesian Biclustering. PLoS Comput. Biol. 2016, 12, e1004791. [Google Scholar] [CrossRef]
- Grossmann, P.; Stringfield, O.; El-Hachem, N.; Bui, M.M.; Rios Velazquez, E.; Parmar, C.; Leijenaar, R.T.; Haibe-Kains, B.; Lambin, P.; Gillies, R.J.; et al. Defining the biological basis of radiomic phenotypes in lung cancer. Elife 2017, 6, e23421. [Google Scholar] [CrossRef]
- Alsahafi, E.; Begg, K.; Amelio, I.; Raulf, N.; Lucarelli, P.; Sauter, T.; Tavassoli, M. Clinical update on head and neck cancer: Molecular biology and ongoing challenges. Cell Death Dis. 2019, 10, 540. [Google Scholar] [CrossRef]
- Mountzios, G.; Rampias, T.; Psyrri, A. The mutational spectrum of squamous-cell carcinoma of the head and neck: Targetable genetic events and clinical impact. Ann. Oncol. 2014, 25, 1889–1900. [Google Scholar] [CrossRef]
- Orhan, C.; Bakir, B.; Dalay, N.; Buyru, N. ZNF703 is an important player in head and neck cancer. Clin. Otolaryngol. 2019, 44, 1080–1086. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.H.; Liu, R.; Lin, X.R.; Luo, J.Q.; Cao, L.J.; Zhang, Q.J.; Lin, S.R.; Geng, L.; Sun, Z.Y.; Ye, S.K.; et al. LRP1B mutation is associated with tumor HPV status and promotes poor disease outcomes with a higher mutation count in HPV-related cervical carcinoma and head & neck squamous cell carcinoma. Int. J. Biol. Sci. 2021, 17, 1744–1756. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Wu, Y.; Feng, M.; Xue, X.; Fan, Y. A novel sevenmiRNA prognostic model to predict overall survival in head and neck squamous cell carcinoma patients. Mol. Med. Rep. 2019, 20, 4340–4348. [Google Scholar] [CrossRef]
- Deville, S.S.; Luft, S.; Kaufmann, M.; Cordes, N. Keap1 inhibition sensitizes head and neck squamous cell carcinoma cells to ionizing radiation via impaired non-homologous end joining and induced autophagy. Cell Death Dis. 2020, 11, 887. [Google Scholar] [CrossRef]
- Lu, W.C.; Liu, C.J.; Tu, H.F.; Chung, Y.T.; Yang, C.C.; Kao, S.Y.; Chang, K.W.; Lin, S.C. miR-31 targets ARID1A and enhances the oncogenicity and stemness of head and neck squamous cell carcinoma. Oncotarget 2016, 7, 57254–57267. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Xu, H.; Wang, W.; Li, S.; Li, H.; Li, T.; Zhang, W.; Yu, X.; Liu, L. The role of collagen in cancer: From bench to bedside. J. Transl. Med. 2019, 17, 309. [Google Scholar] [CrossRef] [PubMed]
- Psyrri, A.; Gkotzamanidou, M.; Papaxoinis, G.; Krikoni, L.; Economopoulou, P.; Kotsantis, I.; Anastasiou, M.; Souliotis, V.L. The DNA damage response network in the treatment of head and neck squamous cell carcinoma. ESMO Open 2021, 6, 100075. [Google Scholar] [CrossRef]
- Wurster, S.; Hennes, F.; Parplys, A.C.; Seelbach, J.I.; Mansour, W.Y.; Zielinski, A.; Petersen, C.; Clauditz, T.S.; Munscher, A.; Friedl, A.A.; et al. PARP1 inhibition radiosensitizes HNSCC cells deficient in homologous recombination by disabling the DNA replication fork elongation response. Oncotarget 2016, 7, 9732–9741. [Google Scholar] [CrossRef]
- Wu, J.; Galvan, K.J.; Bogard, R.D.; Peterson, C.E.; Shergill, A.; Crowe, D.L. DNA Double-strand Break Signaling Is a Therapeutic Target in Head and Neck Cancer. Anticancer Res. 2021, 41, 5393–5403. [Google Scholar] [CrossRef]
- Zhang, Z.; Cui, R.; Li, H.; Li, J. miR-500 promotes cell proliferation by directly targetting LRP1B in prostate cancer. Biosci. Rep. 2019, 39, BSR20181854. [Google Scholar] [CrossRef]
- Beer, A.G.; Zenzmaier, C.; Schreinlechner, M.; Haas, J.; Dietrich, M.F.; Herz, J.; Marschang, P. Expression of a recombinant full-length LRP1B receptor in human non-small cell lung cancer cells confirms the postulated growth-suppressing function of this large LDL receptor family member. Oncotarget 2016, 7, 68721–68733. [Google Scholar] [CrossRef] [PubMed]
- Tarakji, B. Immunohistochemical expression of pRb in pleomorphic adenoma and carcinoma ex pleomorphic adenoma. Med Oral Patol Oral Cir Bucal 2011, 16, e323–e329. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Lin, J.; Rong, L.; Wu, S.; Deng, Z.; Fatkhutdinov, N.; Zundell, J.; Fukumoto, T.; Liu, Q.; Kossenkov, A.; et al. ARID1A promotes genomic stability through protecting telomere cohesion. Nat. Commun. 2019, 10, 4067. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.H.; Bellon, M.; Nicot, C. FBXW7: A critical tumor suppressor of human cancers. Mol. Cancer 2018, 17, 115. [Google Scholar] [CrossRef] [PubMed]
- Pan, C.; Izreig, S.; Yarbrough, W.G.; Issaeva, N. NSD1 mutations by HPV status in head and neck cancer: Differences in survival and response to DNA-damaging agents. Cancers Head Neck 2019, 4, 3. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Cai, C.; Ma, X.; Liu, J.; Chen, L.; Lui, V.W.Y.; Cooper, G.F.; Lu, X. A Novel Bayesian Framework Infers Driver Activation States and Reveals Pathway-Oriented Molecular Subtypes in Head and Neck Cancer. Cancers 2022, 14, 4825. https://doi.org/10.3390/cancers14194825
Liu Z, Cai C, Ma X, Liu J, Chen L, Lui VWY, Cooper GF, Lu X. A Novel Bayesian Framework Infers Driver Activation States and Reveals Pathway-Oriented Molecular Subtypes in Head and Neck Cancer. Cancers. 2022; 14(19):4825. https://doi.org/10.3390/cancers14194825
Chicago/Turabian StyleLiu, Zhengping, Chunhui Cai, Xiaojun Ma, Jinling Liu, Lujia Chen, Vivian Wai Yan Lui, Gregory F. Cooper, and Xinghua Lu. 2022. "A Novel Bayesian Framework Infers Driver Activation States and Reveals Pathway-Oriented Molecular Subtypes in Head and Neck Cancer" Cancers 14, no. 19: 4825. https://doi.org/10.3390/cancers14194825
APA StyleLiu, Z., Cai, C., Ma, X., Liu, J., Chen, L., Lui, V. W. Y., Cooper, G. F., & Lu, X. (2022). A Novel Bayesian Framework Infers Driver Activation States and Reveals Pathway-Oriented Molecular Subtypes in Head and Neck Cancer. Cancers, 14(19), 4825. https://doi.org/10.3390/cancers14194825




