Relevance of Abnormal KCNN1 Expression and Osmotic Hypersensitivity in Ewing Sarcoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Growth Conditions
2.2. RNA Isolation, RT-PCR, RT-qPCR and Capillary Sequencing
2.3. RNA In Situ Hybridization of Patient Tissue Microarrays
2.4. KCNN1 Knockdown
2.5. KCNN1 and EWSR1-FLI1 Overexpression
2.6. Analysis of RNA-Sequencing and ChIP-Sequencing Data
2.7. Patch-Clamp Electrophysiology
2.8. Intracellular Ca2+ Measurement
2.9. Migration Assays
2.10. Cell Viability Assays
2.11. Cell Volume Measurements
3. Results
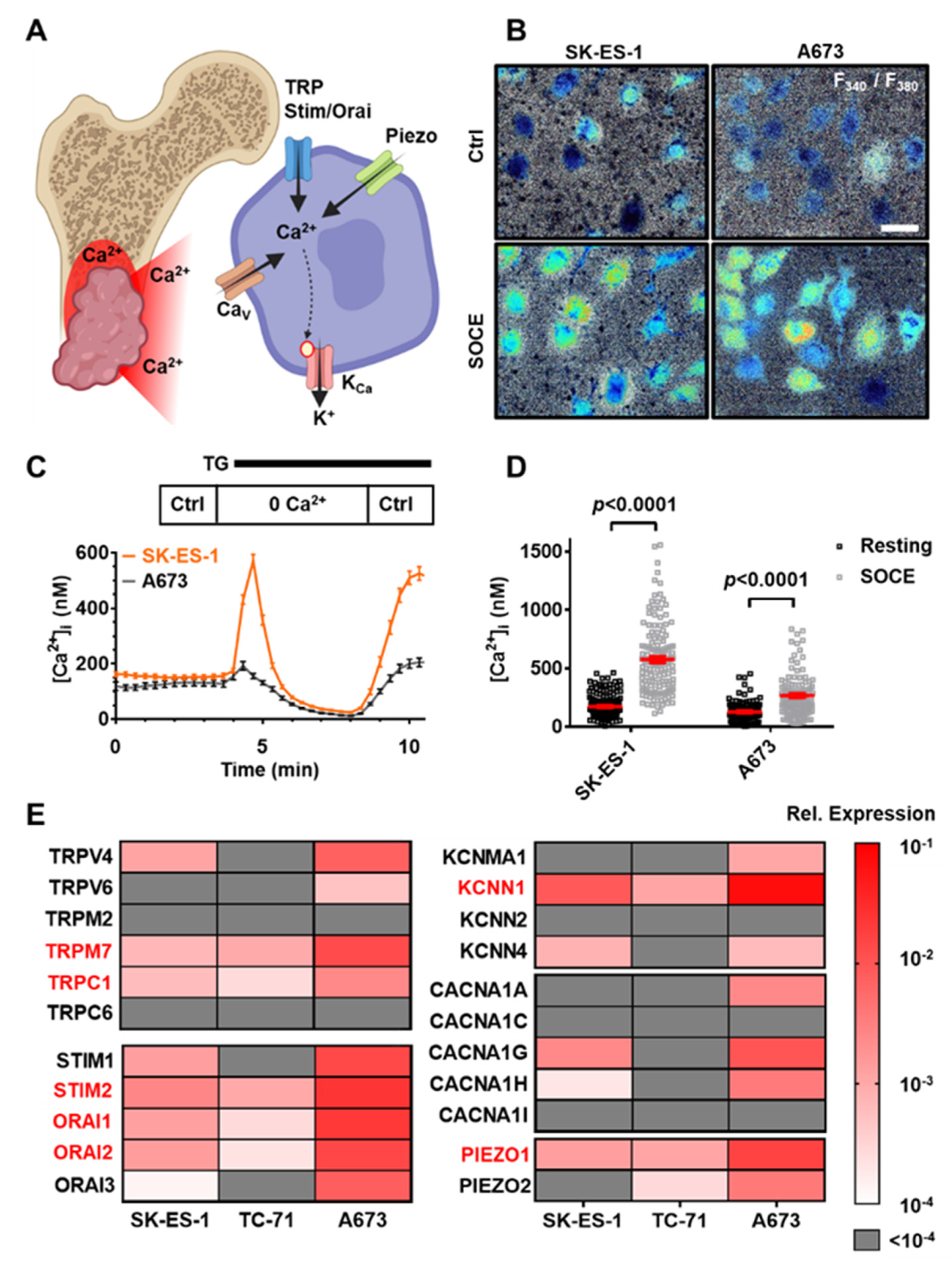
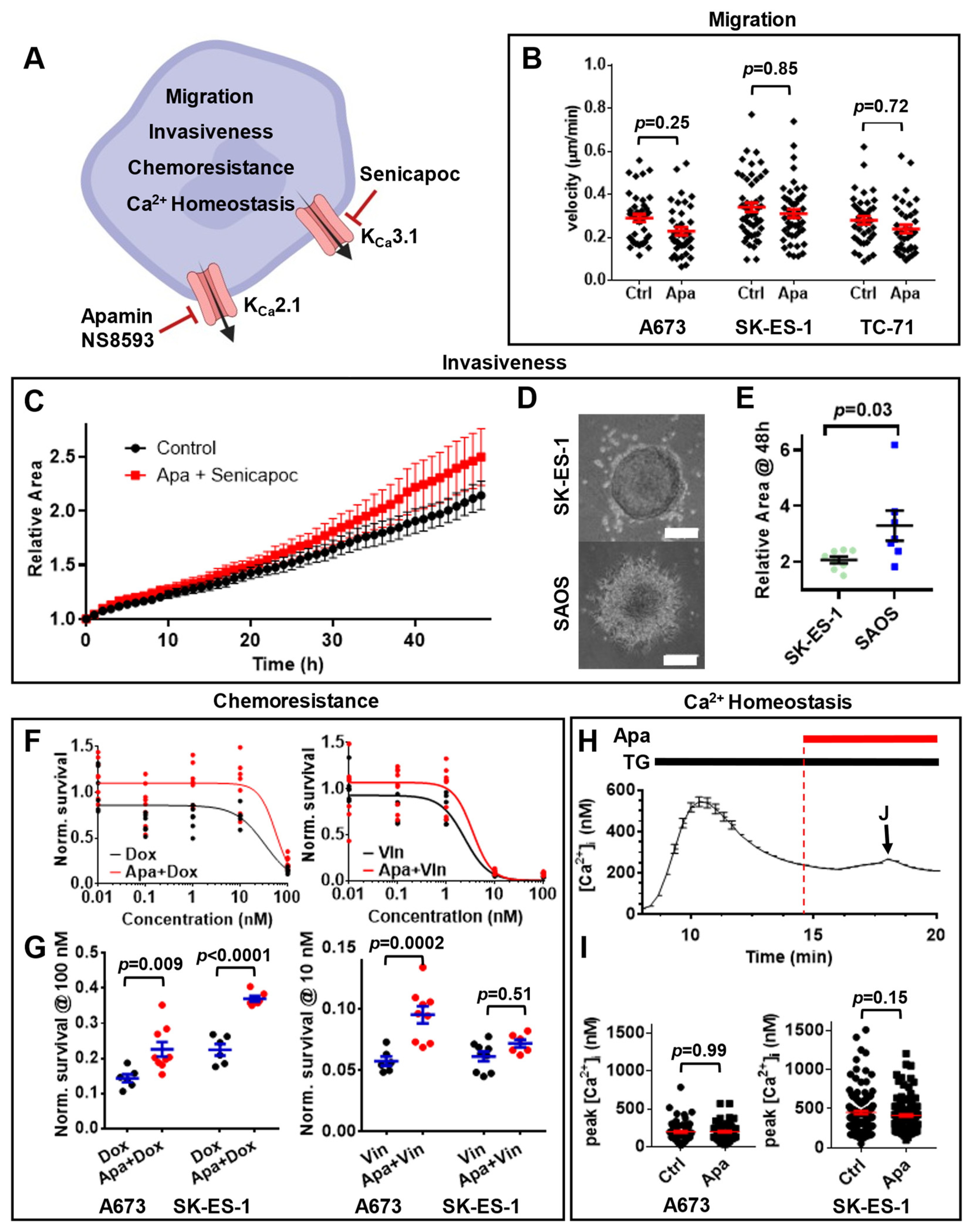
3.1. Multiple Ca2+-Modulated Ion Channels Are Functional in Ewing Sarcoma
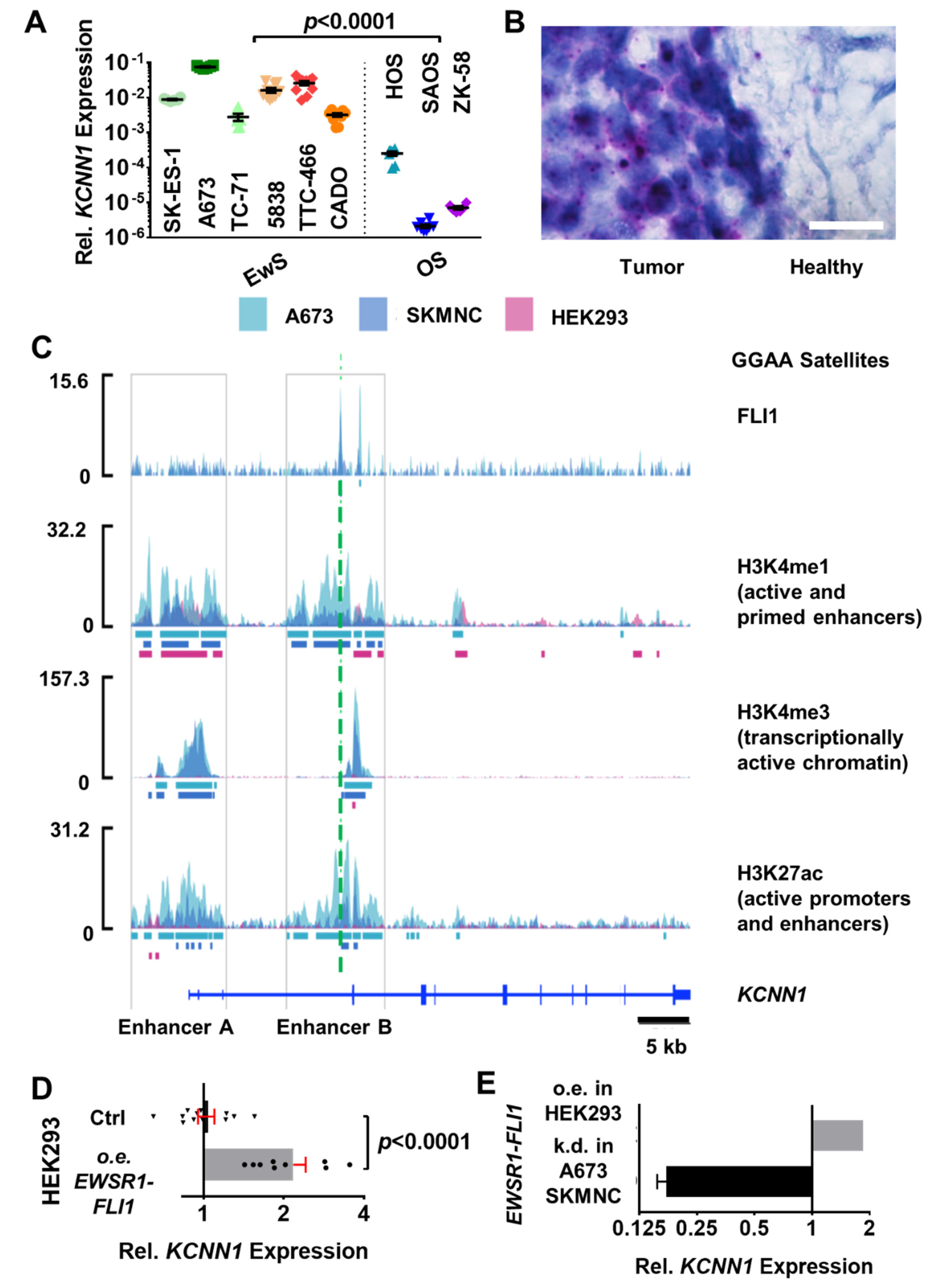
3.2. EWSR1-FLI1 Drives the Expression of an Alternative KCNN1 Transcript Variant in Ewing Sarcoma
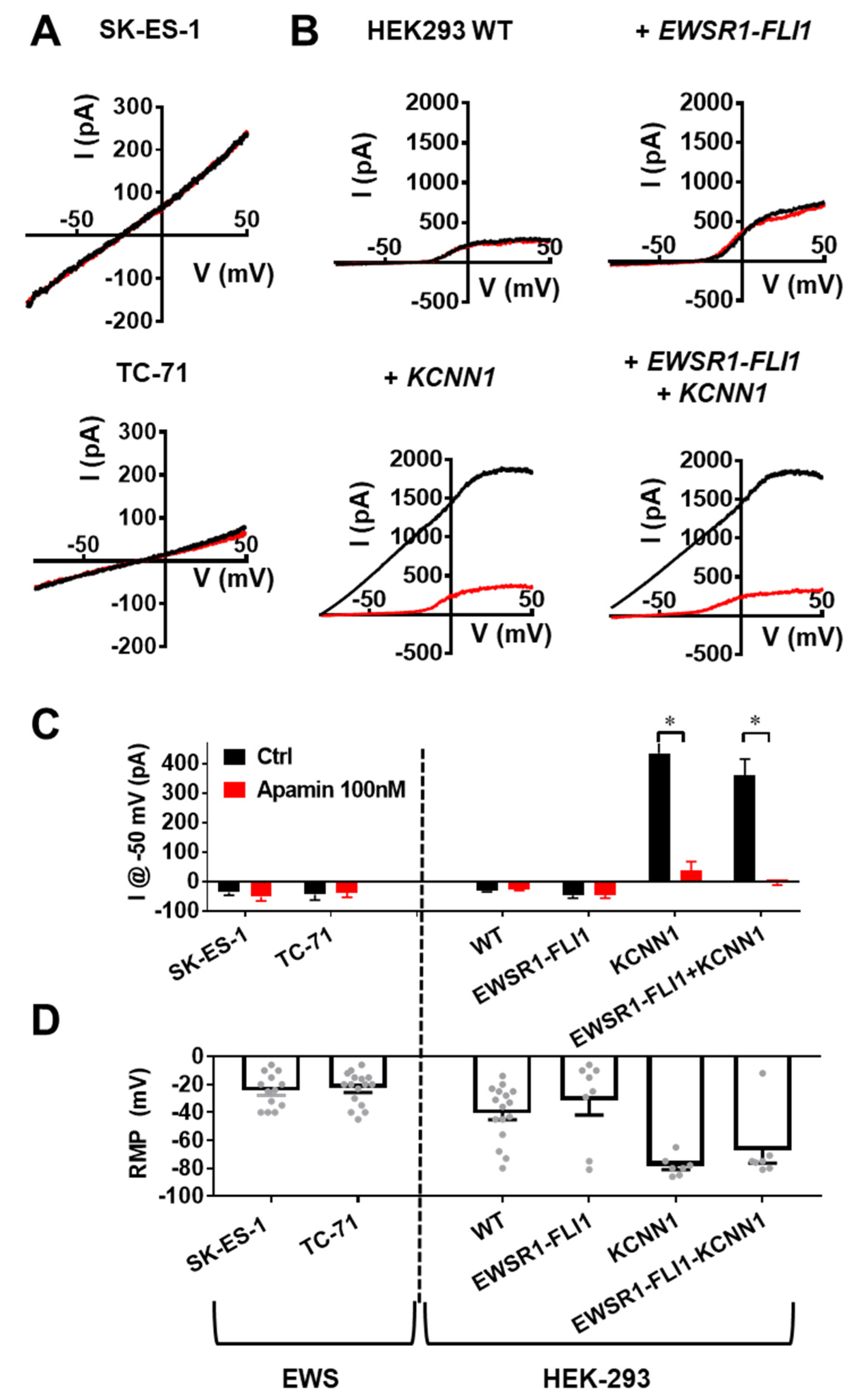
3.3. KCNN1 Expression Does Not Lead to KCa2.1 Function in Ewing Sarcoma
3.4. KCa2.1 Is Non-Functional in EWS Cells
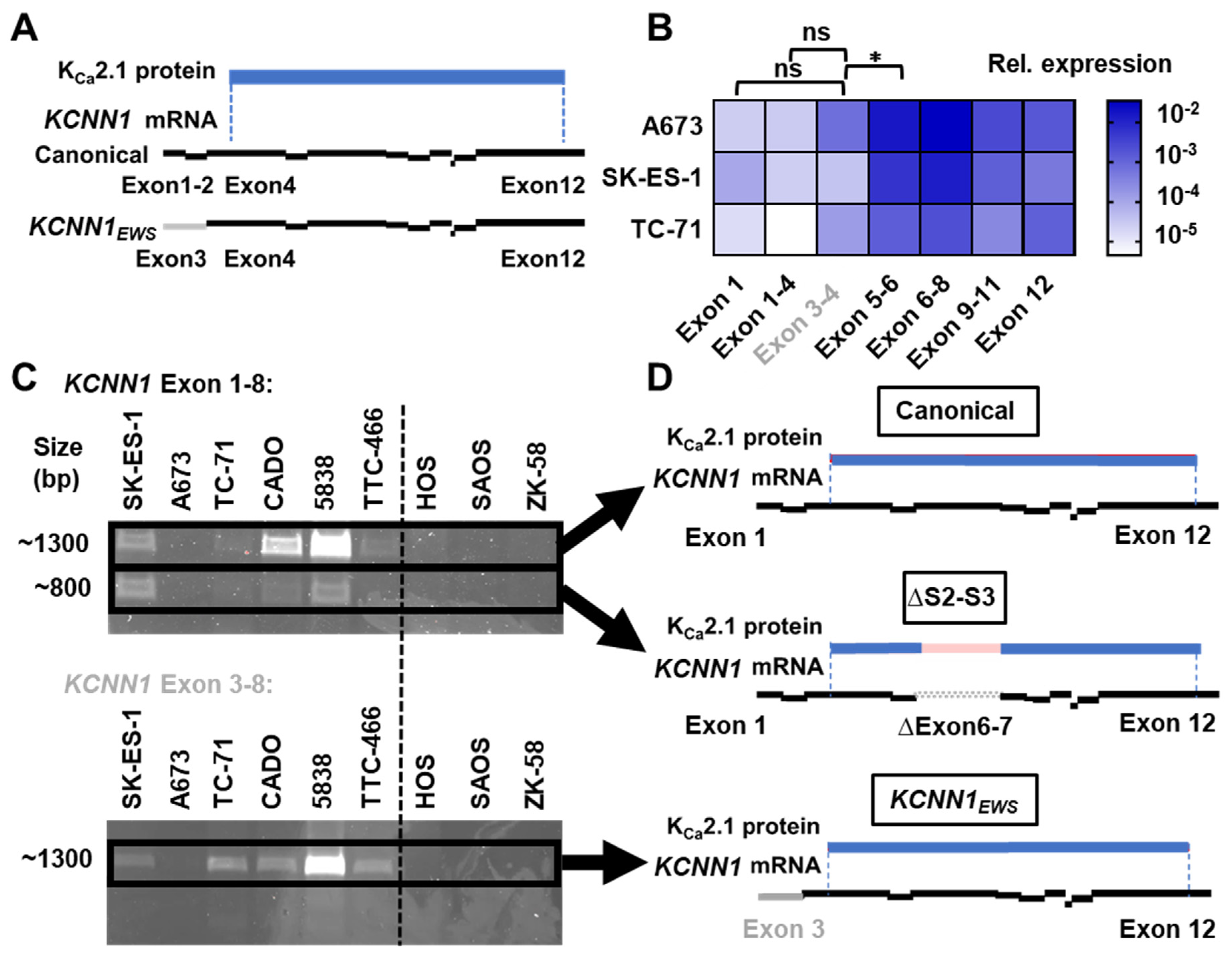
3.5. KCNN1 Transcripts in EwS Are Non-Conductive
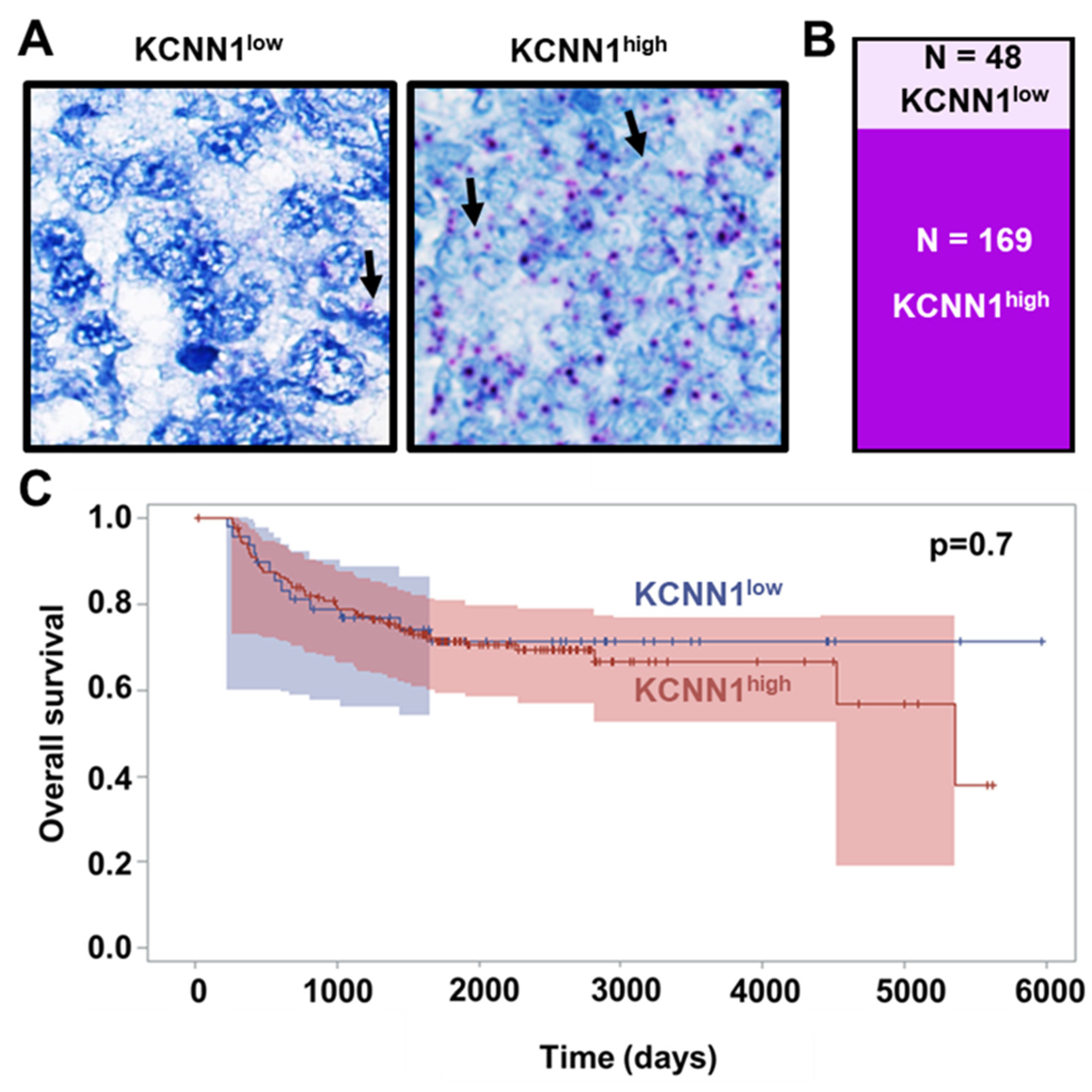
3.6. Lack of KCa2.1 Channel Function Sensitizes EwS Cells to Hypoosmotic Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aran, V.; Devalle, S.; Meohas, W.; Heringer, M.; Cunha Caruso, A.; Pinheiro Aguiar, D.; Leite Duarte, M.E.; Moura Neto, V. Osteosarcoma, Chondrosarcoma and Ewing Sarcoma: Clinical Aspects, Biomarker Discovery and Liquid Biopsy. Crit. Rev. Oncol./Hematol. 2021, 162, 103340. [Google Scholar] [CrossRef] [PubMed]
- Zöllner, S.K.; Amatruda, J.F.; Bauer, S.; Collaud, S.; de Álava, E.; Dubois, S.G.; Hardes, J.; Hartmann, W.; Kovar, H.; Metzler, M.; et al. Ewing Sarcoma—Diagnosis, Treatment, Clinical Challenges and Future Perspectives. J. Clin. Med. 2021, 10, 1685. [Google Scholar] [CrossRef]
- Grünewald, T.G.P.; Bernard, V.; Gilardi-Hebenstreit, P.; Raynal, V.; Surdez, D.; Aynaud, M.M.; Mirabeau, O.; Cidre-Aranaz, F.; Tirode, F.; Zaidi, S.; et al. Chimeric EWSR1-FLI1 Regulates the Ewing Sarcoma Susceptibility Gene EGR2 via a GGAA Microsatellite. Nat. Genet. 2015, 47, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Gaspar, N.; Hawkins, D.S.; Dirksen, U.; Lewis, I.J.; Ferrari, S.; le Deley, M.C.; Kovar, H.; Grimer, R.; Whelan, J.; Claude, L.; et al. Ewing Sarcoma: Current Management and Future Approaches Through Collaboration. J. Clin. Oncol. 2015, 33, 3036–3046. [Google Scholar] [CrossRef] [PubMed]
- Clarke, B. Normal Bone Anatomy and Physiology. Clin. J. Am. Soc. Nephrol. 2008, 3, S131–S139. [Google Scholar] [CrossRef] [PubMed]
- Corre, I.; Verrecchia, F.; Crenn, V.; Redini, F.; Trichet, V. The Osteosarcoma Microenvironment: A Complex but Targetable Ecosystem. Cells 2020, 9, 976. [Google Scholar] [CrossRef]
- Prevarskaya, N.; Skryma, R.; Shuba, Y. Ion Channels in Cancer: Are Cancer Hallmarks Oncochannelopathies? Physiol. Rev. 2018, 98, 559–621. [Google Scholar] [CrossRef]
- Hoffmann, E.K.; Lambert, I.H.; Pedersen, S.F. Physiology of Cell Volume Regulation in Vertebrates. Physiol. Rev. 2009, 89, 193–277. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A Tool to Design Target-Specific Primers for Polymerase Chain Reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Ohmura, S.; Marchetto, A.; Orth, M.F.; Li, J.; Jabar, S.; Ranft, A.; Vinca, E.; Ceranski, K.; Carreño-Gonzalez, M.J.; Romero-Pérez, L.; et al. Translational Evidence for RRM2 as a Prognostic Biomarker and Therapeutic Target in Ewing Sarcoma. Mol. Cancer 2021, 20, 97. [Google Scholar] [CrossRef] [PubMed]
- Steinestel, K.; Trautmann, M.; Jansen, E.P.; Dirksen, U.; Rehkämper, J.; Mikesch, J.H.; Gerke, J.S.; Orth, M.F.; Sannino, G.; Arteaga, M.F.; et al. Focal Adhesion Kinase Confers Pro-Migratory and Antiapoptotic Properties and Is a Potential Therapeutic Target in Ewing Sarcoma. Mol. Oncol. 2020, 14, 248–260. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon Provides Fast and Bias-Aware Quantification of Transcript Expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential Analyses for RNA-Seq: Transcript-Level Estimates Improve Gene-Level Inferences. F1000Res 2016, 4, 1521. [Google Scholar] [CrossRef] [PubMed]
- Lun, A.T.L.; Chen, Y.; Smyth, G.K. It’s DE-Licious: A Recipe for Differential Expression Analyses of RNA-Seq Experiments Using Quasi-Likelihood Methods in EdgeR. Methods Mol. Biol. 2016, 1418, 391–416. [Google Scholar] [CrossRef] [PubMed]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Morgan, M.A.J.; Rickels, R.A.; Collings, C.K.; He, X.; Cao, K.; Herz, H.M.; Cozzolino, K.A.; Abshiru, N.A.; Marshall, S.A.; Rendleman, E.J.; et al. A Cryptic Tudor Domain Links BRWD2/PHIP to COMPASS-Mediated Histone H3K4 Methylation. Genes. Dev. 2017, 31, 2003–2014. [Google Scholar] [CrossRef]
- Bushnell, B. BBMap. Available online: Sourceforge.net/projects/bbmap/ (accessed on 21 April 2022).
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nussbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-Based Analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A Flexible Suite of Utilities for Comparing Genomic Features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Amemiya, H.M.; Kundaje, A.; Boyle, A.P. The ENCODE Blacklist: Identification of Problematic Regions of the Genome. Sci. Rep. 2019, 9, 9354. [Google Scholar] [CrossRef]
- Benson, G. Tandem Repeats Finder: A Program to Analyze DNA Sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Orth, M.F.; Surdez, D.; Marchetto, A.; Grossetête, S.; Gerke, J.S.; Zaidi, S.; Alonso, J.; Sastre, A.; Baulande, S.; Sill, M.; et al. Systematic Multi-Omics Cell Line Profiling Uncovers Principles of Ewing Sarcoma Fusion Oncogene-Mediated Gene Regulation. bioRxiv 2021. [Google Scholar] [CrossRef]
- Kurtenbach, S.; Harbour, J.W. SparK: A Publication-Quality NGS Visualization Tool. bioRxiv 2019. [Google Scholar] [CrossRef]
- Grynkiewicz, G.; Poenie, M.; Tsien, R.Y. A New Generation of Ca2+ Indicators with Greatly Improved Fluorescence Properties. J. Biol. Chem. 1985, 260, 3440–3450. [Google Scholar] [CrossRef]
- Kuntze, A.; Goetsch, O.; Fels, B.; Najder, K.; Unger, A.; Wilhelmi, M.; Sargin, S.; Schimmelpfennig, S.; Neumann, I.; Schwab, A.; et al. Protonation of Piezo1 Impairs Cell-Matrix Interactions of Pancreatic Stellate Cells. Front. Physiol. 2020, 11, 89. [Google Scholar] [CrossRef]
- Dieterich, P.; Klages, R.; Preuss, R.; Schwab, A. Anomalous Dynamics of Cell Migration. Proc. Natl. Acad. Sci. USA 2008, 105, 459–463. [Google Scholar] [CrossRef]
- Orellana, E.; Kasinski, A. Sulforhodamine B (SRB) Assay in Cell Culture to Investigate Cell Proliferation. Bio Protoc. 2016, 6, e1984. [Google Scholar] [CrossRef]
- Cadart, C.; Zlotek-Zlotkiewicz, E.; Venkova, L.; Thouvenin, O.; Racine, V.; le Berre, M.; Monnier, S.; Piel, M. Fluorescence EXclusion Measurement of Volume in Live Cells. Methods Cell Biol. 2017, 139, 103–120. [Google Scholar] [CrossRef] [PubMed]
- Hell, S.; Reiner, G.; Cremer, C.; Stelzer, E.H.K. Aberrations in Confocal Fluorescence Microscopy Induced by Mismatches in Refractive Index. J. Microsc. 1993, 169, 391–405. [Google Scholar] [CrossRef]
- Town, J.; Pais, H.; Harrison, S.; Stead, L.F.; Bataille, C.; Bunjobpol, W.; Zhang, J.; Rabbitts, T.H.; Witte, O.N. Exploring the Surfaceome of Ewing Sarcoma Identifies a New and Unique Therapeutic Target. Proc. Natl. Acad. Sci. USA 2016, 113, 3603–3608. [Google Scholar] [CrossRef] [PubMed]
- Bilke, S.; Schwentner, R.; Yang, F.; Kauer, M.; Jug, G.; Walker, R.L.; Davis, S.; Zhu, Y.J.; Pineda, M.; Meltzer, P.S.; et al. Oncogenic ETS Fusions Deregulate E2F3 Target Genes in Ewing Sarcoma and Prostate Cancer. Genome Res. 2013, 23, 1797–1809. [Google Scholar] [CrossRef]
- Zöllner, S.K.; Selvanathan, S.P.; Graham, G.T.; Commins, R.M.T.; Hong, S.H.; Moseley, E.; Parks, S.; Haladyna, J.N.; Erkizan, H.V.; Dirksen, U.; et al. Inhibition of the Oncogenic Fusion Protein EWS-FLI1 Causes G 2-M Cell Cycle Arrest and Enhanced Vincristine Sensitivity in Ewing’s Sarcoma. Sci. Signal 2017, 10, eaam8429. [Google Scholar] [CrossRef] [PubMed]
- Riggi, N.; Knoechel, B.; Gillespie, S.M.; Rheinbay, E.; Boulay, G.; Suvà, M.L.; Rossetti, N.E.; Boonseng, W.E.; Oksuz, O.; Cook, E.B.; et al. EWS-FLI1 Utilizes Divergent Chromatin Remodeling Mechanisms to Directly Activate or Repress Enhancer Elements in Ewing Sarcoma. Cancer Cell 2014, 26, 668–681. [Google Scholar] [CrossRef]
- Boone, M.A.; Taslim, C.; Crow, J.C.; Selich-Anderson, J.; Byrum, A.K.; Showpnil, I.A.; Sunkel, B.D.; Wang, M.; Stanton, B.Z.; Theisen, E.R.; et al. The FLI Portion of EWS/FLI Contributes a Transcriptional Regulatory Function That Is Distinct and Separable from Its DNA-Binding Function in Ewing Sarcoma. Oncogene 2021, 40, 4759–4769. [Google Scholar] [CrossRef]
- Boulay, G.; Sandoval, G.J.; Riggi, N.; Iyer, S.; Buisson, R.; Naigles, B.; Awad, M.E.; Rengarajan, S.; Volorio, A.; McBride, M.J.; et al. Cancer-Specific Retargeting of BAF Complexes by a Prion-like Domain. Cell 2017, 171, 163–178.e19. [Google Scholar] [CrossRef]
- Paulussen, M.; Craft, A.W.; Lewis, I.; Hackshaw, A.; Douglas, C.; Dunst, J.; Schuck, A.; Winkelmann, W.; Köhler, G.; Poremba, C.; et al. Results of the EICESS-92 Study: Two Randomized Trials of Ewing’s Sarcoma Treatment—Cyclophosphamide Compared with Ifosfamide in Standard-Risk Patients and Assessment of Benefit of Etoposide Added to Standard Treatment in High-Risk Patients. J. Clin. Oncol. 2008, 26, 4385–4393. [Google Scholar] [CrossRef]
- Ladenstein, R.; Pötschger, U.; Le Deley, M.C.; Whelan, J.; Paulussen, M.; Oberlin, O.; Van Den Berg, H.; Dirksen, U.; Hjorth, L.; Michon, J.; et al. Primary Disseminated Multifocal Ewing Sarcoma: Results of the Euro-EWING 99 Trial. J. Clin. Oncol. 2010, 28, 3284–3291. [Google Scholar] [CrossRef]
- Dirksen, U.; Brennan, B.; Le Deley, M.C.; Cozic, N.; Van Den Berg, H.; Bhadri, V.; Brichard, B.; Claude, L.; Craft, A.; Amler, S.; et al. High-Dose Chemotherapy Compared With Standard Chemotherapy and Lung Radiation in Ewing Sarcoma with Pulmonary Metastases: Results of the European Ewing Tumour Working Initiative of National Groups, 99 Trial and EWING 2008. J. Clin. Oncol. 2019, 37, 3192–3202. [Google Scholar] [CrossRef]
- Pedarzani, P.; Stocker, M. Molecular and Cellular Basis of Small- and Intermediate-Conductance, Calcium-Activated Potassium Channel Function in the Brain. Cell. Mol. Life Sci. 2008, 65, 3196. [Google Scholar] [CrossRef] [PubMed]
- Krabbendam, I.E.; Honrath, B.; Culmsee, C.; Dolga, A.M. Mitochondrial Ca2+-Activated K+ Channels and Their Role in Cell Life and Death Pathways. Cell Calcium 2018, 69, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, D.P.; Strøbæk, D.; Hougaard, C.; Jensen, M.L.; Hummel, R.; Sørensen, U.S.; Christophersen, P.; Wulff, H. Negative Gating Modulation by (R)-N-(Benzimidazol-2-Yl)-1,2,3,4-Tetrahydro-1-Naphthylamine (NS8593) Depends on Residues in the Inner Pore Vestibule: Pharmacological Evidence of Deep-Pore Gating of KCa2 Channels. Mol. Pharmacol. 2011, 79, 899–909. [Google Scholar] [CrossRef] [PubMed]
- Altamura, C.; Gavazzo, P.; Pusch, M.; Desaphy, J.F. Ion Channel Involvement in Tumor Drug Resistance. J. Pers. Med. 2022, 12, 210. [Google Scholar] [CrossRef]
- Glaser, F.; Hundehege, P.; Bulk, E.; Todesca, L.M.; Schimmelpfennig, S.; Nass, E.; Budde, T.; Meuth, S.G.; Schwab, A. KCa Channel Blockers Increase Effectiveness of the EGF Receptor TK Inhibitor Erlotinib in Non-Small Cell Lung Cancer Cells (A549). Sci. Rep. 2021, 11, 18330. [Google Scholar] [CrossRef] [PubMed]
- Higham, J.; Sahu, G.; Wazen, R.M.; Colarusso, P.; Gregorie, A.; Harvey, B.S.J.; Goudswaard, L.; Varley, G.; Sheppard, D.N.; Turner, R.W.; et al. Preferred Formation of Heteromeric Channels between Coexpressed SK1 and IKCa Channel Subunits Provides a Unique Pharmacological Profile of Ca2+-Activated Potassium Channels. Mol. Pharmacol. 2019, 96, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Ryland, K.E.; Hawkins, A.G.; Weisenberger, D.J.; Punj, V.; Borinstein, S.C.; Laird, P.W.; Martens, J.R.; Lawlor, E.R. Promoter Methylation Analysis Reveals That KCNA5 Ion Channel Silencing Supports Ewing Sarcoma Cell Proliferation. Mol. Cancer Res. 2016, 14, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Bulk, E.; Ay, A.S.; Hammadi, M.; Ouadid-Ahidouch, H.; Schelhaas, S.; Hascher, A.; Rohde, C.; Thoennissen, N.H.; Wiewrodt, R.; Schmidt, E.; et al. Epigenetic Dysregulation of KCa3.1 Channels Induces Poor Prognosis in Lung Cancer. Int. J. Cancer 2015, 137, 1306–1317. [Google Scholar] [CrossRef] [PubMed]
- Lamy, C.; Goodchild, S.J.; Weatherall, K.L.; Jane, D.E.; Liégeois, J.F.; Seutin, V.; Marrion, N.V. Allosteric Block of KCa2 Channels by Apamin. J. Biol. Chem. 2010, 285, 27067–27077. [Google Scholar] [CrossRef]
- Dolga, A.M.; Netter, M.F.; Perocchi, F.; Doti, N.; Meissner, L.; Tobaben, S.; Grohm, J.; Zischka, H.; Plesnila, N.; Decher, N.; et al. Mitochondrial Small Conductance SK2 Channels Prevent Glutamate-Induced Oxytosis and Mitochondrial Dysfunction. J. Biol. Chem. 2013, 288, 10792–10804. [Google Scholar] [CrossRef]
- Yang, M.Y.; Camara, A.K.S.; Aldakkak, M.; Kwok, W.M.; Stowe, D.F. Identity and Function of a Cardiac Mitochondrial Small Conductance Ca2+-Activated K+ Channel Splice Variant. Biochim. Biophys. Acta Bioenerg. 2017, 1858, 442–458. [Google Scholar] [CrossRef]
- Weaver, A.K.; Bomben, V.C.; Sontheimer, H. Expression and Function of Calcium-Activated Potassium Channels in Human Glioma Cells. Glia 2006, 54, 223–233. [Google Scholar] [CrossRef]
- Mohr, C.J.; Steudel, F.A.; Gross, D.; Ruth, P.; Lo, W.Y.; Hoppe, R.; Schroth, W.; Brauch, H.; Huber, S.M.; Lukowski, R. Cancer-Associated Intermediate Conductance Ca2+-Activated K+ Channel KCa3.1. Cancers 2019, 11, 109. [Google Scholar] [CrossRef]
- Wen, J.; Rusch, M.; Brady, S.W.; Shao, Y.; Edmonson, M.N.; Shaw, T.I.; Powers, B.B.; Tian, L.; Easton, J.; Mullighan, C.G.; et al. The Landscape of Coding RNA Editing Events in Pediatric Cancer. BMC Cancer 2021, 21, 1233. [Google Scholar] [CrossRef]
- Zhou, Y.; Xia, C.; Yin, M.; Wang, X.; Wu, H.; Ji, Y. Distribution and Functional Characteristics of Voltage-Gated Sodium Channels in Immature Cochlear Hair Cells. Neurosci. Bull. 2020, 36, 49–65. [Google Scholar] [CrossRef]
- Wu, D.; Zang, Y.Y.; Shi, Y.Y.; Ye, C.; Cai, W.M.; Tang, X.H.; Zhao, L.; Liu, Y.; Gan, Z.; Chen, G.Q.; et al. Distant Coupling between RNA Editing and Alternative Splicing of the Osmosensitive Cation Channel Tmem63b. J. Biol. Chem. 2020, 295, 18199–18212. [Google Scholar] [CrossRef]
- Kuzmenkov, A.I.; Nekrasova, O.V.; Kudryashova, K.S.; Peigneur, S.; Tytgat, J.; Stepanov, A.V.; Kirpichnikov, M.P.; Grishin, E.V.; Feofanov, A.V.; Vassilevski, A.A. Fluorescent Protein-Scorpion Toxin Chimera Is a Convenient Molecular Tool for Studies of Potassium Channels. Sci. Rep. 2016, 6, 33314. [Google Scholar] [CrossRef]
- Tomita, H.; Shakkottai, V.G.; Gutman, G.A.; Sun, G.; Bunney, W.E.; Cahalan, M.D.; Chandy, K.G.; Gargus, J.J. Novel Truncated Isoform of SK3 Potassium Channel Is a Potent Dominant-Negative Regulator of SK Currents: Implications in Schizophrenia. Mol. Psychiatry 2003, 8, 524–535. [Google Scholar] [CrossRef]
- Zhou, S.J.; Zhang, E.L.; Liang, B.Y.; Zhang, Z.Y.; Chen, X.P.; Huang, Z.Y. Distilled Water Lavage During Surgery Improves Long-Term Outcomes of Patients with Ruptured Hepatocellular Carcinoma. J. Gastrointest. Surg. 2015, 19, 1262–1270. [Google Scholar] [CrossRef]
- Shiozaki, A.; Ichikawa, D.; Kosuga, T.; Marunaka, Y.; Otsuji, E. Regulation of Osmolality for Cancer Treatment. J. Physiol. Sci. 2017, 67, 353–360. [Google Scholar] [CrossRef]
- Zhou, Y.; Mokhtari, R.B.; Pan, J.; Cutz, E.; Yeger, H. Carbonic Anhydrase II Mediates Malignant Behavior of Pulmonary Neuroendocrine Tumors. Am. J. Respir. Cell Mol. Biol. 2015, 52, 183–192. [Google Scholar] [CrossRef]
- Weber, R.; Bertoni, A.P.S.; Bessestil, L.W.; Brasil, B.M.D.A.A.; Brum, L.S.; Furlanetto, T.W. Validation of Reference Genes for Normalization Gene Expression in Reverse Transcription Quantitative PCR in Human Normal Thyroid and Goiter Tissue. BioMed Res. Int. 2014, 2014, 198582. [Google Scholar] [CrossRef]
- Pigozzi, D.; Ducret, T.; Tajeddine, N.; Gala, J.L.; Tombal, B.; Gailly, P. Calcium Store Contents Control the Expression of TRPC1, TRPC3 and TRPV6 Proteins in LNCaP Prostate Cancer Cell Line. Cell Calcium. 2006, 39, 401–415. [Google Scholar] [CrossRef]
- Zheng, D.H.; Guo, W.; Sun, F.J.; Xu, G.Z.; Zang, Z.L.; Shu, H.F.; Yang, H. Expression of TRPC6 and BDNF in Cortical Lesions From Patients With Focal Cortical Dysplasia. J. Neuropathol. Exp. Neurol. 2016, 75, 718. [Google Scholar] [CrossRef]
- Chen, S.J.; Hoffman, N.E.; Shanmughapriy, S.; Bao, L.; Keefer, K.; Conrad, K.; Merali, S.; Takahashi, Y.; Abraham, T.; Hirschler-Laszkiewicz, I.; et al. A Splice Variant of the Human Ion Channel TRPM2 Modulates Neuroblastoma Tumor Growth through Hypoxia-Inducible Factor (HIF)-1/2α. J. Biol. Chem. 2014, 289, 36284–36302. [Google Scholar] [CrossRef]
- O’Grady, S.; Morgan, M.P. Deposition of Calcium in an in Vitro Model of Human Breast Tumour Calcification Reveals Functional Role for ALP Activity, Altered Expression of Osteogenic Genes and Dysregulation of the TRPM7 Ion Channel. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Sugimoto, A.; Miyazaki, A.; Kawarabayashi, K.; Shono, M.; Akazawa, Y.; Hasegawa, T.; Ueda-Yamaguchi, K.; Kitamura, T.; Yoshizaki, K.; Fukumoto, S.; et al. Piezo Type Mechanosensitive Ion Channel Component 1 Functions as a Regulator of the Cell Fate Determination of Mesenchymal Stem Cells. Sci. Rep. 2017, 7, 17696. [Google Scholar] [CrossRef]
- Hu, S.; Li, L.; Huang, W.; Liu, J.; Lan, G.; Yu, S.; Peng, L.; Xie, X.; Yang, L.; Nian, Y.; et al. CAV3.1 Knockdown Suppresses Cell Proliferation, Migration and Invasion of Prostate Cancer Cells by Inhibiting AKT. Cancer Manag. Res. 2018, 10, 4603. [Google Scholar] [CrossRef]
- Daniil, G.; Fernandes-Rosa, F.L.; Chemin, J.; Blesneac, I.; Beltrand, J.; Polak, M.; Jeunemaitre, X.; Boulkroun, S.; Amar, L.; Strom, T.M.; et al. CACNA1H Mutations Are Associated With Different Forms of Primary Aldosteronism. EBioMedicine 2016, 13, 225. [Google Scholar] [CrossRef]
- Chin-Smith, E.C.; Slater, D.M.; Johnson, M.R.; Tribe, R.M. STIM and Orai Isoform Expression in Pregnant Human Myometrium: A Potential Role in Calcium Signaling during Pregnancy. Front. Physiol. 2014, 5, 169. [Google Scholar] [CrossRef][Green Version]
- Elzi, D.J.; Song, M.; Houghton, P.J.; Chen, Y.; Shiio, Y. The Role of FLI-1-EWS, a Fusion Gene Reciprocal to EWS-FLI-1, in Ewing Sarcoma. Genes Cancer 2015, 6, 452–461. [Google Scholar] [CrossRef]

| Cell Line | Diagnosis | Tissue of Origin | EWSR1-Rearrengement | RRID |
|---|---|---|---|---|
| SK-ES-1 | Ewing Sarcoma | Bone | EWS-FLI1 | CVCL_0627 |
| TC-71 | Ewing Sarcoma | Bone, Humerus | EWS-FLI1 | CVCL_2213 |
| A673 | Ewing Sarcoma | Muscle | EWS-FLI1 | CVCL_0080 |
| TTC-466 | Ewing Sarcoma | Lung metastasis | EWS-ERG | CVCL_A444 |
| SAOS | Osteosarcoma | Bone | none | CVCL_0548 |
| HOS | Osteosarcoma | Bone | none | CVCL_0312 |
| ZK-58 | Osteosarcoma | Bone | none | CVCL_9920 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fuest, S.; Post, C.; Balbach, S.T.; Jabar, S.; Neumann, I.; Schimmelpfennig, S.; Sargin, S.; Nass, E.; Budde, T.; Kailayangiri, S.; et al. Relevance of Abnormal KCNN1 Expression and Osmotic Hypersensitivity in Ewing Sarcoma. Cancers 2022, 14, 4819. https://doi.org/10.3390/cancers14194819
Fuest S, Post C, Balbach ST, Jabar S, Neumann I, Schimmelpfennig S, Sargin S, Nass E, Budde T, Kailayangiri S, et al. Relevance of Abnormal KCNN1 Expression and Osmotic Hypersensitivity in Ewing Sarcoma. Cancers. 2022; 14(19):4819. https://doi.org/10.3390/cancers14194819
Chicago/Turabian StyleFuest, Sebastian, Christoph Post, Sebastian T. Balbach, Susanne Jabar, Ilka Neumann, Sandra Schimmelpfennig, Sarah Sargin, Elke Nass, Thomas Budde, Sareetha Kailayangiri, and et al. 2022. "Relevance of Abnormal KCNN1 Expression and Osmotic Hypersensitivity in Ewing Sarcoma" Cancers 14, no. 19: 4819. https://doi.org/10.3390/cancers14194819
APA StyleFuest, S., Post, C., Balbach, S. T., Jabar, S., Neumann, I., Schimmelpfennig, S., Sargin, S., Nass, E., Budde, T., Kailayangiri, S., Altvater, B., Ranft, A., Hartmann, W., Dirksen, U., Rössig, C., Schwab, A., & Pethő, Z. (2022). Relevance of Abnormal KCNN1 Expression and Osmotic Hypersensitivity in Ewing Sarcoma. Cancers, 14(19), 4819. https://doi.org/10.3390/cancers14194819








