Simple Summary
NRF2-activated or NRF2-addicted cancers show high incidence, especially in esophageal squamous cell carcinoma (ESCC). ESCC with high NRF2 expression is largely resistant to the current major treatments for ESCC and therefore shows a very poor prognosis. In order to develop effective treatments for NRF2-addicted esophageal cancers, the elucidation and understanding of the mechanistic basis of NRF2 function in NRF2-addicted cancer cells are critically important. This review summarizes the current knowledge of the KEAP1-NRF2 system and proposes three distinct approaches for the treatment of NRF2-addicted ESCC.
Abstract
NRF2 (nuclear factor erythroid 2-related factor 2) is a transcription factor that regulates the expression of many cytoprotective genes. NRF2 activation is mainly regulated by KEAP1 (kelch-like ECH-associated protein 1) through ubiquitination and proteasome degradation. Esophageal cancer is classified histologically into two major types: esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC). ESCC harbors more genetic alterations in the KEAP-NRF2 system than EAC does, which results in NRF2 activation in these cancers. NRF2-addicted ESCC exhibits increased malignancy and acquisition of resistance to chemoradiotherapy. Therefore, it has been recognized that the development of drugs targeting the KEAP1-NRF2 system based on the molecular dissection of NRF2 function is important and urgent for the treatment of ESCC, along with efficient clinical screening for NRF2-addicted ESCC patients. Recently, the fate of NRF2-activated cells in esophageal tissues, which was under the influence of strong cell competition, and its relationship to the pathogenesis of ESCC, was clarified. In this review, we will summarize the current knowledge of the KEAP1-NRF2 system and the treatment of ESCC. We propose three main strategies for the treatment of NRF2-addicted cancer: (1) NRF2 inhibitors, (2) synthetic lethal drugs for NRF2-addicted cancers, and (3) NRF2 inducers of the host defense system.
1. Regulatory Mechanisms of NRF2 Activation
Nuclear factor erythroid 2-related factor 2 (NRF2 or NFE2L2) is a master transcription factor and regulates the expression of many cytoprotective genes [1]. The NRF2 protein level is regulated by proteasomal degradation through two E3 ubiquitin ligase complexes, kelch-like ECH-associated protein 1 (KEAP1)-Cullin 3 (CUL3) [2,3,4] and β-transducin repeat containing protein ligase (βTrCP)-Cullin 1 (CUL1) (Figure 1) [5,6,7]. In unstressed conditions, the NRF2 protein is maintained at a low level through these regulations utilizing protein degradation [3,8]. NRF2 activity can be induced in response to various stimuli, and the induction is predominantly exploited by the derepression mechanisms of protein degradation regulation processes [3]. Stresses originating from reactive oxygen species (ROS) and toxic chemicals (mostly electrophiles) are sensed by KEAP1. Under unstressed conditions, the KEAP1 homodimer binds NRF2 at two sites within the Neh2 domain, i.e., the DLG and ETGE motifs. Subsequently, the KEAP1-CUL3 ubiquitin E3 ligase complex ubiquitinates NRF2 quite efficiently. This process has been shown to occur in the cytoplasm [9], and ubiquitinated NRF2 is rapidly degraded through the 26S proteasome system. Upon exposure to oxidative or electrophilic stresses, specific reactive cysteine residues within KEAP1 are modified, leading to the attenuating of the ubiquitin ligase activity of the KEAP1-CUL3 complex and the ubiquitination of NRF2. Thus, NRF2 is stabilized, translocates into the nucleus and activates a set of cytoprotective genes [10]. The KEAP1-NRF2 system has been established as a major regulatory machine engaged in the response against oxidative and electrophilic stresses.
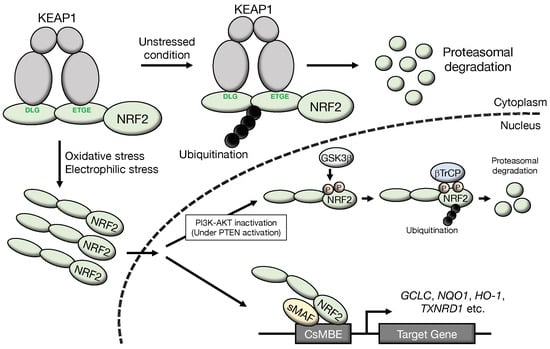
Figure 1.
Molecular mechanisms of NRF2 activation: NRF2 degradation is accelerated through ubiquitination of NRF2 by the KEAP1-CUL3 complex and βTrCP-CUL1 complex. Under unstressed conditions, the KEAP1 homodimer binds NRF2 at two sites within the Neh2 domain, i.e., the DLG and ETGE motifs, and NRF2 is ubiquitinated efficiently. Because NRF2 is rapidly degraded by the proteasome system, the NRF2 protein is maintained at a very low level in unstressed conditions. Upon exposure to oxidative or electrophilic stresses, specific reactive cysteine residues within KEAP1 are modified. This modification attenuates the ubiquitin ligase activity of KEAP1. Thus, NRF2 is stabilized, translocates into the nucleus, and activates a set of cytoprotective genes. On the other hand, the βTrCP-CUL1 complex also ubiquitinates NRF2. To be recognized by βTrCP, NRF2 is phosphorylated by GSK3β at two specific serine residues in the Neh6 domain. Phosphorylated NRF2 is ubiquitinated by the βTrCP-CUL1 ubiquitin E3 ligase complex and degraded through the proteasome system. A cascade of signal transduction has been clarified in which GSK3β is inhibited/phosphorylated by the activated PI3K-AKT pathway, which is under the negative regulation of PTEN. CsMBE, CNC-sMAF binding element.
The βTrCP-CUL1 complex has also been identified to regulate cellular NRF2 levels [5,6]. This complex mainly operates within the nucleus. To be recognized by βTrCP, NRF2 needs to be phosphorylated by glycogen synthase kinase 3β (GSK3β) [5] at two specific serine residues in the Neh6 domain. Phosphorylated NRF2 is ubiquitinated and degraded through the proteasome system. A cascade of signal transduction has been clarified in which GSK3β is inhibited/phosphorylated by the activated phosphatidylinositol 3-kinase (PI3K)-AKT serine/threonine kinase (AKT) pathway, and PI3K/AKT activity is under the negative regulation of phosphatase and tensin homolog (PTEN).
The relative contributions of the KEAP1 pathway and βTrCP pathway have been elucidated. NRF2 does not accumulate in Pten-deleted mouse liver [11], indicating that βTrCP inactivation alone is insufficient to significantly activate NRF2. Analyses of NRF2 accumulation in Pten::Keap1 double knockout mouse livers suggest that regulation by the KEAP1 pathway is predominant, while regulation by the βTrCP pathway is auxiliary and supportive in the regulation of NRF2. This model is nicely supported by an analysis of the newly established Nrf2SA mutant mouse [12], which harbors replacements of two Ser residues at residues 335 and 338 by Ala. While the Nrf2SA/SA mutation does not give rise to strong NRF2 activation in mouse tissues in vivo, simultaneous Keap1 deletion markedly induces NRF2 activity. Severe growth retardation is provoked by the abnormal hyperkeratinization of the esophagus and forestomach.
In addition, the WD40 repeat-containing protein 23 (WDR23) is reported to directly bind the Neh2 domain of NRF2 and ubiquitinate NRF2 independently of KEAP1 by forming a complex with damage-specific DNA binding protein 1 (DDB1) and Cullin 4 (CUL4) [13]. This observation suggests that the WDR23-DDB1-CUL4 complex serves as the regulator of NRF2. Intriguingly, WDR23 was originally known to regulate basic-leucine zipper (b-ZIP) domain-containing protein skn-1 (skinhead-1 protein), which is the C. elegans ortholog of NRF2. The contribution of WDR23 to NRF2 regulation in human tissues needs to be clarified with further studies.
NRF2 forms a heterodimer with one of the three small musculoaponeurotic fibrosarcoma proteins (sMAF), i.e., MAFF, MAFG and MAFK [14,15]. The NRF2-sMAF heterodimer binds to the CNC-sMAF binding element (CsMBE), which is also known as the antioxidant responsive element (ARE) [16,17] or electrophile responsive element (EpRE) [18], in the regulatory regions of the target genes, such as glutamate-cysteine ligase catalytic subunit (GCLC), heme oxygenase-1 (HO-1 or HMOX1), NAD(P)H quinone dehydrogenase 1 (NQO1), and thioredoxin reductase 1 (TXNRD1) [17]. In this way, upon exposure to oxidative or electrophilic stresses, the NRF2 protein is highly induced and exerts key regulatory influences over the cytoprotective function of the cells.
2. The Double-Edged Sword of NRF2 Activation in Cancer Cells
It is known that NRF2 protects the body from oxidative and electrophilic stresses. In this context, a breakthrough in the NRF2 field occurred in 2006. Two laboratories independently discovered somatic mutations in the KEAP1 gene in lung cancer cells that disrupt the protein–protein interaction of KEAP1-NRF2, leading to the constitutive activation of NRF2 [19,20]. Soon after this discovery, somatic mutations in the NRF2 gene that led to the constitutive activation of NRF2 were identified [21]. Since then, the constitutive activation of NRF2 has been found at a very high incidence in many cancers derived from various tissues, driven not only by somatic mutations but also by various other mechanisms (see below) [22,23,24]. We refer to these cancer cells with high NRF2 activity levels as NRF2-activated cancers or NRF2-addicted cancers [25]. Owing to the cytoprotective and metabolic reprogramming functions of NRF2, NRF2-addicted cancers acquire a marked advantage in proliferation and resilience against chemoradiotherapy, resulting in a poor prognosis [21,26]. Therefore, NRF2 activation is often referred to as a ‘double-edged sword’ or by similar phrases [27,28,29]. As described in Section 9.3, the strategy targeting NRF2 activation has been proposed for the NRF2-addicted cancer treatments [30].
From a mechanistic viewpoint, NRF2 activation in cancer cells is mainly elicited by the dysfunction of protein–protein interactions in the NRF2-KEAP1-CUL3 ubiquitin E3 ligase complex. Somatic mutations in the NRF2, KEAP1 or CUL3 genes have been well characterized [31]. Intriguingly, mutations in NRF2 accumulate in two KEAP1-interacting motifs, i.e., the DLG and ETGE motifs within the N-terminal Neh2 domain [21,25,32,33]. The extended DLG (DLGex) motif spans amino acid positions 17 to 51, while the ETGE motif is located at positions 77 to 82 [34]. As described above, these somatic mutations are gain-of-function mutations of NRF2.
In addition to somatic mutations, several other causes of NRF2 activation have been identified. Exon skipping of NRF2 exon 2 and/or exon 3 has been reported in lung cancer and head and neck cancers [35]. The removal of NRF2 exon 2 abolishes its ability to bind to KEAP1 and activates NRF2. Epigenetic modification, i.e., hypermethylation in the promoter region of the KEAP1 gene, has also been identified to constitutively activate NRF2 in lung and renal cancers [36,37]. The modification of KEAP1 cysteine residues by oncometabolites such as fumarate and itaconate has also been reported to activate NRF2 [38,39,40]. All of these factors lead to constitutive NRF2 activation and contribute to growth, malignant transformation and chemoradiotherapy resistance.
An intriguing and unexpected mechanism is the disruption of the KEAP1-NRF2 interaction through the accumulation of the autophagy chaperone p62 [41]. p62 is encoded by the sequestosome 1 (SQSTM1) gene [42] and retains a phylogenetically conserved STGE motif within the KEAP1-interacting domain [41]. After the phosphorylation of serine residues in the motif, the pSTGE motif mimics the ETGE motif in NRF2, and its affinity for KEAP1 is comparable to that of ETGE and higher affinity than that of the DLGex motif [43]. p62 accumulation causes NRF2 activation by inhibiting the KEAP1-NRF2 interaction via a Hinge-and-Latch mechanism [10]. This mechanism has been verified using a mouse model with knockout of Atg7, an essential gene for autophagy [44]. Notably, it has been found that hepatocellular adenoma is provoked by p62-mediated NRF2 activation in liver-specific Atg7 knockout mice [45]. Consistent with the findings in the mouse model, in human hepatocellular carcinoma, p62- and KEAP1-positive aggregates are abundant, suggesting that successive NRF2 activation contributes to the development of hepatocellular carcinoma [45]. It has also been reported that in human esophageal squamous cell carcinoma (ESCC) patients, NRF2 activation and phospho-p62 accumulation are associated with radioresistance [46].
3. Functions of NRF2 in the Esophagus and Esophageal Cancers
NRF2 appears to be related to the differentiation [47], development [48], barrier function [49] and metabolism [50] of the esophagus. The comparison of NRF2 expression in various tissues revealed that the esophagus expresses NRF2 at the highest level (Human Protein Atlas; https://www.proteinatlas.org, accessed on 6 May 2022) [51]. In detail, NRF2 expression in the esophagus was 5.4-fold higher than that in the lowest-expressing tissue (pancreas) and 1.5-fold higher than that in the second-highest-expressing tissues (stomach and liver) (Figure 2a). In contrast, the KEAP1 level is extremely high in skeletal muscle compared with other human tissues (Figure 2b).
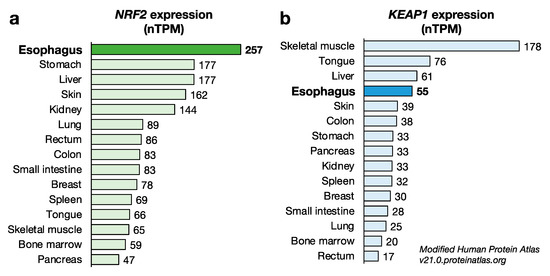
Figure 2.
Expression levels of NRF2 and KEAP1 protein-coding transcripts in various tissues. (a) Expression level of NRF2 protein-coding transcripts in various tissues. Of note, the NRF2 expression level in the esophagus was the highest among the tissues examined. (b) Expression level of KEAP1 protein-coding transcripts in various tissues. The KEAP1 expression level in the esophagus is comparable to that in the tongue and the liver. Data were obtained from Human Protein Atlas v21.0.proteinatlas.org. nTPM: normalized protein-coding transcripts per million.
The KEAP1 level in the esophagus is comparable to that in the tongue and liver. While NRF2 activation is meticulously regulated at the protein level, the high NRF2 mRNA level in the esophagus suggests that the esophageal epithelium has the ability to express high levels of NRF2 at the protein level. In fact, severe hyperkeratosis in the esophageal epithelium is observed as the most characteristic phenotype in Keap1 systemic knockout (Keap1–/–) mice [52].
Apart from its function in the normal esophageal epithelium, NRF2’s contribution to the development and progression of esophageal cancer has been a topic of particular interest in recent years. Esophageal cancer is one of the most aggressive tumors in humans and is the sixth leading cause of cancer-related death [53]. Esophageal cancer is classified histologically into esophageal adenocarcinoma (EAC) and ESCC, which show distinct epidemiological and genomic characteristics [54] (Figure 3).
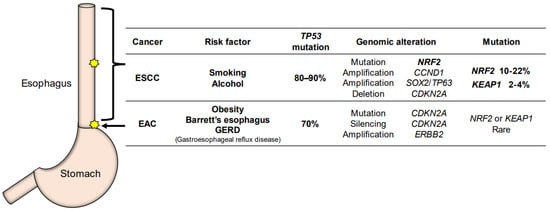
Figure 3.
The characteristic difference between ESCC (esophageal squamous cell carcinoma) and EAC (esophageal adenocarcinoma). Most cases of ESCC occur in the middle thoracic esophagus, and ESCC is highly associated with smoking and alcohol consumption. Somatic mutations of NRF2 are often found in ESCC. EAC occurs in the lower thoracic esophagus, down to the esophagogastric junction. EAC originates from Barrett’s esophagus and GERD (gastroesophageal reflux disease). Genomic alterations in the KEAP1-NRF2 system are rarely found in EAC.
EAC is predominant in North America and Europe [55]. Obesity increases the risk of Barrett’s esophagus and EAC due to the increase in intragastric pressure or hiatal hernia [56]. In Barrett’s esophagus caused by gastroesophageal reflux disease (GERD), normal squamous mucosa in the lower thoracic to abdominal esophagus is replaced with metaplastic columnar epithelium, a precancerous condition preceding EAC (Figure 3). In EAC, TP53 mutations are found at a high level, approximately 70% [57]. Genomic alterations in the KEAP1-NRF2 system are rarely found in EAC [57], but mutation/silencing of CDKN2A and amplification of ERBB2 and VEGFA are found frequently. EAC shows genomic characteristics similar to the most common chromosomal instability type of gastric adenocarcinoma [58].
In contrast, two studies reported that NRF2 is constitutively activated in EAC in the absence of somatic mutations in NRF2 or KEAP1 [59,60]. This finding is interesting if we consider the finding that NRF2 is activated in the esophageal epithelium with GERD, which suggests that NRF2 plays a cytoprotective function in GERD [49]. Thus, the contribution of NRF2 activation to EAC development is currently unclear. Further examinations utilizing sophisticated in vivo models of EAC are necessary for this purpose.
4. Frequently Activated NRF2 in ESCC
The incidence of ESCC is more prevalent in Asia, Africa, and South America than in Europe and North America [53]. Most cases of ESCC occur in the middle thoracic esophagus region, and ESCC is found to be highly associated with smoking and alcohol consumption [61] (Figure 3). The genomic characteristics of ESCC resemble those of squamous cell carcinoma of the lung and head-and-neck [54]. TP53 mutations are commonly found in esophageal cancers. In particular, TP53 mutations are found in more than 80% of ESCC cases, while TP53 mutations are found in approximately 70% of EAC cases.
NRF2 is one of the frequently mutated genes in ESCC, and 10–22% of ESCC cases retain somatic mutations in the NRF2 gene [21,62]. In addition to NRF2, somatic mutations are also found in MLL2, ZNF750, NOTCH1 and TGFBR2 [63,64,65,66,67,68]. Genomic amplifications of CCND1 and SOX2/TP63 and deletions of CDKN2A have also been reported in ESCC cases. For NRF2 activation, a variety of genetic alterations in the members of the KEAP1-NRF2 system or related molecules should also be considered. These abnormalities include somatic mutations of KEAP1, somatic mutations or deletions of CUL3, and deletions of ATG7, which occur in 2–4%, 1–5% and 6% of ESCC cases, respectively [54]. Furthermore, the activation of the PI3K-AKT pathway is frequently found in ESCC. For instance, PTEN, one of the members of this pathway, harbors somatic mutations and deletions in 2–5% and 3% of cases, respectively. Another member, PIK3CA, exhibits somatic mutations, amplifications and overexpression in 5–10%, 10–41%, and 50% of cases, respectively [63,64,66,69,70]. Activating mutations or abnormalities in the PI3K-AKT pathway in ESCC are particularly intriguing, as these contribute to the inactivation of the downstream GSK3β, and GSK3β is known to induce NRF2 activation [5].
Of the many intriguing observations that can be extracted from the TCGA Pan-Cancer Atlas using cBioPortal [71,72] (http://cancergenome.nih.gov/, accessed on 6 May 2022), one of the most surprising findings is that ESCC is the cancer with the highest frequency of genomic alterations, which include somatic mutations, amplifications and deletions (Figure 4). NRF2 somatic mutations were originally found in squamous cell carcinoma (SCC) in the lung [19], and it had previously been considered that lung SCC must be the cancer type with the highest prevalence of NRF2 somatic mutations.
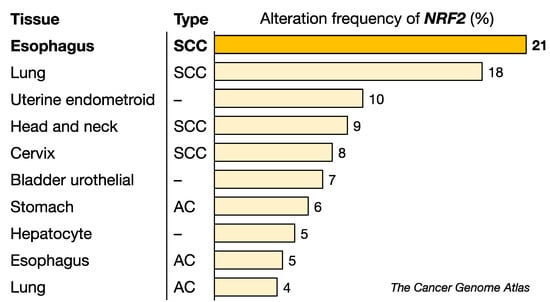
Figure 4.
Alteration frequency of the NRF2 gene in various cancers. ESCC has the highest alteration frequency among the cancers examined. NRF2 mutations appear to be more common in SCC than in AC. SCC, squamous cell carcinoma, AC, adenocarcinoma. Data were obtained from the TCGA dataset.
Based on comprehensive analyses of DNA methylation, mRNA expression, copy number variation and microRNA expression data by TCGA Research Network [54], ESCC is clustered into 3 subtypes: ESCC1, ESCC2 and ESCC3. ESCC1 is the major subtype (56%) and frequently harbors genomic alterations in the KEAP1-NRF2 system and related members. For instance, 30% of ESCC1 cases harbor genomic alterations in NRF2, 6% harbor alterations in KEAP1, and 10% harbor alterations of CUL3. ESCC2 and EAC retain, albeit at a low frequency, genomic alterations in the KEAP1-NRF2 system.
The frequency of NRF2-activated cancers may be much higher than that of cancers bearing gene alterations in the KEAP1-NRF2 system and related members. Additional mechanisms may lead to NRF2 activation in ESCC. Since NRF2 activation confers chemoradiotherapy resistance and poor prognosis to ESCC cases [21,26,73,74,75], it is important to clarify the contribution of NRF2 to cancer biology and medicine.
5. NRF2-Addicted ESCC Cell Lines
Two series of ESCC cell lines have been established in Japan. One is the TE series of ESCC cell lines. The first paper describing the TE cell line was published in 1979 by Nishihira et al. at Tohoku University [76]. The other ESCC cell line is the KYSE series, which was reported in 1992 by Shimada et al. at Kyoto University [77]. Other ESCC cell lines have also been established and used to study various aspects of ESCC. Somatic mutations eliciting NRF2 activation and their consequences have been studied utilizing these cell lines.
We evaluated ESCC cell lines for NRF2 expression levels and the presence of somatic mutations in related genes. The results are summarized in Table 1. ESCC cell lines can be classified into two groups based on the levels of NRF2 activation status, i.e., a high-level NRF2-expressing group and a normal-level NRF2-expressing group [21,73,78,79,80,81]. This classification can be seen in clustering analysis using RNA-sequencing (RNA-seq) data [82] as well as somatic mutations in the COSMIC database (Catalogue of Somatic Mutations in Cancers; https://cancer.sanger.ac.uk/cell_lines) [83]. In this search, the TE and KYSE series of cell lines were found to contain almost comparable numbers of NRF2 high-level and normal-level cell lines (Table 1). Because of the clinical observations summarized above, we expected that NRF2 high-level cell lines would be derived more from poorly differentiated ESCC than from highly differentiated ESCC. However, the NRF2 activity status appears not to be closely related to the cancer cell differentiation stage of the original ESCC (Table 1).

Table 1.
Classification of ESCC cell lines in NRF2 activation. n.d., not determined.
6. Esophageal Phenotype in Mouse Models with Genetic Modification of Nrf2 or Keap1
Studies on the physiological and pathological contributions of the KEAP1-NRF2 system in the esophagus have been developed using genetically modified mice (Table 2). Two lines of Nrf2 systemic knockout (Nrf2–/–) mice were generated in the 1990s [85,86] and have been utilized in various analyses. Nrf2–/– mice are sensitive to chemical carcinogenesis, as phase II detoxication enzymes are under NRF2 regulation, so their expression is downregulated and not induced in response to carcinogens [86]. In fact, Nrf2–/– mice develop many more tumors in the upper aerodigestive tract than wild-type mice upon the administration of a chemical carcinogen, 4-nitroquinoline-1-oxide (4NQO) [87].

Table 2.
Esophageal phenotypes of genetically modified mouse models targeting Nrf2 and Keap1.
We also generated Keap1 systemic knockout (Keap1–/–) mice that serve as an NRF2-activated genetic model by inserting the LacZ gene into Keap1 exon 2 (Figure 5a) [52]. Unexpectedly, Keap1–/– mice develop hyperkeratosis of the esophageal and forestomach epithelium and die before weaning because of the lack of food intake. The lethality and hyperkeratosis is nicely rescued by simultaneous systemic Nrf2 knockout (Keap1–/–::Nrf2–/–) [52] or keratinocyte-specific Nrf2 knockout (Keap1–/–::Nrf2flox/flox::Keratin5-Cre) [90], clearly demonstrating that esophageal hyperkeratosis is provoked by constitutive and massive NRF2 activation as a consequence of Keap1 deletion. Keap1–/– mice are rescued from juvenile lethality, and hyperkeratosis is largely avoided in Nrf2 heterozygous compound mutant mice (Keap1–/–::Nrf2+/–) [89], indicating that massive Nrf2 expression is required to provoke severe hyperkeratosis of the esophagus (Table 2).
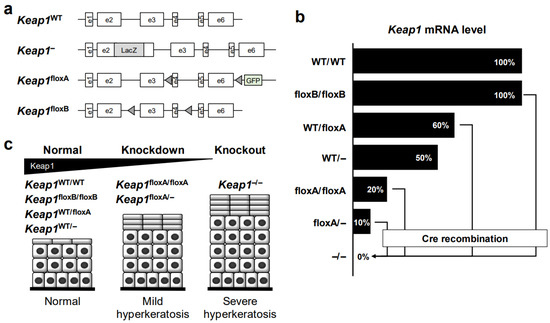
Figure 5.
Genetically modified Keap1 and esophageal phenotypes in mice. (a) Construction of Keap1-modified genes in mice. Diagrams of wild-type (WT), systemic knockout (–/–), and Keap1flox mice (two lines) are shown. (b) The expression level of Keap1 mRNAs in Keap1 genetically modified mice. The Keap1 mRNA expression from the Keap1floxB allele is comparable with that of the WT allele in the absence of Cre recombination. The expression in the homozygous Keap1floxA allele is decreased to 20% of that of the WT allele, even in the absence of Cre recombination. Keap1 in floxA/floxA, floxB/floxB or floxA/– appears to be substantially deleted in the presence of Cre recombination. (c) Keap1-dependent hyperkeratosis in the esophageal epithelium of Keap1 genetically modified mice.
We found that while the wild-type Keap1 transgene (Tg-Keap1WT) is able to rescue esophageal hyperkeratosis in Keap1–/– mice [91], Keap1 point mutant transgenes (Tg-Keap1 mutant), including C273&288A, G364C and G430C, cannot rescue hyperkeratosis and lethality. These results indicate that these amino acid residues are critical for KEAP1 function to repress NRF2 [91,92]. In fact, cysteine 273 (C273) of KEAP1 serves as a sensor for various electrophiles, and C288 serves as a sensor for 15d-prostaglandin J2, 4-hydroxynonenal and arsenite [96]. G364C and G430C mutations were originally identified in human lung cancer patients [19] but have not been found in ESCC to date. Intriguingly, Keap1–/–::Tg-Keap1WT::Tg-Keap1mutant mice show hyperkeratosis and juvenile lethality (Table 2), whereas Keap1 heterozygous knockout mice (Keap1+/–) have no obvious changes in the esophagus [52,93]. These data suggest an intriguing mechanism. Since KEAP1 forms a homodimer and binds one molecule of NRF2, mutant KEAP1 may form a heterodimer with wild-type KEAP1, which lacks ubiquitin ligase activity. Therefore, KEAP1 mutants may have a dominant-negative effect in ESCC cell lines harboring heterozygous KEAP1 mutations (Table 1).
To further analyze the consequences of Keap1 deletion more specifically, two lines of conditional knockout mice were generated: the Keap1floxA mouse line [97] and the Keap1floxB mouse line [98] (Figure 5a). The Keap1floxA allele has two loxP sites on both sides of Keap1 exons 4–6 [97] and green fluorescent protein (GFP) downstream of exon 6 (Figure 5a). From this allele, we found that Keap1 mRNA expression is decreased to 20% of that from the wild-type allele, even in the absence of Cre recombination [93,98] (Figure 5b). As Keap1floxA/– mice show milder hyperkeratosis than Keap1–/– mice, Keap1floxA/– mice can survive with the remaining Keap1 mRNA expression, which is approximately 10% of the level of wild-type mice [93]. We have been capitalizing this genetically hypomorphic expression of KEAP1 and utilizing the Keap1floxA/floxA or Keap1floxA/– lines of mice as Keap1 knockdown models, which are viable models showing the systematic expression of high-level NRF2.
Consistent with the phenotype in Keap1–/– mice, the hyperkeratosis of the esophagus and forestomach in Keap1floxA/– mice is also diminished by the transgenic expression of wild-type KEAP1 (Tg-Keap1WT) [93]. Meanwhile, Keap1 mRNA expression in Keap1floxA/floxA mice completely disappeared in the presence of K5-Cre recombinase (Figure 5b), so keratinocyte-specific Keap1 knockout (Keap1floxA/floxA::K5Cre) mice died before weaning due to severe hyperkeratosis of the esophagus, similar to Keap1–/– mice [93]. As expected, Keap1+/floxA::K5Cre mice showed no obvious change in the esophagus, similar to Keap1+/– mice. Upon exposure to 4NQO, a chemical carcinogen, Keap1floxA/– mice develop many fewer tumors than wild-type mice [87]. We surmise that this is likely due to the high level of expression of cytoprotective genes, but it is possible that mechanical protection by hyperkeratosis or thickened epithelium in the esophagus also plays a role.
Biswal and colleagues generated the Keap1floxB line of mice in which two loxP sites flank exons 3–4 of Keap1 (Figure 5a). Unlike the Keap1floxA allele, the Keap1floxB allele retains intact Keap1 mRNA expression levels in the absence of Cre recombinase [94,98]. After Cre recombination, Keap1 expression is almost completely knocked out. These two Keap1flox mouse lines exhibit intriguing phenotypic differences based on graded Keap1 expression. Thus, we are able to impose subtle changes in Keap1 gene expression depending on the purpose.
7. Cell Competition in Esophageal Epithelium and the Fate of NRF2-Deleted Cells
The existence of stem cells in the esophageal epithelia is still controversial [99]. While one group insists that the esophageal epithelium is maintained by uniformly distributed progenitor cells in the basal layer [100], other groups propose that specific esophageal epithelial stem cells exist in the basal layer, which is composed of one to four cell layers in humans and only one layer in mice. In the former theory, each progenitor cell attaches to the basement membrane and shows equal self-reproducibility and the ability to contribute to the maintenance of esophageal epithelium homeostasis by supplying differentiated epithelial cells. The latter theory assumes the existence of esophageal stem cells with high-level expression of integrin α6 (ITGA6), ITGB4 and CD73 [101], KRT15 [102] or COL17A1 and KRT15 [103].
Related to this issue, the presence of a cell competition mechanism within the esophageal epithelium has been revealed by means of lineage tracing experiments using fluorescent reporter mice and mathematical models [100,104,105,106,107,108,109,110]. This mechanism is particularly interesting if we consider the fact that various somatic mutations accumulate and cell clusters harboring those mutations emerge in a patchy or island-like manner in the homogenous population of the esophageal squamous epithelium. In this model, ‘winner’ clones that acquire a competitive advantage to the surrounding ‘loser’ clones expand their cell population. In fact, it has been shown that epidermal cells with high expression of COL17A1 become ‘winners’ [111]. An intriguing observation here is that the expression of hemidesmosome constituents, including COL17A1, ITGA6 and ITGB4, is closely related to both esophageal stemness [101,103] and cell competition [105].
Given the high incidence of somatic mutations in the NRF2 gene in ESCC, we hypothesized that the loss of NRF2 activity might influence cell competition in the esophageal epithelium. To address this hypothesis, we designed a CreERT2-based conditional deletion of the Nrf2 gene. The CreERT2 system has been used in numerous lineage tracing studies to induce the tissue-specific and tamoxifen-inducible expression of Cre recombinase [112,113]. The recombination efficiency of this system is limited within 30–80%, which also varies among Cre-expressing vector constructs, animals, genotypes, tissues, cell types and tamoxifen doses [114,115]. Owing to these characteristics, we successfully established an in vivo model in which NRF2-deleted cells coexisted with NRF2-normal cells [88]. Features of these coexistent cells were monitored after NRF2 deletion in the mouse esophageal epithelium by crossing Keratin5-CreERT2 (K5CreERT2) mice and administering tamoxifen.
In Nrf2flox/flox::K5CreERT2 mice, approximately 50% of Nrf2 DNA in the esophageal epithelium is recombined, and NRF2-deleted cells emerge segmentally [88]. While NRF2-deleted cells resided and were maintained in the epithelium under normal conditions, we found that these cells were selectively eliminated from the epithelium upon exposure to the chemical carcinogen 4NQO (Figure 6a). That is, NRF2-normal cells survive, while NRF2-deleted cells disappear in the presence of the chemical carcinogen 4NQO. One plausible interpretation of this result is that NRF2-deleted cells are lost in competition with NRF2-normal cells in the esophageal epithelium in the presence of the chemical carcinogen 4NQO.
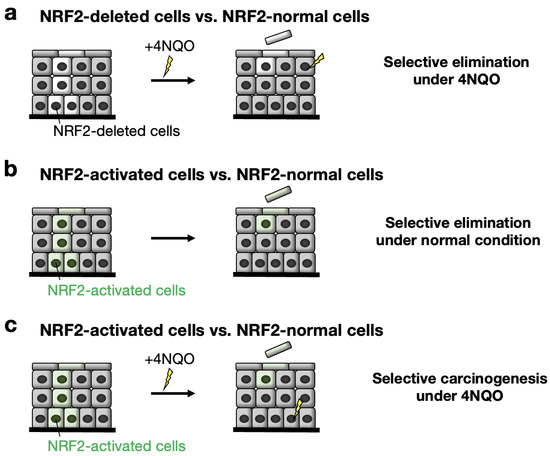
Figure 6.
Fate of NRF2-deleted and NRF2-activated cells in the esophageal epithelium when colocalized with NRF2 and KEAP1 normal epithelial cells. The fate of these esophageal cells was examined after treatment with the carcinogen 4NQO (4-nitroquinoline-1-oxide). The results are well explained by the cell competition model (see text). (a) NRF2-deleted cells vs. NRF2-normal cells under 4NQO. Note that NRF2-deleted cells are eliminated from the esophageal epithelium under 4NQO administration. (b) NRF2-activated cells vs. NRF2-normal cells under 4NQO untreated conditions. Note that NRF2-activated cells are eliminated from the esophageal epithelium. (c) NRF2-activated cells vs. NRF2-normal cells under 4NQO. Note that NRF2-activated cells are eliminated from the esophageal epithelium. NRF2-normal cells are an origin of carcinogenesis under 4NQO administration.
8. Fate of NRF2-Activated Cells in the Esophageal Epithelium
We also hypothesized that a gain of NRF2 activity may influence cell competition in the esophageal epithelium. To address this hypothesis, we designed a CreERT2-based conditional deletion of the Keap1 gene and successfully established an in vivo model in which KEAP1-deleted (i.e., NRF2-activated) epithelial cells coexist with KEAP1-normal (i.e., NRF2-normal) cells in the esophageal epithelium [95]. The features of these coexistent cells were monitored after KEAP1 deletion/NRF2 activation in the mouse esophageal epithelium after the administration of tamoxifen. To our surprise, in the esophagus of Keap1floxB/floxB::K5CreERT2 mice, KEAP1-deleted (i.e., NRF2-activated) cells were selectively and rapidly eliminated from the esophageal epithelium even under normal conditions (Figure 6b). KEAP1-deleted/NRF2-activated cells exhibit decreased COL17A1 expression and commit to cell differentiation. Thus, the cells lose their competitive advantage against KEAP1-normal/NRF2-normal cells in the epithelium. An intriguing hypothesis that emerges from these observations is that KEAP1-deleted/NRF2-activated cells may be routinely generated in the epithelium due to somatic mutations caused by the toxic substances passing through the esophagus but then be eliminated regularly through cell competition in the esophageal epithelium.
An important question here is the mechanism by which NRF2-activated or NRF2-addicted cancers are generated in light of the cell active competition model. We surmise that multiple pathways or events are involved in the formation of NRF2-addicted esophageal cancers in addition to NRF2 activation. The study showing that NRF2 mutation is a late event during ESCC evolution supports this hypothesis [116]. As described in Section 3 and Section 4, TP53 mutations are the most common mutations in ESCC. In fact, mice cells with the Tp53R245W mutation (equivalent to the TP53R248W mutation in humans) are shown to become winners and expand clonally in the competition between the esophageal epithelium and epidermal cells [107,108]. Esophageal carcinoma retains a variety of driver candidates in addition to TP53. Therefore, somatic mutations in these genes, including TP53, may be the tumor initiation mutations required for NRF2-activated cells to become winner cells and ultimately develop into cancer cells. We hypothesize that NRF2 activation may play a role in favoring the survival/expansion of winner TP53- or other mutation-bearing cells.
9. NRF2 as a Therapeutic Target for Cancers
It has been shown that NRF2-addicted cancers in various tissues or organs, including NRF2-addicted ESCC, are resistant to current mainstream cancer therapies. Some studies have reported that NRF2-addicted ESCC is resistant to chemoradiotherapy using cisplatin and fluorouracil, the major treatments for ESCC [26,46]. Even immune checkpoint inhibitors, including nivolumab and pembrolizumab, are not effective, especially in NRF2-addicted non-small cell lung cancer (NSCLC). NRF2-addicted NSCLC shows a poorer prognosis than NRF2-normal cancer [117]. To overcome NRF2-addicted cancers that are resistant to general or mainstream treatments, new therapies targeting NRF2 appear to be necessary. To this end, we propose three strategies that target either NRF2-addicted cancer cells or host defenses for the effective treatment of NRF2-addicted cancers (Figure 7).
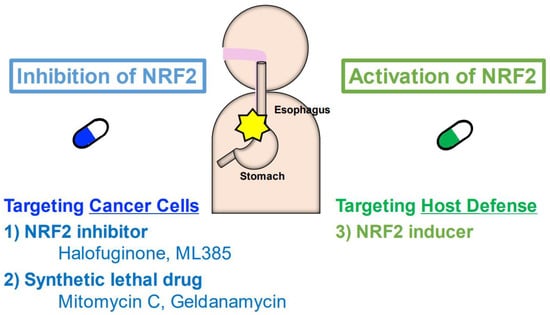
Figure 7.
Three new strategies for the treatment of NRF2-addicted cancer. First, NRF2 inhibitors, including halofuginone and ML385, can act to target NRF2-addicted cancer cells. Second, synthetic lethal drugs will target NRF2-addicted cancer cells; this group includes mitomycin C and geldanamycin. Third, NRF2 inducers can activate the host defense system surrounding NRF2-addicted cancers.
9.1. NRF2 Inhibitors to Treat NRF2-Addicted Cancers
It seems reasonable to consider the development of NRF2 inhibitors as an approach against NRF2-addicted cancers. Through chemical library screening utilizing an ARE-Luc reporter, we identified halofuginone, a derivative of the plant alkaloid febrifugine, as a specific inhibitor of NRF2 [80]. Mechanistic studies have revealed that halofuginone induces the amino acid starvation response by inhibiting prolyl-tRNA synthetase activity [118], resulting in the depletion of the NRF2 protein, as NRF2 shows very rapid turnover with a half-life of less than 18 min [3,8]. Halofuginone has been widely used as an antibiotic in animals [119]. In fact, halofuginone enhanced the anticancer effects of cisplatin in both in vitro and in vivo experiments using KYSE70, a cell line derived from NRF2-addicted ESCC [80]. Halofuginone has already been used in phase II human trials as a therapy for AIDS-related Kaposi sarcoma [120]. While halofuginone exhibits some adverse effects, including a reduction in red blood cells and white blood cells, we have developed a nanomedicine technology in which halofuginone is encapsulated into micelles [121]. Halofuginone-micelles show much reduced side effects with regard to suppressing hematopoietic and immune cells [121], demonstrating that halofuginone-micelles are promising NRF2 inhibitors for the treatment of NRF2-addicted ESCC.
Along this line, Biswal and colleagues identified ML385 as an NRF2 inhibitor through high-throughput screening [122]. ML385 is a small molecule that directly inhibits NRF2 DNA binding through the Neh1 domain and blocks NRF2 transcriptional activity. ML385 shows a specific antitumor effect for NRF2-addicted NSCLC with KEAP1 mutation both in vitro and in vivo [122,123], indicating that ML385 is another important drug candidate for the treatment of NRF2-addicted ESCC. In addition, several other phytochemicals have been reported as NRF2 inhibitors [124,125,126]. Further development of these drugs is expected.
9.2. Synthetic Lethal Drugs to Treat NRF2-Addicted Cancers
The development of synthetic lethal anticancer drugs is an alternative approach for the treatment of NRF2-addicted cancers. This approach exploits the ability of NRF2 to induce a set of drug metabolizing enzymes [127,128]. Therefore, in this scenario, a set of drug-metabolizing enzymes that are highly activated in NRF2-addicted cancers transform anticancer prodrugs into active anticancer drugs. This approach and the identified synthetic lethal drugs harbor several critical advantages. For instance, nonmutant cells within the patient will be insensitive to the treatment, so the drugs should have a large therapeutic window with limited adverse effects.
Through synthetic lethal drug screening of an off-patent drug library utilizing isogenic cell lines, mitomycin C was identified as a promising candidate [81]. Mitomycin C is a DNA alkylating agent and shows enhanced toxicity specific to NRF2-addicted cancer cells. Mitomycin C contains a quinone structure that is bioactivated through cytochrome P450 reductase (P450R or CYPOR) and NQO1, which are NRF2-target gene products [81]. Therefore, mitomycin C is specifically activated by aberrant NRF2 activation. As an already approved clinical drug for cancers in the head and neck, stomach, colon and anus, pancreas, lung, cervix, bladder, and breast [129], mitomycin C is expected to be the first practical and economical drug for NRF2-addicted ESCC.
The geldanamycin-derived HSP90 inhibitors 17-AAG, 17-DMAG and IPI-505 have also been identified as synthetic lethal drugs for NRF-addicted cancer cells [127]. For this group of drugs, NQO1 acts to metabolize quinone compounds to hydroquinone and convert less-toxic prodrugs into actively toxic anticancer drugs specifically in NRF2-activated cells. These drugs have been used in many clinical trials but were withdrawn primarily due to toxicity issues [130,131,132]. While position 19-substituted geldanamycins have been developed to ameliorate this toxicity by blocking the reaction of the compounds with biological nucleophiles [133], we would like to propose an alternative approach in which geldanamycins are used for the treatment of NRF2-addicted cancers based on the synthetic lethal mechanism.
9.3. NRF2 Inducers to Target the Host Defense System
As a completely distinct strategy for treating NRF2-addicted cancers, we propose NRF2 inducers targeting the host microenvironmental defense. In a series of analyses utilizing chemical carcinogenesis of the mouse lung and esophagus, we identified that systemic activation of NRF2 in mice, both genetic and pharmacological, improves the survival of the mice with NRF2-addicted cancers [87,98,134]. These analyses converge to the conclusion that NRF2 activation in microenvironment cells suppresses tumor progression and is beneficial for the treatment of NRF2-addicted cancers. The concept of this strategy may be symbolized as ‘Fighting Fire with Fire’. The activation of NRF2 in microenvironmental cells may contribute to cells not only overcoming the adverse effects of anticancer drugs but also protecting the anticancer immunity of the host from the immune suppressing effect elicited by NRF2-activated cells. In this regard, it should be noted that NRF2 inducers are used in humans. For instance, sulforaphane [135,136,137] is used as a cytoprotective dietary supplement, and dimethyl fumarate [138] is used for the treatment of relapsing multiple sclerosis. Bardoxolone methyl [139,140] is now in clinical trials for diabetic kidney disease and other diseases.
9.4. Others
Several studies have focused on metabolic activities in NRF2-addicted cancers. The dual inhibition of glycolysis and glutaminolysis by the mTOR inhibitor sapanisertib (also named MLN0128, INK128 or TAK-228) and the glutaminase inhibitor telaglenastat (CB-839) to target NRF2-addicted NSCLC is currently in a clinical trial [141,142]. The suppression of glycolysis by mTOR inhibitors elicits metabolic adaptation thorough glutaminolysis in cancer cells that consume abundant glucose [143]. As NRF2-addicted cancers exhibit enhanced glutamine dependence, the inhibition of glutaminolysis is expected to suppress the proliferation of NRF2-addicted cancers. Therefore, the combination of sapanisertib and telaglenastat may become a therapy in NRF2-addicted cancers.
The development of microRNAs (miRNAs) may become a potential therapy for cancers. It has been shown that NRF2 in ESCC is downregulated by miR-507, -634, -450a, and -129-5p, or activated through the downregulation of KEAP1 by miR-432-3p [144,145]. Therefore, these miRNAs targeting NRF2 may emerge as a potential therapy to treat NRF2-addicted ESCC.
10. Toward Clinical Use of NRF2 Target Drugs
As described above, genetic alterations in the KEAP1-NRF2 system occur in more than 30% of ESCC cases. One of the important issues remaining to be addressed to achieve progress in NRF2-addicted cancer-specific treatments is how to efficiently and precisely identify the NRF2-activated status in clinical sites. NRF2 activation is generally evaluated according to the nuclear accumulation of NRF2 proteins and the upregulation of target genes. Therefore, multiple approaches to detect NRF2 activation have been used for human ESCC samples at the experimental stage.
One classic approach is the immunohistochemical analysis of tumor tissue specimens for the nuclear NRF2 protein [26,46,74,75,78,146] In the immunohistochemical analysis, higher NRF2 expression in ESCC than in the matched normal tissue in the same case is generally considered to signify the detection of an NRF2-addicted cancer. Utilizing this approach, it has been identified that NRF2 expression in cancers is associated with tumor stage and clinical outcome [73]. Similarly, accumulating lines of evidence support the relationship between nuclear NRF2 expression and survival [26,74,147,148].
An alternative approach is to use cancer genome analysis and challenge the detection of NRF2 gene alterations in cancers [21,33,62,75]. In fact, somatic mutations in the NRF2 gene are shown to be associated with NRF2 accumulation in the nucleus [75] and poor prognosis in ESCC patients [21]. However, there are causes of NRF2 activation other than NRF2 genomic alterations, and these causes need to be examined. Such alternate causes include exon skipping, epigenetic modification and oncometabolites (see Section 2), which should not be overlooked in the analysis of NRF2-addicted cancers.
Comprehensive examination of NRF2 target gene mRNA expression by quantitative PCR or RNA sequencing analysis is an attractive approach toward identifying NRF2-addicted ESCC [73,78]. However, there remain clinical hurdles for the routine examination of RNA expression in tumor specimens. In this regard, the establishment of clinical phenome centers as well as further technological advances are anticipated. Most of all, as a less invasive approach, liquid biopsy using a small volume of blood or urine samples for the detection of specific molecules derived from NRF2-addicted cancers is highly anticipated, although the development of these technologies has been quite challenging to date.
11. Conclusions
It has been emerging that NRF2 activation is one of the major phenotypes that is intimately related to poor prognosis in ESCC. Three main approaches have emerged in the treatment of the NRF2-addicted cancers. Further clinical research and/or trials with these approaches will tell us how we can overcome these NRF2-activated cancers. We fully expect that new treatments for NRF2-addicted ESCC will improve the prognosis of the ESCC patients in the near future.
Author Contributions
W.H., H.O., K.T. and M.Y.: writing—original draft preparation, reviewing and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by MEXT/JSPS KAKENHI [19H05649 (M.Y.), 19K07395 (K.T.)] and AMED-P-CREATE [JP20cm0106101 (M.Y.)].
Acknowledgments
The authors appreciate the members of the Medical Biochemistry and Surgery departments for their continuous discussions.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Yamamoto, M.; Kensler, T.W.; Motohashi, H. The KEAP1-NRF2 system: A thiol-based sensor-effector apparatus for maintaining redox homeostasis. Physiol. Rev. 2018, 98, 1169–1203. [Google Scholar] [CrossRef]
- Itoh, K.; Wakabayashi, N.; Katoh, Y.; Ishii, T.; Igarashi, K.; Engel, J.D.; Yamamoto, M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999, 13, 76–86. [Google Scholar] [CrossRef]
- Kobayashi, A.; Kang, M.I.; Okawa, H.; Ohtsuji, M.; Zenke, Y.; Chiba, T.; Igarashi, K.; Yamamoto, M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell Biol. 2004, 24, 7130–7139. [Google Scholar] [CrossRef]
- Katoh, Y.; Iida, K.; Kang, M.I.; Kobayashi, A.; Mizukami, M.; Tong, K.I.; McMahon, M.; Hayes, J.D.; Itoh, K.; Yamamoto, M. Evolutionary conserved N-terminal domain of Nrf2 is essential for the Keap1-mediated degradation of the protein by proteasome. Arch. Biochem. Biophys. 2005, 433, 342–350. [Google Scholar] [CrossRef]
- Rada, P.; Rojo, A.I.; Chowdhry, S.; McMahon, M.; Hayes, J.D.; Cuadrado, A. SCF/β-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol. Cell Biol. 2011, 31, 1121–1133. [Google Scholar] [CrossRef] [PubMed]
- Rada, P.; Rojo, A.I.; Evrard-Todeschi, N.; Innamorato, N.G.; Cotte, A.; Jaworski, T.; Tobon-Velasco, J.C.; Devijver, H.; Garcia-Mayoral, M.F.; Van Leuven, F.; et al. Structural and functional characterization of Nrf2 degradation by the glycogen synthase kinase 3/beta-TrCP axis. Mol. Cell Biol. 2012, 32, 3486–3499. [Google Scholar] [CrossRef] [PubMed]
- Chowdhry, S.; Zhang, Y.; McMahon, M.; Sutherland, C.; Cuadrado, A.; Hayes, J.D. Nrf2 is controlled by two distinct beta-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene 2013, 32, 3765–3781. [Google Scholar] [CrossRef] [PubMed]
- Itoh, K.; Wakabayashi, N.; Katoh, Y.; Ishii, T.; O’Connor, T.; Yamamoto, M. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells 2003, 8, 379–391. [Google Scholar] [CrossRef]
- Watai, Y.; Kobayashi, A.; Nagase, H.; Mizukami, M.; McEvoy, J.; Singer, J.D.; Itoh, K.; Yamamoto, M. Subcellular localization and cytoplasmic complex status of endogenous Keap1. Genes Cells 2007, 12, 1163–1178. [Google Scholar] [CrossRef]
- Horie, Y.; Suzuki, T.; Inoue, J.; Iso, T.; Wells, G.; Moore, T.W.; Mizushima, T.; Dinkova-Kostova, A.T.; Kasai, T.; Kamei, T.; et al. Molecular basis for the disruption of Keap1-Nrf2 interaction via Hinge & Latch mechanism. Commun. Biol. 2021, 4, 576. [Google Scholar] [CrossRef]
- Taguchi, K.; Hirano, I.; Itoh, T.; Tanaka, M.; Miyajima, A.; Suzuki, A.; Motohashi, H.; Yamamoto, M. Nrf2 enhances cholangiocyte expansion in Pten-deficient livers. Mol. Cell Biol. 2014, 34, 900–913. [Google Scholar] [CrossRef] [PubMed]
- Kuga, A.; Tsuchida, K.; Panda, H.; Horiuchi, M.; Otsuki, A.; Taguchi, K.; Katsuoka, F.; Suzuki, M.; Yamamoto, M. The β-TrCP-Mediated Pathway Cooperates with the Keap1-Mediated Pathway in Nrf2 Degradation In Vivo. Mol. Cell Biol. 2022, 42, e0056321. [Google Scholar] [CrossRef] [PubMed]
- Lo, J.Y.; Spatola, B.N.; Curran, S.P. WDR23 regulates NRF2 independently of KEAP1. PLoS Genet. 2017, 13, e1006762. [Google Scholar] [CrossRef]
- Otsuki, A.; Suzuki, M.; Katsuoka, F.; Tsuchida, K.; Suda, H.; Morita, M.; Shimizu, R.; Yamamoto, M. Unique cistrome defined as CsMBE is strictly required for Nrf2-sMaf heterodimer function in cytoprotection. Free Radic. Biol. Med. 2016, 91, 45–57. [Google Scholar] [CrossRef]
- Motohashi, H.; Katsuoka, F.; Engel, J.D.; Yamamoto, M. Small Maf proteins serve as transcriptional cofactors for keratinocyte differentiation in the Keap1-Nrf2 regulatory pathway. Proc. Natl. Acad. Sci. USA 2004, 101, 6379–6384. [Google Scholar] [CrossRef] [PubMed]
- Rushmore, T.H.; Morton, M.R.; Pickett, C.B. The antioxidant responsive element. Activation by oxidative stress and identification of the DNA consensus sequence required for functional activity. J. Biol. Chem. 1991, 266, 11632–11639. [Google Scholar] [CrossRef]
- Hirotsu, Y.; Katsuoka, F.; Funayama, R.; Nagashima, T.; Nishida, Y.; Nakayama, K.; Engel, J.D.; Yamamoto, M. Nrf2-MafG heterodimers contribute globally to antioxidant and metabolic networks. Nucleic Acids Res. 2012, 40, 10228–10239. [Google Scholar] [CrossRef]
- Friling, R.S.; Bensimon, A.; Tichauer, Y.; Daniel, V. Xenobiotic-inducible expression of murine glutathione S-transferase Ya subunit gene is controlled by an electrophile-responsive element. Proc. Natl. Acad. Sci. USA 1990, 87, 6258–6262. [Google Scholar] [CrossRef]
- Padmanabhan, B.; Tong, K.I.; Ohta, T.; Nakamura, Y.; Scharlock, M.; Ohtsuji, M.; Kang, M.I.; Kobayashi, A.; Yokoyama, S.; Yamamoto, M. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell 2006, 21, 689–700. [Google Scholar] [CrossRef]
- Singh, A.; Misra, V.; Thimmulappa, R.K.; Lee, H.; Ames, S.; Hoque, M.O.; Herman, J.G.; Baylin, S.B.; Sidransky, D.; Gabrielson, E.; et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006, 3, e420. [Google Scholar] [CrossRef]
- Shibata, T.; Kokubu, A.; Saito, S.; Narisawa-Saito, M.; Sasaki, H.; Aoyagi, K.; Yoshimatsu, Y.; Tachimori, Y.; Kushima, R.; Kiyono, T.; et al. NRF2 mutation confers malignant potential and resistance to chemoradiation therapy in advanced esophageal squamous cancer. Neoplasia 2011, 13, 864–873. [Google Scholar] [CrossRef]
- Noman, A.S.M.; Parag, R.R.; Rashid, M.I.; Rahman, M.Z.; Chowdhury, A.A.; Sultana, A.; Jerin, C.; Siddiqua, A.; Rahman, L.; Shirin, A.; et al. Widespread expression of Sonic hedgehog (Shh) and Nrf2 in patients treated with cisplatin predicts outcome in resected tumors and are potential therapeutic targets for HPV-negative head and neck cancer. Ther. Adv. Med. Oncol. 2020, 12, 1758835920911229. [Google Scholar] [CrossRef] [PubMed]
- Ooi, A.; Dykema, K.; Ansari, A.; Petillo, D.; Snider, J.; Kahnoski, R.; Anema, J.; Craig, D.; Carpten, J.; Teh, B.T.; et al. CUL3 and NRF2 mutations confer an NRF2 activation phenotype in a sporadic form of papillary renal cell carcinoma. Cancer Res. 2013, 73, 2044–2051. [Google Scholar] [CrossRef]
- Konstantinopoulos, P.A.; Spentzos, D.; Fountzilas, E.; Francoeur, N.; Sanisetty, S.; Grammatikos, A.P.; Hecht, J.L.; Cannistra, S.A. Keap1 mutations and Nrf2 pathway activation in epithelial ovarian cancer. Cancer Res. 2011, 71, 5081–5089. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, K.; Yamamoto, M. The KEAP1-NRF2 system in cancer. Front. Oncol. 2017, 7, 85. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, Y.; Okumura, H.; Uchikado, Y.; Kita, Y.; Sasaki, K.; Owaki, T.; Ishigami, S.; Natsugoe, S. Nrf2 is useful for predicting the effect of chemoradiation therapy on esophageal squamous cell carcinoma. Ann. Surg. Oncol. 2014, 21, 2347–2352. [Google Scholar] [CrossRef]
- Hayes, J.D.; McMahon, M. The double-edged sword of Nrf2: Subversion of redox homeostasis during the evolution of cancer. Mol. Cell 2006, 21, 732–734. [Google Scholar] [CrossRef]
- Menegon, S.; Columbano, A.; Giordano, S. The Dual Roles of NRF2 in Cancer. Trends Mol. Med. 2016, 22, 578–593. [Google Scholar] [CrossRef]
- Sporn, M.B.; Liby, K.T. NRF2 and cancer: The good, the bad and the importance of context. Nat. Rev. Cancer 2012, 12, 564–571. [Google Scholar] [CrossRef]
- Emanuelli, M.; Sartini, D.; Molinelli, E.; Campagna, R.; Pozzi, V.; Salvolini, E.; Simonetti, O.; Campanati, A.; Offidani, A. The Double-Edged Sword of Oxidative Stress in Skin Damage and Melanoma: From Physiopathology to Therapeutical Approaches. Antioxidants 2022, 11, 612. [Google Scholar] [CrossRef]
- Cloer, E.W.; Goldfarb, D.; Schrank, T.P.; Weissman, B.E.; Major, M.B. NRF2 Activation in Cancer: From DNA to Protein. Cancer Res. 2019, 79, 889–898. [Google Scholar] [CrossRef] [PubMed]
- Ma, S.; Paiboonrungruan, C.; Yan, T.; Williams, K.P.; Major, M.B.; Chen, X.L. Targeted therapy of esophageal squamous cell carcinoma: The NRF2 signaling pathway as target. Ann. N. Y. Acad. Sci. 2018, 1434, 164–172. [Google Scholar] [CrossRef]
- Kerins, M.J.; Ooi, A. A catalogue of somatic NRF2 gain-of-function mutations in cancer. Sci. Rep. 2018, 8, 12846. [Google Scholar] [CrossRef] [PubMed]
- Fukutomi, T.; Takagi, K.; Mizushima, T.; Ohuchi, N.; Yamamoto, M. Kinetic, thermodynamic, and structural characterizations of the association between Nrf2-DLGex degron and Keap1. Mol. Cell Biol. 2014, 34, 832–846. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, L.D.; Lee, J.; Gnad, F.; Klijn, C.; Schaub, A.; Reeder, J.; Daemen, A.; Bakalarski, C.E.; Holcomb, T.; Shames, D.S.; et al. Recurrent Loss of NFE2L2 Exon 2 Is a Mechanism for Nrf2 Pathway Activation in Human Cancers. Cell Rep. 2016, 16, 2605–2617. [Google Scholar] [CrossRef]
- Wang, R.; An, J.; Ji, F.; Jiao, H.; Sun, H.; Zhou, D. Hypermethylation of the Keap1 gene in human lung cancer cell lines and lung cancer tissues. Biochem. Biophys. Res. Commun. 2008, 373, 151–154. [Google Scholar] [CrossRef]
- Fabrizio, F.P.; Costantini, M.; Copetti, M.; la Torre, A.; Sparaneo, A.; Fontana, A.; Poeta, L.; Gallucci, M.; Sentinelli, S.; Graziano, P.; et al. Keap1/Nrf2 pathway in kidney cancer: Frequent methylation of KEAP1 gene promoter in clear renal cell carcinoma. Oncotarget 2017, 8, 11187–11198. [Google Scholar] [CrossRef]
- Ooi, A.; Wong, J.C.; Petillo, D.; Roossien, D.; Perrier-Trudova, V.; Whitten, D.; Min, B.W.; Tan, M.H.; Zhang, Z.; Yang, X.J.; et al. An antioxidant response phenotype shared between hereditary and sporadic type 2 papillary renal cell carcinoma. Cancer Cell 2011, 20, 511–523. [Google Scholar] [CrossRef]
- Adam, J.; Hatipoglu, E.; O’Flaherty, L.; Ternette, N.; Sahgal, N.; Lockstone, H.; Baban, D.; Nye, E.; Stamp, G.W.; Wolhuter, K.; et al. Renal cyst formation in Fh1-deficient mice is independent of the Hif/Phd pathway: Roles for fumarate in KEAP1 succination and Nrf2 signaling. Cancer Cell 2011, 20, 524–537. [Google Scholar] [CrossRef]
- Mills, E.L.; Ryan, D.G.; Prag, H.A.; Dikovskaya, D.; Menon, D.; Zaslona, Z.; Jedrychowski, M.P.; Costa, A.S.H.; Higgins, M.; Hams, E.; et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 2018, 556, 113–117. [Google Scholar] [CrossRef]
- Komatsu, M.; Kurokawa, H.; Waguri, S.; Taguchi, K.; Kobayashi, A.; Ichimura, Y.; Sou, Y.S.; Ueno, I.; Sakamoto, A.; Tong, K.I.; et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010, 12, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Yanagawa, T.; Kawane, T.; Yuki, K.; Seita, J.; Yoshida, H.; Bannai, S. Murine peritoneal macrophages induce a novel 60-kDa protein with structural similarity to a tyrosine kinase p56lck-associated protein in response to oxidative stress. Biochem. Biophys. Res. Commun. 1996, 226, 456–460. [Google Scholar] [CrossRef] [PubMed]
- Ichimura, Y.; Waguri, S.; Sou, Y.S.; Kageyama, S.; Hasegawa, J.; Ishimura, R.; Saito, T.; Yang, Y.; Kouno, T.; Fukutomi, T.; et al. Phosphorylation of p62 activates the Keap1-Nrf2 pathway during selective autophagy. Mol. Cell 2013, 51, 618–631. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, M.; Waguri, S.; Koike, M.; Sou, Y.S.; Ueno, T.; Hara, T.; Mizushima, N.; Iwata, J.; Ezaki, J.; Murata, S.; et al. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell 2007, 131, 1149–1163. [Google Scholar] [CrossRef]
- Inami, Y.; Waguri, S.; Sakamoto, A.; Kouno, T.; Nakada, K.; Hino, O.; Watanabe, S.; Ando, J.; Iwadate, M.; Yamamoto, M.; et al. Persistent activation of Nrf2 through p62 in hepatocellular carcinoma cells. J. Cell Biol. 2011, 193, 275–284. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, J.; Li, M.; Kong, L.; Yu, J. The expression of p-p62 and nuclear Nrf2 in esophageal squamous cell carcinoma and association with radioresistance. Thorac. Cancer 2020, 11, 130–139. [Google Scholar] [CrossRef]
- Jiang, M.; Ku, W.Y.; Zhou, Z.; Dellon, E.S.; Falk, G.W.; Nakagawa, H.; Wang, M.L.; Liu, K.; Wang, J.; Katzka, D.A.; et al. BMP-driven NRF2 activation in esophageal basal cell differentiation and eosinophilic esophagitis. J. Clin. Investig. 2015, 125, 1557–1568. [Google Scholar] [CrossRef]
- Chen, H.; Li, J.; Li, H.; Hu, Y.; Tevebaugh, W.; Yamamoto, M.; Que, J.; Chen, X. Transcript profiling identifies dynamic gene expression patterns and an important role for Nrf2/Keap1 pathway in the developing mouse esophagus. PLoS ONE 2012, 7, e36504. [Google Scholar] [CrossRef][Green Version]
- Chen, H.; Hu, Y.; Fang, Y.; Djukic, Z.; Yamamoto, M.; Shaheen, N.J.; Orlando, R.C.; Chen, X. Nrf2 deficiency impairs the barrier function of mouse oesophageal epithelium. Gut 2014, 63, 711–719. [Google Scholar] [CrossRef]
- Fu, J.; Xiong, Z.; Huang, C.; Li, J.; Yang, W.; Han, Y.; Paiboonrungruan, C.; Major, M.B.; Chen, K.N.; Kang, X.; et al. Hyperactivity of the transcription factor Nrf2 causes metabolic reprogramming in mouse esophagus. J. Biol. Chem. 2019, 294, 327–340. [Google Scholar] [CrossRef]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Wakabayashi, N.; Itoh, K.; Wakabayashi, J.; Motohashi, H.; Noda, S.; Takahashi, S.; Imakado, S.; Kotsuji, T.; Otsuka, F.; Roop, D.R.; et al. Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat. Genet. 2003, 35, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research, N.; Analysis Working Group; Asan, U.; Agency, B.C.C.; Brigham Women’s, H.; Broad, I.; Brown, U.; Case Western Reserve, U.; Dana-Farber Cancer, I.; Duke, U.; et al. Integrated genomic characterization of oesophageal carcinoma. Nature 2017, 541, 169–175. [Google Scholar] [CrossRef]
- Edgren, G.; Adami, H.O.; Weiderpass, E.; Nyrén, O. A global assessment of the oesophageal adenocarcinoma epidemic. Gut 2013, 62, 1406–1414. [Google Scholar] [CrossRef] [PubMed]
- Emerenziani, S.; Rescio, M.P.; Guarino, M.P.; Cicala, M. Gastro-esophageal reflux disease and obesity, where is the link? World J. Gastroenterol. 2013, 19, 6536–6539. [Google Scholar] [CrossRef] [PubMed]
- Dulak, A.M.; Stojanov, P.; Peng, S.; Lawrence, M.S.; Fox, C.; Stewart, C.; Bandla, S.; Imamura, Y.; Schumacher, S.E.; Shefler, E.; et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat. Genet 2013, 45, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef]
- Sriramajayam, K.; Peng, D.; Lu, H.; Zhou, S.; Bhat, N.; McDonald, O.G.; Que, J.; Zaika, A.; El-Rifai, W. Activation of NRF2 by APE1/REF1 is redox-dependent in Barrett’s related esophageal adenocarcinoma cells. Redox. Biol. 2021, 43, 101970. [Google Scholar] [CrossRef]
- Peng, D.; Zaika, A.; Que, J.; El-Rifai, W. The antioxidant response in Barrett’s tumorigenesis: A double-edged sword. Redox. Biol. 2021, 41, 101894. [Google Scholar] [CrossRef]
- Rustgi, A.K.; El-Serag, H.B. Esophageal Carcinoma. N. Engl. J. Med. 2014, 371, 2499–2509. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.R.; Oh, J.E.; Kim, M.S.; Kang, M.R.; Park, S.W.; Han, J.Y.; Eom, H.S.; Yoo, N.J.; Lee, S.H. Oncogenic NRF2 mutations in squamous cell carcinomas of oesophagus and skin. J. Pathol. 2010, 220, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.B.; Chen, Z.L.; Li, J.G.; Hu, X.D.; Shi, X.J.; Sun, Z.M.; Zhang, F.; Zhao, Z.R.; Li, Z.T.; Liu, Z.Y.; et al. Genetic landscape of esophageal squamous cell carcinoma. Nat. Genet. 2014, 46, 1097–1102. [Google Scholar] [CrossRef]
- Chang, J.; Tan, W.; Ling, Z.; Xi, R.; Shao, M.; Chen, M.; Luo, Y.; Zhao, Y.; Liu, Y.; Huang, X.; et al. Genomic analysis of oesophageal squamous-cell carcinoma identifies alcohol drinking-related mutation signature and genomic alterations. Nat. Commun. 2017, 8, 15290. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.D.; Liao, X.Y.; Chen, Y.B.; Huang, S.Y.; Xue, W.Q.; Li, F.F.; Ge, X.S.; Liu, D.Q.; Cai, Q.; Long, J.; et al. Genomic Characterization of Esophageal Squamous Cell Carcinoma Reveals Critical Genes Underlying Tumorigenesis and Poor Prognosis. Am. J. Hum. Genet. 2016, 98, 709–727. [Google Scholar] [CrossRef] [PubMed]
- Sawada, G.; Niida, A.; Uchi, R.; Hirata, H.; Shimamura, T.; Suzuki, Y.; Shiraishi, Y.; Chiba, K.; Imoto, S.; Takahashi, Y.; et al. Genomic Landscape of Esophageal Squamous Cell Carcinoma in a Japanese Population. Gastroenterology 2016, 150, 1171–1182. [Google Scholar] [CrossRef]
- Cui, Y.; Chen, H.; Xi, R.; Cui, H.; Zhao, Y.; Xu, E.; Yan, T.; Lu, X.; Huang, F.; Kong, P.; et al. Whole-genome sequencing of 508 patients identifies key molecular features associated with poor prognosis in esophageal squamous cell carcinoma. Cell Res. 2020, 30, 902–913. [Google Scholar] [CrossRef]
- Lin, D.C.; Wang, M.R.; Koeffler, H.P. Genomic and Epigenomic Aberrations in Esophageal Squamous Cell Carcinoma and Implications for Patients. Gastroenterology 2018, 154, 374–389. [Google Scholar] [CrossRef]
- Lin, D.C.; Hao, J.J.; Nagata, Y.; Xu, L.; Shang, L.; Meng, X.; Sato, Y.; Okuno, Y.; Varela, A.M.; Ding, L.W.; et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nat. Genet. 2014, 46, 467–473. [Google Scholar] [CrossRef]
- Song, Y.; Li, L.; Ou, Y.; Gao, Z.; Li, E.; Li, X.; Zhang, W.; Wang, J.; Xu, L.; Zhou, Y.; et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature 2014, 509, 91–95. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Kitano, Y.; Baba, Y.; Nakagawa, S.; Miyake, K.; Iwatsuki, M.; Ishimoto, T.; Yamashita, Y.I.; Yoshida, N.; Watanabe, M.; Nakao, M.; et al. Nrf2 promotes oesophageal cancer cell proliferation via metabolic reprogramming and detoxification of reactive oxygen species. J. Pathol. 2018, 244, 346–357. [Google Scholar] [CrossRef]
- Zhang, J.; Jiao, Q.; Kong, L.; Yu, J.; Fang, A.; Li, M.; Yu, J. Nrf2 and Keap1 abnormalities in esophageal squamous cell carcinoma and association with the effect of chemoradiotherapy. Thorac. Cancer 2018, 9, 726–735. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Zhou, X.; Yu, X.; Chen, X.; Hu, X.; Lu, J.; Zhao, H.; Cao, Q.; Gu, Y.; Yang, Y.; et al. High expression of nuclear NRF2 combined with NFE2L2 alterations predicts poor prognosis in esophageal squamous cell carcinoma patients. Mod. Pathol. 2022, 35, 929–937. [Google Scholar] [CrossRef] [PubMed]
- Nishihira, T.; Kasai, M.; Mori, S.; Watanabe, T.; Kuriya, Y.; Suda, M.; Kitamura, M.; Hirayama, K.; Akaishi, T.; Sasaki, T. Characteristics of two cell lines (TE-1 and TE-2) derived from human squamous cell carcinoma of the esophagus. Gan 1979, 70, 575–584. [Google Scholar]
- Shimada, Y.; Imamura, M.; Wagata, T.; Yamaguchi, N.; Tobe, T. Characterization of 21 newly established esophageal cancer cell lines. Cancer 1992, 69, 277–284. [Google Scholar] [CrossRef]
- Feng, L.; Zhao, K.; Sun, L.; Yin, X.; Zhang, J.; Liu, C.; Li, B. SLC7A11 regulated by NRF2 modulates esophageal squamous cell carcinoma radiosensitivity by inhibiting ferroptosis. J. Transl. Med. 2021, 19, 367. [Google Scholar] [CrossRef]
- Paiboonrungruang, C.; Simpson, E.; Xiong, Z.; Huang, C.; Li, J.; Li, Y.; Chen, X. Development of targeted therapy of NRF2(high) esophageal squamous cell carcinoma. Cell Signal 2021, 86, 110105. [Google Scholar] [CrossRef]
- Odera, J.O.; Xiong, Z.; Huang, C.; Gu, N.; Yang, W.; Githang’a, J.; Odera, E.; Paiboonrungruang, C.; Chen, X. NRF2/ACSS2 axis mediates the metabolic effect of alcohol drinking on esophageal squamous cell carcinoma. Biochem. J. 2020, 477, 3075–3089. [Google Scholar] [CrossRef]
- Tsuchida, K.; Tsujita, T.; Hayashi, M.; Ojima, A.; Keleku-Lukwete, N.; Katsuoka, F.; Otsuki, A.; Kikuchi, H.; Oshima, Y.; Suzuki, M.; et al. Halofuginone enhances the chemo-sensitivity of cancer cells by suppressing NRF2 accumulation. Free Radic. Biol. Med. 2017, 103, 236–247. [Google Scholar] [CrossRef]
- Baird, L.; Yamamoto, M. NRF2-Dependent Bioactivation of Mitomycin C as a Novel Strategy to Target KEAP1-NRF2 Pathway Activation in Human Cancer. Mol. Cell Biol. 2021, 41, e00473-20. [Google Scholar] [CrossRef]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Liu, N.; Yang, X.; Ji, G.; Li, M. Polygalacin D suppresses esophageal squamous cell carcinoma growth and metastasis through regulating miR-142-5p/Nrf2 axis. Free Radic. Biol. Med. 2021, 164, 58–75. [Google Scholar] [CrossRef]
- Chan, K.; Lu, R.; Chang, J.C.; Kan, Y.W. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc. Natl. Acad. Sci. USA 1996, 93, 13943–13948. [Google Scholar] [CrossRef]
- Itoh, K.; Chiba, T.; Takahashi, S.; Ishii, T.; Igarashi, K.; Katoh, Y.; Oyake, T.; Hayashi, N.; Satoh, K.; Hatayama, I.; et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem. Biophys. Res. Commun. 1997, 236, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Ohkoshi, A.; Suzuki, T.; Ono, M.; Kobayashi, T.; Yamamoto, M. Roles of Keap1-Nrf2 system in upper aerodigestive tract carcinogenesis. Cancer Prev. Res. 2013, 6, 149–159. [Google Scholar] [CrossRef]
- Horiuchi, M.; Taguchi, K.; Hirose, W.; Tsuchida, K.; Suzuki, M.; Taniyama, Y.; Kamei, T.; Yamamoto, M. Cellular Nrf2 levels determine cell fate during chemical carcinogenesis in esophageal epithelium. Mol. Cell Biol. 2021, 41, e00536-20. [Google Scholar] [CrossRef]
- Suzuki, T.; Shibata, T.; Takaya, K.; Shiraishi, K.; Kohno, T.; Kunitoh, H.; Tsuta, K.; Furuta, K.; Goto, K.; Hosoda, F.; et al. Regulatory nexus of synthesis and degradation deciphers cellular Nrf2 expression levels. Mol. Cell Biol. 2013, 33, 2402–2412. [Google Scholar] [CrossRef]
- Suzuki, T.; Seki, S.; Hiramoto, K.; Naganuma, E.; Kobayashi, E.H.; Yamaoka, A.; Baird, L.; Takahashi, N.; Sato, H.; Yamamoto, M. Hyperactivation of Nrf2 in early tubular development induces nephrogenic diabetes insipidus. Nat. Commun. 2017, 8, 14577. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Suzuki, T.; Kobayashi, A.; Wakabayashi, J.; Maher, J.; Motohashi, H.; Yamamoto, M. Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol. Cell Biol. 2008, 28, 2758–2770. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Maher, J.; Yamamoto, M. Select heterozygous Keap1 mutations have a dominant-negative effect on wild-type Keap1 in vivo. Cancer Res. 2011, 71, 1700–1709. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, K.; Maher, J.M.; Suzuki, T.; Kawatani, Y.; Motohashi, H.; Yamamoto, M. Genetic analysis of cytoprotective functions supported by graded expression of Keap1. Mol. Cell Biol. 2010, 30, 3016–3026. [Google Scholar] [CrossRef] [PubMed]
- Blake, D.J.; Singh, A.; Kombairaju, P.; Malhotra, D.; Mariani, T.J.; Tuder, R.M.; Gabrielson, E.; Biswal, S. Deletion of Keap1 in the lung attenuates acute cigarette smoke-induced oxidative stress and inflammation. Am. J. Respir. Cell Mol. Biol. 2010, 42, 524–536. [Google Scholar] [CrossRef] [PubMed]
- Hirose, W.; Horiuchi, M.; Li, D.; Motoike, I.N.; Zhang, L.; Nishi, H.; Taniyama, Y.; Kamei, T.; Suzuki, M.; Kinoshita, K.; et al. Selective Elimination of NRF2-Activated Cells by Competition with Neighboring Cells in the Esophageal Epithelium. Cell Mol. Gastroenterol. Hepatol. 2022; in press. [Google Scholar] [CrossRef]
- Saito, R.; Suzuki, T.; Hiramoto, K.; Asami, S.; Naganuma, E.; Suda, H.; Iso, T.; Yamamoto, H.; Morita, M.; Baird, L.; et al. Characterizations of Three Major Cysteine Sensors of Keap1 in Stress Response. Mol. Cell Biol. 2016, 36, 271–284. [Google Scholar] [CrossRef]
- Okawa, H.; Motohashi, H.; Kobayashi, A.; Aburatani, H.; Kensler, T.W.; Yamamoto, M. Hepatocyte-specific deletion of the keap1 gene activates Nrf2 and confers potent resistance against acute drug toxicity. Biochem. Biophys. Res. Commun. 2006, 339, 79–88. [Google Scholar] [CrossRef]
- Hayashi, M.; Kuga, A.; Suzuki, M.; Panda, H.; Kitamura, H.; Motohashi, H.; Yamamoto, M. Microenvironmental activation of Nrf2 restricts the progression of Nrf2-activated malignant tumors. Cancer Res. 2020, 80, 3331–3344. [Google Scholar] [CrossRef]
- Hayakawa, Y.; Nakagawa, H.; Rustgi, A.K.; Que, J.; Wang, T.C. Stem cells and origins of cancer in the upper gastrointestinal tract. Cell Stem Cell 2021, 28, 1343–1361. [Google Scholar] [CrossRef]
- Piedrafita, G.; Kostiou, V.; Wabik, A.; Colom, B.; Fernandez-Antoran, D.; Herms, A.; Murai, K.; Hall, B.A.; Jones, P.H. A single-progenitor model as the unifying paradigm of epidermal and esophageal epithelial maintenance in mice. Nat. Commun. 2020, 11, 1429. [Google Scholar] [CrossRef]
- DeWard, A.D.; Cramer, J.; Lagasse, E. Cellular heterogeneity in the mouse esophagus implicates the presence of a nonquiescent epithelial stem cell population. Cell Rep. 2014, 9, 701–711. [Google Scholar] [CrossRef]
- Giroux, V.; Lento, A.A.; Islam, M.; Pitarresi, J.R.; Kharbanda, A.; Hamilton, K.E.; Whelan, K.A.; Long, A.; Rhoades, B.; Tang, Q.; et al. Long-lived keratin 15+ esophageal progenitor cells contribute to homeostasis and regeneration. J. Clin. Investig. 2017, 127, 2378–2391. [Google Scholar] [CrossRef] [PubMed]
- Busslinger, G.A.; Weusten, B.L.A.; Bogte, A.; Begthel, H.; Brosens, L.A.A.; Clevers, H. Human gastrointestinal epithelia of the esophagus, stomach, and duodenum resolved at single-cell resolution. Cell Rep. 2021, 34, 108819. [Google Scholar] [CrossRef]
- Doupe, D.P.; Alcolea, M.P.; Roshan, A.; Zhang, G.; Klein, A.M.; Simons, B.D.; Jones, P.H. A single progenitor population switches behavior to maintain and repair esophageal epithelium. Science 2012, 337, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Alcolea, M.P.; Greulich, P.; Wabik, A.; Frede, J.; Simons, B.D.; Jones, P.H. Differentiation imbalance in single oesophageal progenitor cells causes clonal immortalization and field change. Nat. Cell Biol. 2014, 16, 615–622. [Google Scholar] [CrossRef] [PubMed]
- Frede, J.; Greulich, P.; Nagy, T.; Simons, B.D.; Jones, P.H. A single dividing cell population with imbalanced fate drives oesophageal tumour growth. Nat. Cell Biol. 2016, 18, 967–978. [Google Scholar] [CrossRef] [PubMed]
- Murai, K.; Skrupskelyte, G.; Piedrafita, G.; Hall, M.; Kostiou, V.; Ong, S.H.; Nagy, T.; Cagan, A.; Goulding, D.; Klein, A.M.; et al. Epidermal Tissue Adapts to Restrain Progenitors Carrying Clonal p53 Mutations. Cell Stem Cell 2018, 23, 687–699 e688. [Google Scholar] [CrossRef]
- Fernandez-Antoran, D.; Piedrafita, G.; Murai, K.; Ong, S.H.; Herms, A.; Frezza, C.; Jones, P.H. Outcompeting p53-Mutant Cells in the Normal Esophagus by Redox Manipulation. Cell Stem Cell 2019, 25, 329–341 e326. [Google Scholar] [CrossRef]
- Colom, B.; Alcolea, M.P.; Piedrafita, G.; Hall, M.W.J.; Wabik, A.; Dentro, S.C.; Fowler, J.C.; Herms, A.; King, C.; Ong, S.H.; et al. Spatial competition shapes the dynamic mutational landscape of normal esophageal epithelium. Nat. Genet. 2020, 52, 604–614. [Google Scholar] [CrossRef]
- Colom, B.; Herms, A.; Hall, M.W.J.; Dentro, S.C.; King, C.; Sood, R.K.; Alcolea, M.P.; Piedrafita, G.; Fernandez-Antoran, D.; Ong, S.H.; et al. Mutant clones in normal epithelium outcompete and eliminate emerging tumours. Nature 2021, 598, 510–514. [Google Scholar] [CrossRef]
- Liu, N.; Matsumura, H.; Kato, T.; Ichinose, S.; Takada, A.; Namiki, T.; Asakawa, K.; Morinaga, H.; Mohri, Y.; De Arcangelis, A.; et al. Stem cell competition orchestrates skin homeostasis and ageing. Nature 2019, 568, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Brocard, J.; Warot, X.; Wendling, O.; Messaddeq, N.; Vonesch, J.L.; Chambon, P.; Metzger, D. Spatio-temporally controlled site-specific somatic mutagenesis in the mouse. Proc. Natl. Acad. Sci. USA 1997, 94, 14559–14563. [Google Scholar] [CrossRef]
- Indra, A.K.; Warot, X.; Brocard, J.; Bornert, J.M.; Xiao, J.H.; Chambon, P.; Metzger, D. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: Comparison of the recombinase activity of the tamoxifen-inducible Cre-ER(T) and Cre-ER(T2) recombinases. Nucleic Acids Res. 1999, 27, 4324–4327. [Google Scholar] [CrossRef] [PubMed]
- Heffner, C.S.; Herbert Pratt, C.; Babiuk, R.P.; Sharma, Y.; Rockwood, S.F.; Donahue, L.R.; Eppig, J.T.; Murray, S.A. Supporting conditional mouse mutagenesis with a comprehensive cre characterization resource. Nat. Commun. 2012, 3, 1218. [Google Scholar] [CrossRef] [PubMed]
- Kristianto, J.; Johnson, M.G.; Zastrow, R.K.; Radcliff, A.B.; Blank, R.D. Spontaneous recombinase activity of Cre-ERT2 in vivo. Transgenic Res. 2017, 26, 411–417. [Google Scholar] [CrossRef]
- Hao, J.J.; Lin, D.C.; Dinh, H.Q.; Mayakonda, A.; Jiang, Y.Y.; Chang, C.; Jiang, Y.; Lu, C.C.; Shi, Z.Z.; Xu, X.; et al. Spatial intratumoral heterogeneity and temporal clonal evolution in esophageal squamous cell carcinoma. Nat. Genet. 2016, 48, 1500–1507. [Google Scholar] [CrossRef]
- Arbour, K.C.; Jordan, E.; Kim, H.R.; Dienstag, J.; Yu, H.A.; Sanchez-Vega, F.; Lito, P.; Berger, M.; Solit, D.B.; Hellmann, M.; et al. Effects of Co-occurring Genomic Alterations on Outcomes in Patients with KRAS-Mutant Non-Small Cell Lung Cancer. Clin. Cancer Res. 2018, 24, 334–340. [Google Scholar] [CrossRef]
- Keller, T.L.; Zocco, D.; Sundrud, M.S.; Hendrick, M.; Edenius, M.; Yum, J.; Kim, Y.J.; Lee, H.K.; Cortese, J.F.; Wirth, D.F.; et al. Halofuginone and other febrifugine derivatives inhibit prolyl-tRNA synthetase. Nat. Chem. Biol. 2012, 8, 311–317. [Google Scholar] [CrossRef]
- Naciri, M.; Mancassola, R.; Yvore, P.; Peeters, J.E. The effect of halofuginone lactate on experimental Cryptosporidium parvum infections in calves. Vet. Parasitol. 1993, 45, 199–207. [Google Scholar] [CrossRef]
- Koon, H.B.; Fingleton, B.; Lee, J.Y.; Geyer, J.T.; Cesarman, E.; Parise, R.A.; Egorin, M.J.; Dezube, B.J.; Aboulafia, D.; Krown, S.E. Phase II AIDS Malignancy Consortium trial of topical halofuginone in AIDS-related Kaposi sarcoma. J. Acquir. Immune Defic. Syndr. 2011, 56, 64–68. [Google Scholar] [CrossRef]
- Panda, H.; Suzuki, M.; Naito, M.; Saito, R.; Wen, H.; Baird, L.; Uruno, A.; Miyata, K.; Yamamoto, M. Halofuginone micelle nanoparticles eradicate Nrf2-activated lung adenocarcinoma without systemic toxicity. Free Radic. Biol. Med. 2022, 187, 92–104. [Google Scholar] [CrossRef]
- Singh, A.; Venkannagari, S.; Oh, K.H.; Zhang, Y.Q.; Rohde, J.M.; Liu, L.; Nimmagadda, S.; Sudini, K.; Brimacombe, K.R.; Gajghate, S.; et al. Small Molecule Inhibitor of NRF2 Selectively Intervenes Therapeutic Resistance in KEAP1-Deficient NSCLC Tumors. ACS Chem. Biol. 2016, 11, 3214–3225. [Google Scholar] [CrossRef]
- Gong, M.; Li, Y.; Ye, X.; Zhang, L.; Wang, Z.; Xu, X.; Shen, Y.; Zheng, C. Loss-of-function mutations in KEAP1 drive lung cancer progression via KEAP1/NRF2 pathway activation. Cell Commun. Signal 2020, 18, 98. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Guo, Y.; Feng, Z.; Bergan, R.; Li, B.; Qin, Y.; Zhao, L.; Zhang, Z.; Shi, M. Involvement of the PI3K/Akt/Nrf2 Signaling Pathway in Resveratrol-Mediated Reversal of Drug Resistance in HL-60/ADR Cells. Nutr. Cancer 2019, 71, 1007–1018. [Google Scholar] [CrossRef] [PubMed]
- Robledinos-Antón, N.; Fernández-Ginés, R.; Manda, G.; Cuadrado, A. Activators and Inhibitors of NRF2: A Review of Their Potential for Clinical Development. Oxid. Med. Cell Longev. 2019, 2019, 9372182. [Google Scholar] [CrossRef] [PubMed]
- Tossetta, G.; Marzioni, D. Natural and synthetic compounds in Ovarian Cancer: A focus on NRF2/KEAP1 pathway. Pharmacol. Res. 2022, 183, 106365. [Google Scholar] [CrossRef]
- Baird, L.; Suzuki, T.; Takahashi, Y.; Hishinuma, E.; Saigusa, D.; Yamamoto, M. Geldanamycin-Derived HSP90 Inhibitors Are Synthetic Lethal with NRF2. Mol. Cell Biol. 2020, 40, e00377-20. [Google Scholar] [CrossRef]
- O’Neil, N.J.; Bailey, M.L.; Hieter, P. Synthetic lethality and cancer. Nat. Rev. Genet. 2017, 18, 613–623. [Google Scholar] [CrossRef]
- Bradner, W.T. Mitomycin C: A clinical update. Cancer Treat. Rev. 2001, 27, 35–50. [Google Scholar] [CrossRef]
- Jhaveri, K.; Taldone, T.; Modi, S.; Chiosis, G. Advances in the clinical development of heat shock protein 90 (Hsp90) inhibitors in cancers. Biochim. Biophys. Acta 2012, 1823, 742–755. [Google Scholar] [CrossRef]
- Wagner, A.J.; Chugh, R.; Rosen, L.S.; Morgan, J.A.; George, S.; Gordon, M.; Dunbar, J.; Normant, E.; Grayzel, D.; Demetri, G.D. A phase I study of the HSP90 inhibitor retaspimycin hydrochloride (IPI-504) in patients with gastrointestinal stromal tumors or soft-tissue sarcomas. Clin. Cancer Res. 2013, 19, 6020–6029. [Google Scholar] [CrossRef] [PubMed]
- Goetz, M.P.; Toft, D.; Reid, J.; Ames, M.; Stensgard, B.; Safgren, S.; Adjei, A.A.; Sloan, J.; Atherton, P.; Vasile, V.; et al. Phase I trial of 17-allylamino-17-demethoxygeldanamycin in patients with advanced cancer. J. Clin. Oncol. 2005, 23, 1078–1087. [Google Scholar] [CrossRef] [PubMed]
- Kitson, R.R.; Chang, C.H.; Xiong, R.; Williams, H.E.; Davis, A.L.; Lewis, W.; Dehn, D.L.; Siegel, D.; Roe, S.M.; Prodromou, C.; et al. Synthesis of 19-substituted geldanamycins with altered conformations and their binding to heat shock protein Hsp90. Nat. Chem. 2013, 5, 307–314. [Google Scholar] [CrossRef][Green Version]
- Satoh, H.; Moriguchi, T.; Saigusa, D.; Baird, L.; Yu, L.; Rokutan, H.; Igarashi, K.; Ebina, M.; Shibata, T.; Yamamoto, M. NRF2 Intensifies Host Defense Systems to Prevent Lung Carcinogenesis, but After Tumor Initiation Accelerates Malignant Cell Growth. Cancer Res. 2016, 76, 3088–3096. [Google Scholar] [CrossRef] [PubMed]
- Dinkova-Kostova, A.T.; Fahey, J.W.; Wade, K.L.; Jenkins, S.N.; Shapiro, T.A.; Fuchs, E.J.; Kerns, M.L.; Talalay, P. Induction of the phase 2 response in mouse and human skin by sulforaphane-containing broccoli sprout extracts. Cancer Epidemiol. Biomark. Prev. 2007, 16, 847–851. [Google Scholar] [CrossRef]
- Fahey, J.W.; Haristoy, X.; Dolan, P.M.; Kensler, T.W.; Scholtus, I.; Stephenson, K.K.; Talalay, P.; Lozniewski, A. Sulforaphane inhibits extracellular, intracellular, and antibiotic-resistant strains of Helicobacter pylori and prevents benzo[a]pyrene-induced stomach tumors. Proc. Natl. Acad. Sci. USA 2002, 99, 7610–7615. [Google Scholar] [CrossRef]
- Dinkova-Kostova, A.T.; Fahey, J.W.; Kostov, R.V.; Kensler, T.W. KEAP1 and Done? Targeting the NRF2 Pathway with Sulforaphane. Trends Food Sci. Technol. 2017, 69, 257–269. [Google Scholar] [CrossRef]
- Gold, R.; Kappos, L.; Arnold, D.L.; Bar-Or, A.; Giovannoni, G.; Selmaj, K.; Tornatore, C.; Sweetser, M.T.; Yang, M.; Sheikh, S.I.; et al. Placebo-controlled phase 3 study of oral BG-12 for relapsing multiple sclerosis. N. Engl. J. Med. 2012, 367, 1098–1107. [Google Scholar] [CrossRef]
- Szczesny-Malysiak, E.; Stojak, M.; Campagna, R.; Grosicki, M.; Jamrozik, M.; Kaczara, P.; Chlopicki, S. Bardoxolone Methyl Displays Detrimental Effects on Endothelial Bioenergetics, Suppresses Endothelial ET-1 Release, and Increases Endothelial Permeability in Human Microvascular Endothelium. Oxid. Med. Cell Longev. 2020, 2020, 4678252. [Google Scholar] [CrossRef]
- de Zeeuw, D.; Akizawa, T.; Audhya, P.; Bakris, G.L.; Chin, M.; Christ-Schmidt, H.; Goldsberry, A.; Houser, M.; Krauth, M.; Lambers Heerspink, H.J.; et al. Bardoxolone methyl in type 2 diabetes and stage 4 chronic kidney disease. N. Engl. J. Med. 2013, 369, 2492–2503. [Google Scholar] [CrossRef] [PubMed]
- Riess, J.W.; Frankel, P.; Shackelford, D.; Dunphy, M.; Badawi, R.D.; Nardo, L.; Cherry, S.R.; Lanza, I.; Reid, J.; Gonsalves, W.I.; et al. Phase 1 Trial of MLN0128 (Sapanisertib) and CB-839 HCl (Telaglenastat) in Patients with Advanced NSCLC (NCI 10327): Rationale and Study Design. Clin. Lung Cancer 2021, 22, 67–70. [Google Scholar] [CrossRef] [PubMed]
- Romero, R.; Sayin, V.I.; Davidson, S.M.; Bauer, M.R.; Singh, S.X.; LeBoeuf, S.E.; Karakousi, T.R.; Ellis, D.C.; Bhutkar, A.; Sanchez-Rivera, F.J.; et al. Keap1 loss promotes Kras-driven lung cancer and results in dependence on glutaminolysis. Nat. Med. 2017, 23, 1362–1368. [Google Scholar] [CrossRef] [PubMed]
- Momcilovic, M.; Bailey, S.T.; Lee, J.T.; Fishbein, M.C.; Braas, D.; Go, J.; Graeber, T.G.; Parlati, F.; Demo, S.; Li, R.; et al. The GSK3 Signaling Axis Regulates Adaptive Glutamine Metabolism in Lung Squamous Cell Carcinoma. Cancer Cell 2018, 33, 905–921 e905. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Inoue, J.; Kawano, T.; Kozaki, K.; Omura, K.; Inazawa, J. The impact of miRNA-based molecular diagnostics and treatment of NRF2-stabilized tumors. Mol. Cancer Res. 2014, 12, 58–68. [Google Scholar] [CrossRef]
- Akdemir, B.; Nakajima, Y.; Inazawa, J.; Inoue, J. miR-432 Induces NRF2 Stabilization by Directly Targeting KEAP1. Mol. Cancer Res. 2017, 15, 1570–1578. [Google Scholar] [CrossRef]
- Xia, D.; Zhang, X.R.; Ma, Y.L.; Zhao, Z.J.; Zhao, R.; Wang, Y.Y. Nrf2 promotes esophageal squamous cell carcinoma (ESCC) resistance to radiotherapy through the CaMKIIalpha-associated activation of autophagy. Cell Biosci. 2020, 10, 90. [Google Scholar] [CrossRef]
- Akaishi, R.; Fujishima, F.; Ishida, H.; Tsunokake, J.; Yamauchi, T.; Gokon, Y.; Ueki, S.; Fukutomi, T.; Okamoto, H.; Takaya, K.; et al. HO-1 in lymph node metastasis predicted overall survival in patients with esophageal squamous cell carcinoma receiving neoadjuvant chemoradiation therapy. Cancer Rep. 2022, 5, e1477. [Google Scholar] [CrossRef]
- Akaishi, R.; Fujishima, F.; Ishida, H.; Tsunokake, J.; Yamauchi, T.; Gokon, Y.; Ueki, S.; Fukutomi, T.; Okamoto, H.; Takaya, K.; et al. Correlation between TXNRD1/HO-1 expression and response to neoadjuvant chemoradiation therapy in patients with esophageal squamous cell carcinoma. Esophagus 2022, 19, 436–443. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).