Time-Series Clustering of Single-Cell Trajectories in Collective Cell Migration
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Electrospinning
2.2. Cell Culture and Time-Lapse Observation
2.3. Cell Trajectory Data and Its Normalization
2.4. Distance Matrix of Cell Trajectories
2.5. Two-Dimensional Representation of Cell Trajectories Based on a Distance Matrix
2.6. Clustering of Cell Trajectories
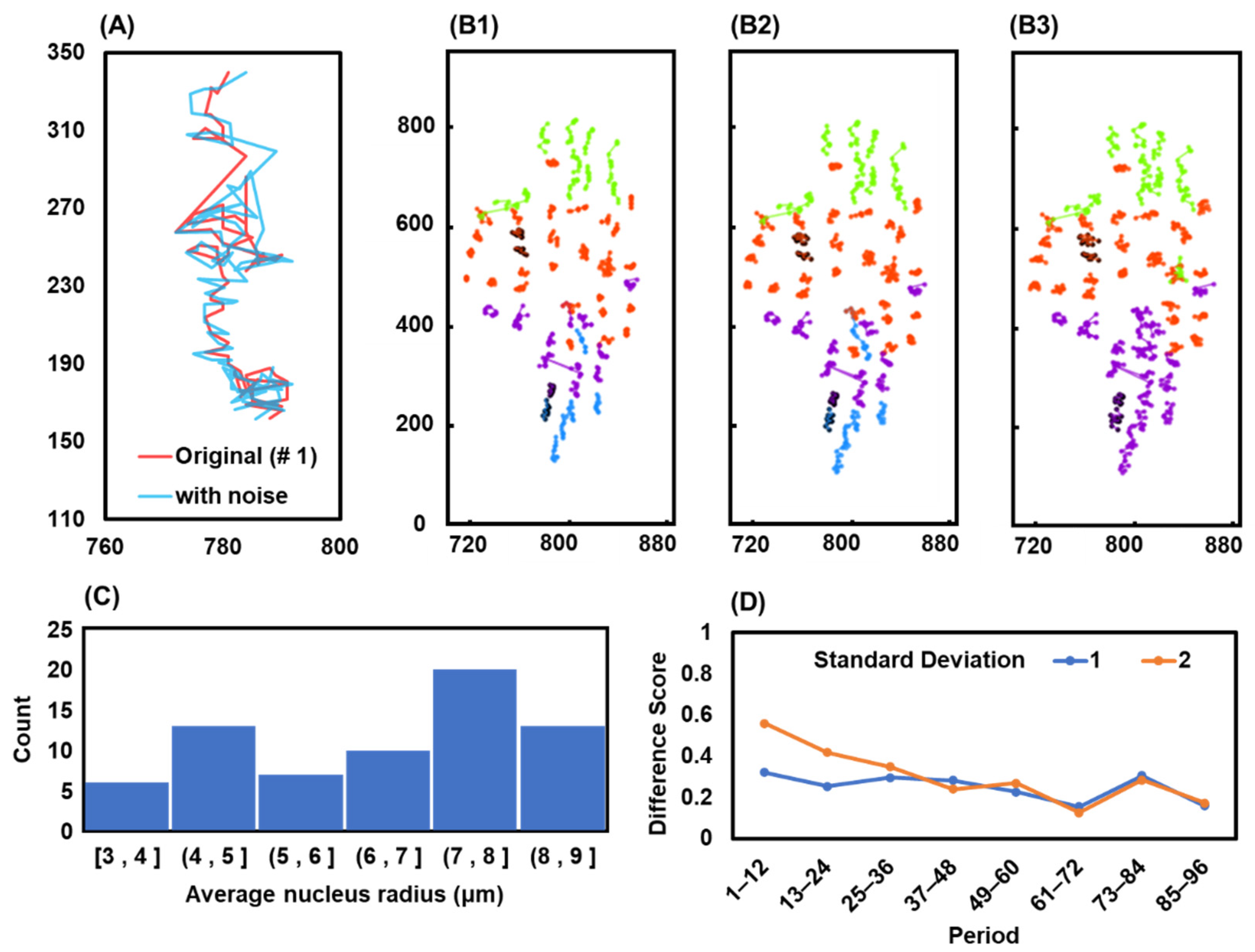
2.7. Robustness of Our Method
3. Results and Discussion
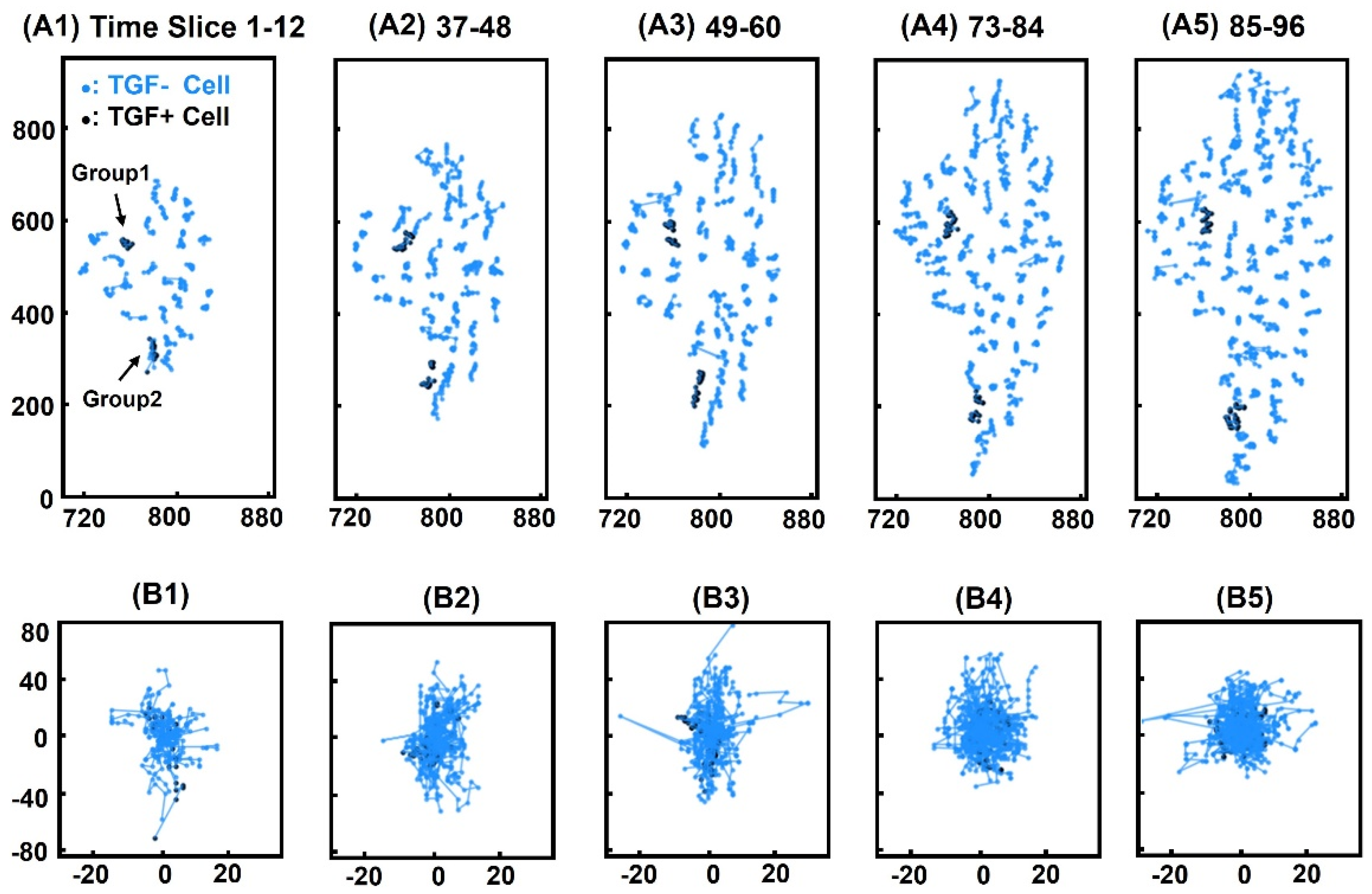
3.1. Cell Tracking
3.2. Dimension Reduction
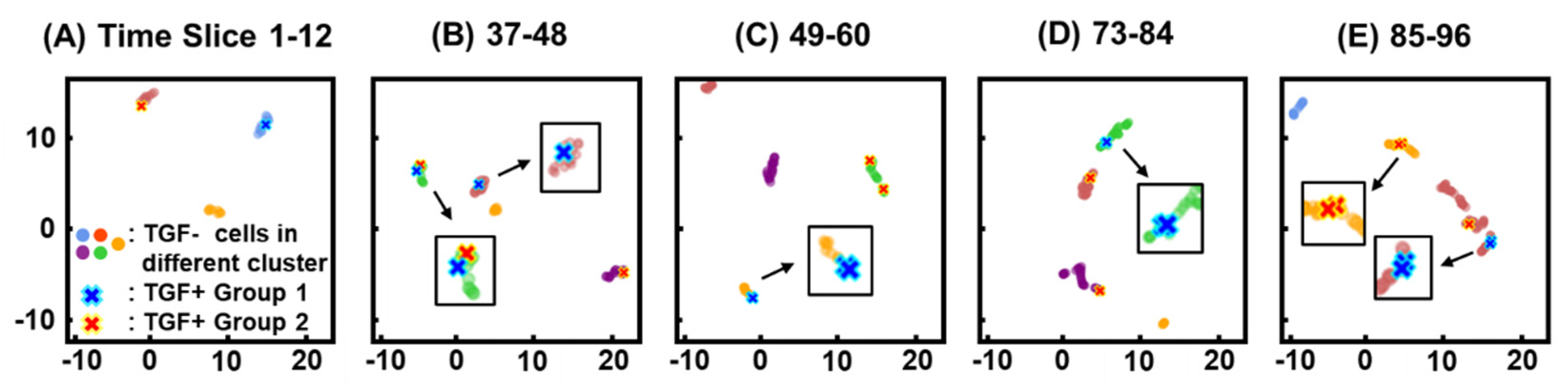
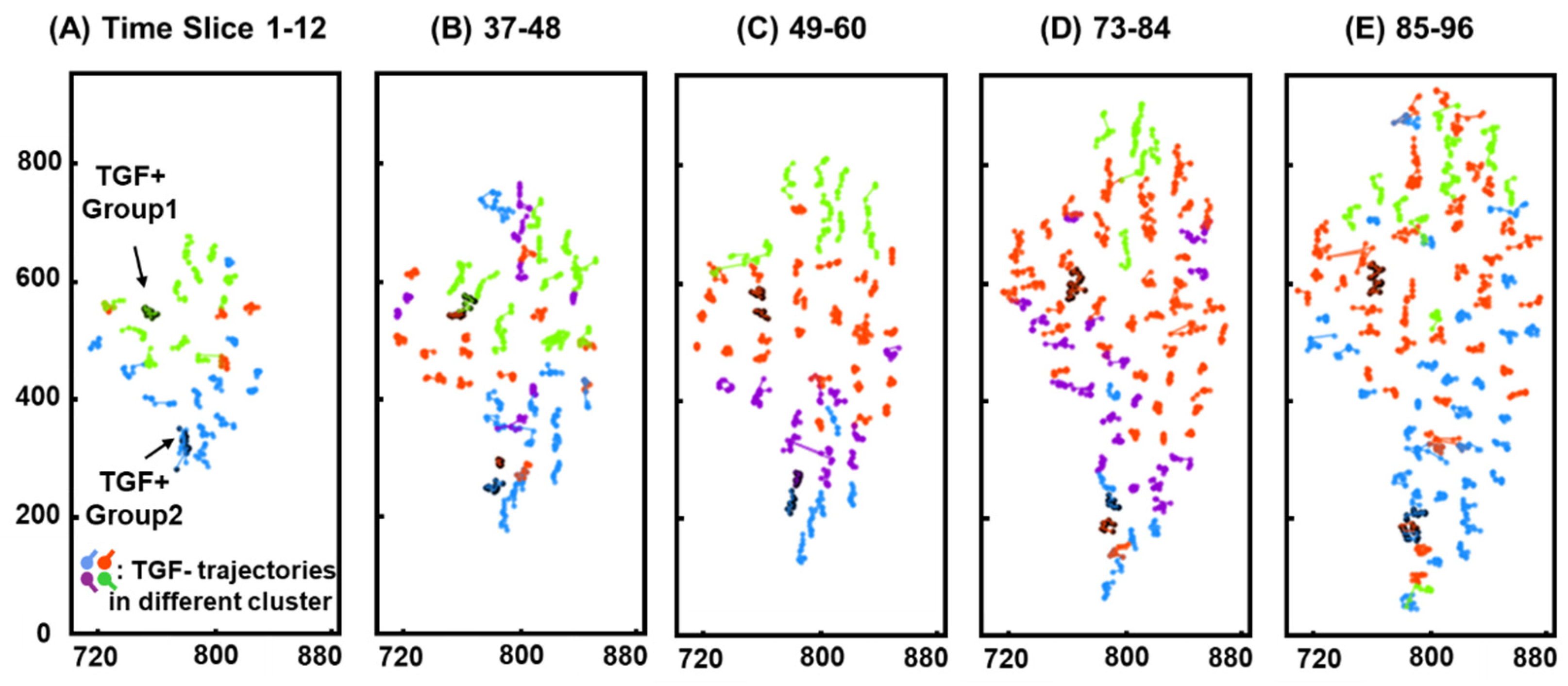
3.3. Clustering
3.4. Similarity of Migration Patterns
3.5. Positional Similarity
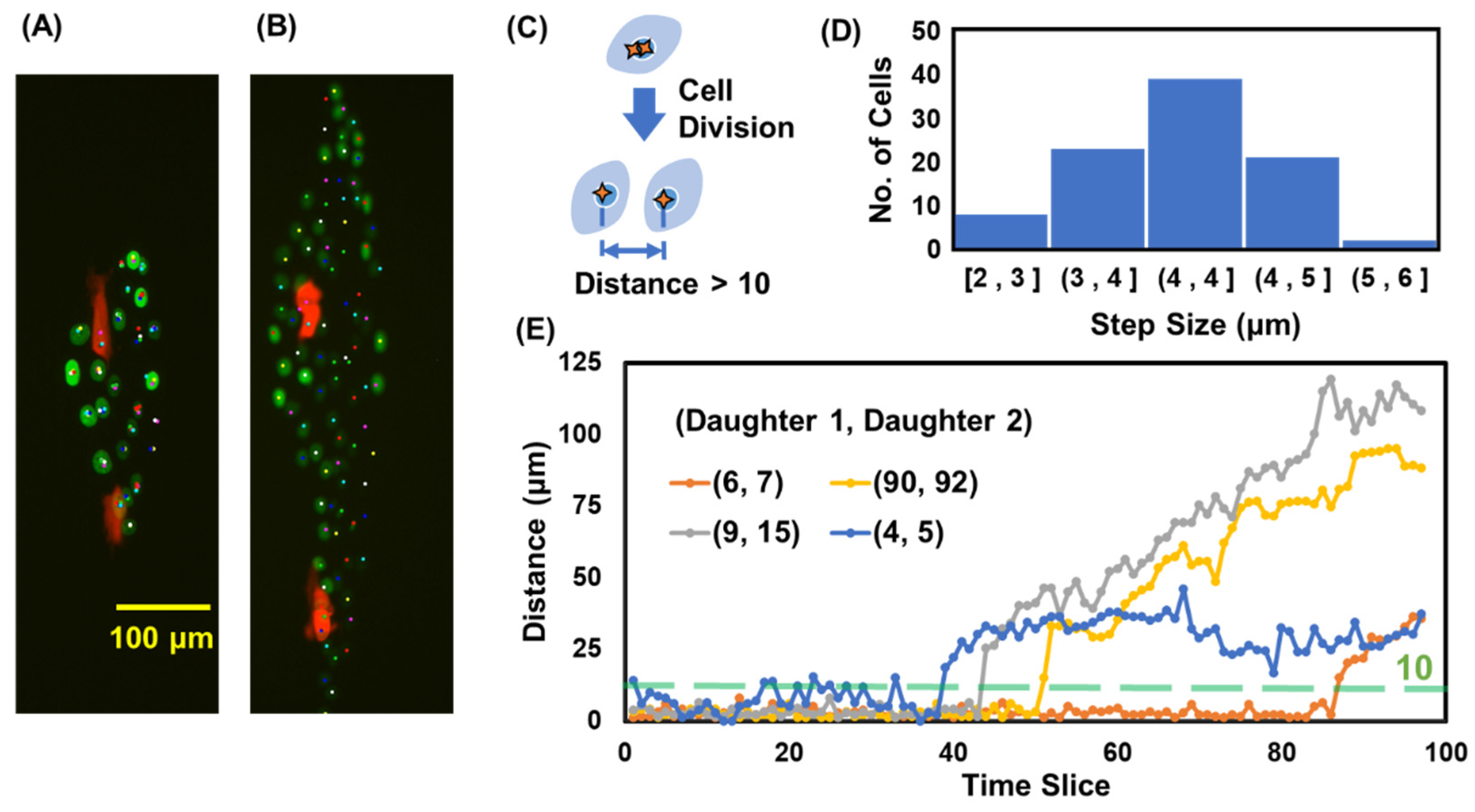
3.6. Cell Division
3.7. Robustness
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jain, S.; Cachoux, V.M.L.; Narayana, G.H.N.S.; de Beco, S.; D’Alessandro, J.; Cellerin, V.; Chen, T.; Heuzé, M.L.; Marcq, P.; Mège, R.M.; et al. The Role of Single-Cell Mechanical Behaviour and Polarity in Driving Collective Cell Migration. Nat. Phys. 2020, 16, 802–809. [Google Scholar] [CrossRef]
- Khalil, A.A.; de Rooij, J. Cadherin Mechanotransduction in Leader-Follower Cell Specification during Collective Migration. Exp. Cell Res. 2019, 376, 86–91. [Google Scholar] [CrossRef]
- Collins, T.A.; Yeoman, B.M.; Katira, P. To Lead or to Herd: Optimal Strategies for 3D Collective Migration of Cell Clusters. Biomech. Model. Mechanobiol. 2020, 19, 1551–1564. [Google Scholar] [CrossRef]
- Saénz-de-Santa-María, I.; Celada, L.; Chiara, M.-D. The Leader Position of Mesenchymal Cells Expressing N-Cadherin in the Collective Migration of Epithelial Cancer. Cells 2020, 9, 731. [Google Scholar] [CrossRef]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The MiR-200 Family and MiR-205 Regulate Epithelial to Mesenchymal Transition by Targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef]
- Burk, U.; Schubert, J.; Wellner, U.; Schmalhofer, O.; Vincan, E.; Spaderna, S.; Brabletz, T. A Reciprocal Repression between ZEB1 and Members of the MiR-200 Family Promotes EMT and Invasion in Cancer Cells. EMBO Rep. 2008, 9, 582–589. [Google Scholar] [CrossRef]
- Dongre, A.; Weinberg, R.A. New Insights into the Mechanisms of Epithelial–Mesenchymal Transition and Implications for Cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef]
- Yang, H.; Ganguly, A.; Cabral, F. Inhibition of Cell Migration and Cell Division Correlates with Distinct Effects of Microtubule Inhibiting Drugs. J. Biol. Chem. 2010, 285, 32242–32250. [Google Scholar] [CrossRef]
- Ganguly, A.; Yang, H.; Sharma, R.; Patel, K.D.; Cabral, F. The Role of Microtubules and Their Dynamics in Cell Migration. J. Biol. Chem. 2012, 287, 43359–43369. [Google Scholar] [CrossRef]
- Doyle, A.D.; Wang, F.W.; Matsumoto, K.; Yamada, K.M. One-Dimensional Topography Underlies Three-Dimensional Fi Brillar Cell Migration. J. Cell Biol. 2009, 184, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Caswell, P.T.; Zech, T. Actin-Based Cell Protrusion in a 3D Matrix. Trends Cell Biol. 2018, 28, 823–834. [Google Scholar] [CrossRef]
- Borgne-Rochet, M.L.; Angevin, L.; Bazellières, E.; Ordas, L.; Comunale, F.; Denisov, E.V.; Tashireva, L.A.; Perelmuter, V.M.; Bièche, I.; Vacher, S.; et al. P-Cadherin-Induced Decorin Secretion Is Required for Collagen Fiber Alignment and Directional Collective Cell Migration. J. Cell Sci. 2019, 132, jcs233189. [Google Scholar] [CrossRef]
- Grünert, S.; Jechlinger, M.; Beug, H. Diverse Cellular and Molecular Mechanisms Contribute to Epithelial Plasticity and Metastasis. Nat. Rev. Mol. Cell Biol. 2003, 4, 657–665. [Google Scholar] [CrossRef]
- Thompson, E.W.; Williams, E.D. EMT and MET in Carcinoma—Clinical Observations, Regulatory Pathways and New Models. Clin. Exp. Metastasis 2008, 25, 591–592. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; Lee, M.; Kim, N.; Heo, W. Do Optogenetic Toolkit Reveals the Role of Ca2+ Sparklets in Coordinated Cell Migration. Proc. Natl. Acad. Sci. USA 2016, 113, 5952–5957. [Google Scholar] [CrossRef]
- Becsky, D.; Szabo, K.; Gyulai-Nagy, S.; Gajdos, T.; Bartos, Z.; Balind, A.; Dux, L.; Horvath, P.; Erdelyi, M.; Homolya, L.; et al. Syndecan-4 Modulates Cell Polarity and Migration by Influencing Centrosome Positioning and Intracellular Calcium Distribution. Front. Cell Dev. Biol. 2020, 8, 575227. [Google Scholar] [CrossRef]
- Morrison, J.A.; McLennan, R.; Wolfe, L.A.; Gogol, M.M.; Meier, S.; McKinney, M.C.; Teddy, J.M.; Holmes, L.; Semerad, C.L.; Box, A.C.; et al. Single-Cell Transcriptome Analysis of Avian Neural Crest Migration Reveals Signatures of Invasion and Molecular Transitions. eLife 2017, 6, e28415. [Google Scholar] [CrossRef] [PubMed]
- Capuana, L.; Boström, A.; Etienne-Manneville, S. Multicellular Scale Front-to-Rear Polarity in Collective Migration. Curr. Opin. Cell Biol. 2020, 62, 114–122. [Google Scholar] [CrossRef]
- Rani, S.; Sikka, G. Recent Techniques of Clustering of Time Series Data: A Survey. Int. J. Comput. Appl. 2012, 52, 1–9. [Google Scholar] [CrossRef]
- Niennattrakul, V.; Ratanamahatana, C.A. Inaccuracies of Shape Averaging Method Using Dynamic Time Warping for Time Series Data. In Proceedings of the Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics); Shi, Y., van Albada, G.D., Dongarra, J., Sloot, P.M.A., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; Volume 4487, pp. 513–520. [Google Scholar]
- Hozumi, Y.; Wang, R.; Yin, C.; Wei, G.W. UMAP-Assisted K-Means Clustering of Large-Scale SARS-CoV-2 Mutation Datasets. Comput. Biol. Med. 2021, 131, 104264. [Google Scholar] [CrossRef]
- Wold, S.; Esbensen, K.; Geladi, P. Principal Component Analysis. Chemom. Intell. Lab. Syst. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Mead, A. Review of the Development of Multidimensional Scaling Methods. Statistician 1992, 41, 27. [Google Scholar] [CrossRef]
- Laurens, V.D.M.; Hinton, G. Visualizing Data Using T-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- McInnes, L.; Healy, J.; Melville, J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv 2018, arXiv:1802.03426. [Google Scholar]
- Pyatnitskiy, M.; Mazo, I.; Shkrob, M.; Schwartz, E.; Kotelnikova, E. Clustering Gene Expression Regulators: New Approach to Disease Subtyping. PLoS ONE 2014, 9, e84955. [Google Scholar] [CrossRef]
- Fujita, A.; Severino, P.; Kojima, K.; Sato, J.R.; Patriota, A.G.; Miyano, S. Functional Clustering of Time Series Gene Expression Data by Granger Causality. BMC Syst. Biol. 2012, 6, 137. [Google Scholar] [CrossRef]
- Schneider, D.; Tarantola, M.; Janshoff, A. Dynamics of TGF-β Induced Epithelial-to-Mesenchymal Transition Monitored by Electric Cell-Substrate Impedance Sensing. Biochim. Biophys. Acta—Mol. Cell Res. 2011, 1813, 2099–2107. [Google Scholar] [CrossRef][Green Version]
- Lonseko, Z.M.; Adjei, P.E.; Du, W.; Luo, C.; Hu, D.; Zhu, L.; Gan, T.; Rao, N. Gastrointestinal Disease Classification in Endoscopic Images Using Attention-Guided Convolutional Neural Networks. Appl. Sci. 2021, 11, 11136. [Google Scholar] [CrossRef]
- Hussain, S.M.; Buongiorno, D.; Altini, N.; Berloco, F.; Prencipe, B.; Moschetta, M.; Bevilacqua, V.; Brunetti, A. Shape-Based Breast Lesion Classification Using Digital Tomosynthesis Images: The Role of Explainable Artificial Intelligence. Appl. Sci. 2022, 12, 6230. [Google Scholar] [CrossRef]
- Althuwaynee, O.F.; Aydda, A.; Hwang, I.T.; Lee, Y.K.; Kim, S.W.; Park, H.J.; Lee, M.S.; Park, Y. Uncertainty Reduction of Unlabeled Features in Landslide Inventory Using Machine Learning T-SNE Clustering and Data Mining Apriori Association Rule Algorithms. Appl. Sci. 2021, 11, 556. [Google Scholar] [CrossRef]
- MacQueen, J. Some Methods for Classification and Analysis of Multivariate Observations. In Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability, Los Angeles, CA, USA, 1 January 1967; Volume 1, pp. 281–297. [Google Scholar]
- Rousseeuw, P.J. Silhouettes: A Graphical Aid to the Interpretation and Validation of Cluster Analysis. J. Comput. Appl. Math. 1987, 20, 53–65. [Google Scholar] [CrossRef]
- Luecken, M.D.; Theis, F.J. Current Best Practices in Single-cell RNA-seq Analysis: A Tutorial. Mol. Syst. Biol. 2019, 15, e8746. [Google Scholar] [CrossRef]
- Moon, K.R.; Stanley, J.S.; Burkhardt, D.; van Dijk, D.; Wolf, G.; Krishnaswamy, S. Manifold Learning-Based Methods for Analyzing Single-Cell RNA-Sequencing Data. Curr. Opin. Syst. Biol. 2018, 7, 36–46. [Google Scholar] [CrossRef]
- Kölsch, V.; Charest, P.G.; Firtel, R.A. The Regulation of Cell Motility and Chemotaxis by Phospholipid Signaling. J. Cell Sci. 2008, 121, 551–559. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xin, Z.; Kajita, M.K.; Deguchi, K.; Suye, S.-i.; Fujita, S. Time-Series Clustering of Single-Cell Trajectories in Collective Cell Migration. Cancers 2022, 14, 4587. https://doi.org/10.3390/cancers14194587
Xin Z, Kajita MK, Deguchi K, Suye S-i, Fujita S. Time-Series Clustering of Single-Cell Trajectories in Collective Cell Migration. Cancers. 2022; 14(19):4587. https://doi.org/10.3390/cancers14194587
Chicago/Turabian StyleXin, Zhuohan, Masashi K. Kajita, Keiko Deguchi, Shin-ichiro Suye, and Satoshi Fujita. 2022. "Time-Series Clustering of Single-Cell Trajectories in Collective Cell Migration" Cancers 14, no. 19: 4587. https://doi.org/10.3390/cancers14194587
APA StyleXin, Z., Kajita, M. K., Deguchi, K., Suye, S.-i., & Fujita, S. (2022). Time-Series Clustering of Single-Cell Trajectories in Collective Cell Migration. Cancers, 14(19), 4587. https://doi.org/10.3390/cancers14194587






