A Simultaneous Multiparametric 18F-FDG PET/MRI Radiomics Model for the Diagnosis of Triple Negative Breast Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
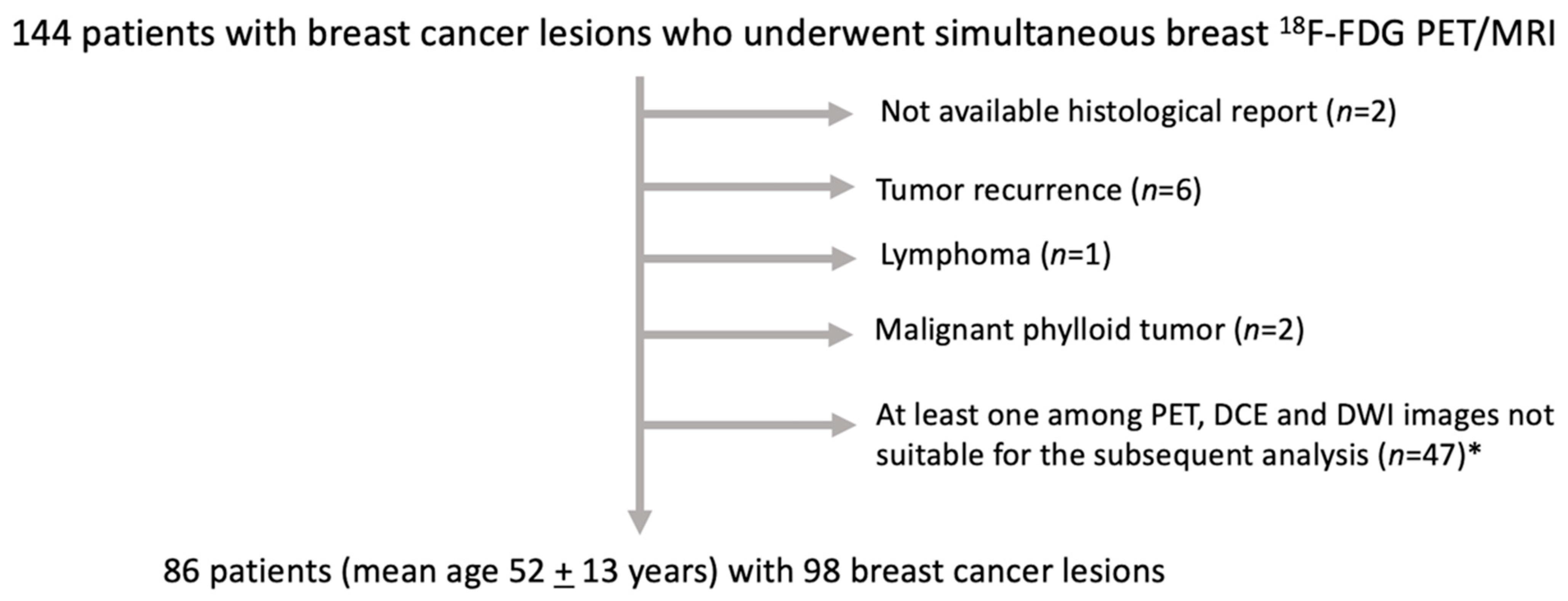
2.1. Patient Sample
2.2. 18F-FDG PET/MRI Acquisition Protocol
2.3. Image Analysis
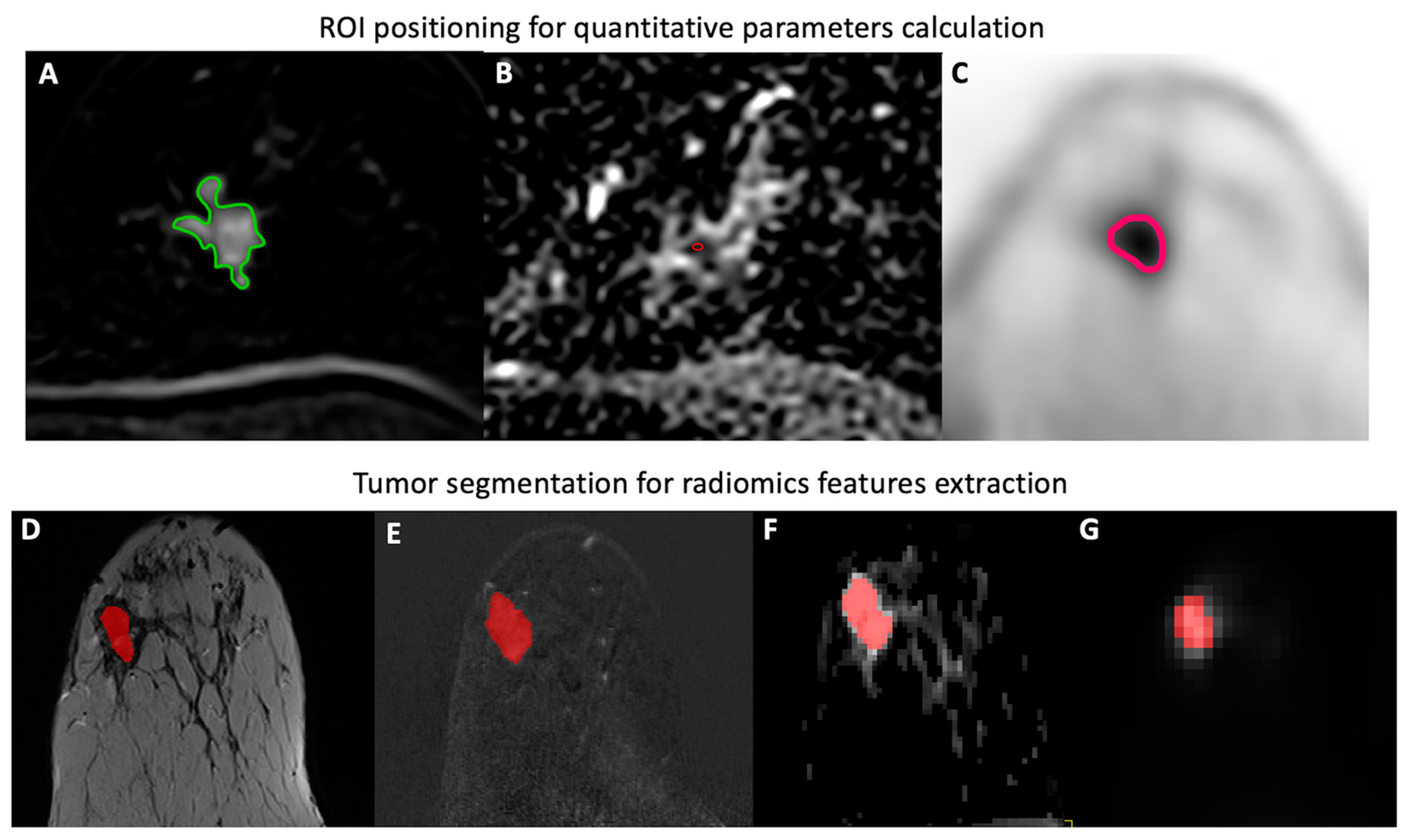
Quantitative Parameters
2.4. Radiomics Analysis
2.4.1. Tumor Segmentation
2.4.2. Radiomic Feature Extraction
2.4.3. Radiomic Feature Selection and Machine Learning
2.5. Reference Standard
2.6. Statistical Analysis
3. Results
3.1. Patient Sample
3.2. Feature Selection and Machine Learning Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, K.; Lu, P.; Beeraka, N.M.; Sukocheva, O.A.; Madhunapantula, S.V.; Liu, J.; Sinelnikov, M.Y.; Nikolenko, V.N.; Bulygin, K.V.; Mikhaleva, L.M.; et al. Mitochondrial mutations and mitoepigenetics: Focus on regulation of oxidative stress-induced responses in breast cancers. Semin. Cancer Biol. 2022, 83, 556–569. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Zhang, J.; Beeraka, N.M.; Tang, C.; Babayeva, Y.V.; Sinelnikov, M.Y.; Zhang, X.; Zhang, J.; Liu, J.; Reshetov, I.V.; et al. Advances in the Prevention and Treatment of Obesity-Driven Effects in Breast Cancers. Front. Oncol. 2022, 12, 820968. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.-S.; Zhao, Z.; Yang, Z.-N.; Xu, F.; Lu, H.-J.; Zhu, Z.-Y.; Shi, W.; Jiang, J.; Yao, P.-P.; Zhu, H.-P. Risk Factors and Preventions of Breast Cancer. Int. J. Biol. Sci. 2017, 13, 1387–1397. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Polyak, K. Heterogeneity in breast cancer. J. Clin. Investig. 2011, 121, 3786–3788. [Google Scholar] [CrossRef]
- Hudis, C.A. Trastuzumab—Mechanism of Action and Use in Clinical Practice. N. Engl. J. Med. 2007, 357, 39–51. [Google Scholar] [CrossRef]
- Dai, X.; Xiang, L.; Li, T.; Bai, Z. Cancer Hallmarks, Biomarkers and Breast Cancer Molecular Subtypes. J. Cancer 2016, 7, 1281–1294. [Google Scholar] [CrossRef]
- Dogan, B.E.; Turnbull, L.W. Imaging of triple-negative breast cancer. Ann. Oncol. 2012, 23, vi23–vi29. [Google Scholar] [CrossRef]
- Turashvili, G.; Brogi, E. Tumor Heterogeneity in Breast Cancer. Front. Med. 2017, 4, 227. [Google Scholar] [CrossRef]
- Rosen, E.L.; Eubank, W.B.; Mankoff, D.A. FDG PET, PET/CT, and Breast Cancer Imaging. RadioGraphics 2007, 27, S215–S229. [Google Scholar] [CrossRef] [PubMed]
- Mann, R.M.; Cho, N.; Moy, L. Breast MRI: State of the Art. Radiology 2019, 292, 520–536. [Google Scholar] [CrossRef]
- Shinde, S.S.; Maschauer, S.; Prante, O. Sweetening Pharmaceutical Radiochemistry by 18F-Fluoroglycosylation: Recent Progress and Future Prospects. Pharmaceuticals 2021, 14, 1175. [Google Scholar] [CrossRef] [PubMed]
- Ulaner, G.A.; Castillo, R.; Goldman, D.A.; Wills, J.; Riedl, C.; Pinker-Domenig, K.; Jochelson, M.S.; Gönen, M. 18F-FDG-PET/CT for systemic staging of newly diagnosed triple-negative breast cancer. Eur. J. Nucl. Med. Mol. Imaging 2016, 43, 1937–1944. [Google Scholar] [CrossRef] [PubMed]
- Li, S.P.; Padhani, A.R.; Taylor, N.J.; Beresford, M.J.; Ah-See, M.-L.W.; Stirling, J.J.; D’Arcy, J.A.; Collins, D.J.; Makris, A. Vascular characterisation of triple negative breast carcinomas using dynamic MRI. Eur. Radiol. 2011, 21, 1364–1373. [Google Scholar] [CrossRef]
- Pujara, A.C.; Kim, E.; Axelrod, D.; Melsaether, A.N. PET/MRI in Breast Cancer. J. Magn. Reson. Imaging 2019, 49, 328–342. [Google Scholar] [CrossRef]
- Krajnc, D.; Papp, L.; Nakuz, T.S.; Magometschnigg, H.F.; Grahovac, M.; Spielvogel, C.P.; Ecsedi, B.; Bago-Horvath, Z.; Haug, A.; Karanikas, G.; et al. Breast Tumor Characterization Using [18F] FDG-PET/CT Imaging Combined with Data Preprocessing and Radiomics. Cancers 2021, 13, 1249. [Google Scholar] [CrossRef]
- Leithner, D.; Mayerhoefer, M.E.; Martinez, D.F.; Jochelson, M.S.; Morris, E.A.; Thakur, S.B.; Pinker, K. Non-Invasive Assessment of Breast Cancer Molecular Subtypes with Multiparametric Magnetic Resonance Imaging Radiomics. J. Clin. Med. 2020, 9, 1853. [Google Scholar] [CrossRef]
- Leithner, D.; Horvat, J.V.; Marino, M.A.; Bernard-Davila, B.; Jochelson, M.S.; Ochoa-Albiztegui, R.E.; Martinez, D.; Morris, E.A.; Thakur, S.; Pinker, K. Radiomic signatures with contrast-enhanced magnetic resonance imaging for the assessment of breast cancer receptor status and molecular subtypes: Initial results. Breast Cancer Res. 2019, 21, 106. [Google Scholar] [CrossRef]
- Umutlu, L.; Kirchner, J.; Bruckmann, N.; Morawitz, J.; Antoch, G.; Ingenwerth, M.; Bittner, A.-K.; Hoffmann, O.; Haubold, J.; Grueneisen, J.; et al. Multiparametric Integrated 18F-FDG PET/MRI-Based Radiomics for Breast Cancer Phenotyping and Tumor Decoding. Cancers 2021, 13, 2928. [Google Scholar] [CrossRef]
- Romeo, V.; Clauser, P.; Rasul, S.; Kapetas, P.; Gibbs, P.; Baltzer, P.A.T.; Hacker, M.; Woitek, R.; Helbich, T.H.; Pinker, K. AI-enhanced simultaneous multiparametric 18F-FDG PET/MRI for accurate breast cancer diagnosis. Eur. J. Nucl. Med. Mol. Imaging 2022, 49, 596–608. [Google Scholar] [CrossRef] [PubMed]
- Rausch, I.; Rust, P.; DiFranco, M.D.; Lassen, M.; Stadlbauer, A.; Mayerhoefer, M.E.; Hartenbach, M.; Hacker, M.; Beyer, T. Reproducibility of MRI Dixon-Based Attenuation Correction in Combined PET/MR with Applications for Lean Body Mass Estimation. J. Nucl. Med. 2016, 57, 1096–1101. [Google Scholar] [CrossRef][Green Version]
- Martinez-Möller, A.; Souvatzoglou, M.; Delso, G.; Bundschuh, R.A.; Chefd’Hotel, C.; Ziegler, S.I.; Navab, N.; Schwaiger, M.; Nekolla, S.G. Tissue Classification as a Potential Approach for Attenuation Correction in Whole-Body PET/MRI: Evaluation with PET/CT Data. J. Nucl. Med. 2009, 50, 520–526. [Google Scholar] [CrossRef] [PubMed]
- Baltzer, P.; Mann, R.M.; Iima, M.; Sigmund, E.E.; Clauser, P.; Gilbert, F.J.; Martincich, L.; Partridge, S.C.; Patterson, A.; Pinker, K.; et al. Diffusion-weighted imaging of the breast—A consensus and mission statement from the EUSOBI International Breast Diffusion-Weighted Imaging working group. Eur. Radiol. 2020, 30, 1436–1450. [Google Scholar] [CrossRef]
- Zöllner, F.G.; Weisser, G.; Reich, M.; Kaiser, S.; Schoenberg, S.O.; Sourbron, S.P.; Schad, L.R. UMMPerfusion: An Open Source Software Tool Towards Quantitative MRI Perfusion Analysis in Clinical Routine. J. Digit. Imaging 2013, 26, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Romeo, V.; Cavaliere, C.; Imbriaco, M.; Verde, F.; Petretta, M.; Franzese, M.; Stanzione, A.; Cuocolo, R.; Aiello, M.; Basso, L.; et al. Tumor segmentation analysis at different post-contrast time points: A possible source of variability of quantitative DCE-MRI parameters in locally advanced breast cancer. Eur. J. Radiol. 2020, 126, 108907. [Google Scholar] [CrossRef]
- Apte, A.P.; Iyer, A.; Crispin-Ortuzar, M.; Pandya, R.; Van Dijk, L.V.; Spezi, E.; Thor, M.; Um, H.; Veeraraghavan, H.; Oh, J.H.; et al. Technical Note: Extension of CERR for computational radiomics: A comprehensive MATLAB platform for reproducible radiomics research. Med. Phys. 2018, 45, 3713–3720. [Google Scholar] [CrossRef]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.J.W.L.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef]
- McNeish, D.M. Using Lasso for Predictor Selection and to Assuage Overfitting: A Method Long Overlooked in Behavioral Sciences. Multivar. Behav. Res. 2015, 50, 471–484. [Google Scholar] [CrossRef]
- Pal, M.; Mather, P.M. Support vector machines for classification in remote sensing. Int. J. Remote Sens. 2005, 26, 1007–1011. [Google Scholar] [CrossRef]
- Goldhirsch, A.; Winer, E.P.; Coates, A.S.; Gelber, R.D.; Piccart-Gebhart, M.; Thürlimann, B.; Senn, H.-J. Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann. Oncol. 2013, 24, 2206–2223. [Google Scholar] [CrossRef] [PubMed]
- Akin, M.; Orguc, S.; Aras, F.; Kandiloglu, A.R. Molecular subtypes of invasive breast cancer: Correlation between PET/computed tomography and MRI findings. Nucl. Med. Commun. 2020, 41, 810–816. [Google Scholar] [CrossRef] [PubMed]
- Jena, A.; Taneja, S.; Singh, A.; Negi, P.; Sarin, R.; Das, P.K.; Singhal, M. Reliability of 18F-FDG PET Metabolic Parameters Derived Using Simultaneous PET/MRI and Correlation with Prognostic Factors of Invasive Ductal Carcinoma: A Feasibility Study. Am. J. Roentgenol. 2017, 209, 662–670. [Google Scholar] [CrossRef] [PubMed]
- Meyer, H.-J.; Wienke, A.; Surov, A. Diffusion-Weighted Imaging of Different Breast Cancer Molecular Subtypes: A Systematic Review and Meta-Analysis. Breast Care 2021, 17, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhu, Y.; Burnside, E.S.; Huang, E.; Drukker, K.; Hoadley, K.; Fan, C.; Conzen, S.D.; Zuley, M.; Net, J.M.; et al. Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. NPJ Breast Cancer 2016, 2, 16012. [Google Scholar] [CrossRef]
- Polyak, K. Breast cancer: Origins and evolution. J. Clin. Investig. 2007, 117, 3155–3163. [Google Scholar] [CrossRef]
- Fan, M.; Chen, J.; Gao, J.; Xue, W.; Wang, Y.; Li, W.; Zhou, L.; Li, X.; Jiang, C.; Sun, Y.; et al. Triggering a switch from basal- to luminal-like breast cancer subtype by the small-molecule diptoindonesin G via induction of GABARAPL1. Cell Death Dis. 2020, 11, 635. [Google Scholar] [CrossRef]
- Feng, Q.; Hu, Q.; Liu, Y.; Yang, T.; Yin, Z. Diagnosis of triple negative breast cancer based on radiomics signatures extracted from preoperative contrast-enhanced chest computed tomography. BMC Cancer 2020, 20, 579. [Google Scholar] [CrossRef]
- Kohavi, R. A Study of Cross-Validation and Bootstrap for Accuracy Estimation and Model Selection. In Proceedings of the Appears in the International Joint Conference on Articial Intelligence (IJCAI), Montreal, Quebec, Canada, 20–25 August 1995; Volume 2. [Google Scholar]
| Radiomic Model | PET/MR Images | Selected Features/Quantitative Parameters |
|---|---|---|
| Quantitative parameters | DCE, ADC, PET parameters (Model 1) | SUVmax, PF, ADCmean contralateral breast, ADCmean tumor lesion, MTT |
| Radiomic features extracted from single 18F-FDG PET/MR images | ADCr(Model 2) | cluster shade (GLCM) |
| strength (NGTDM) | ||
| Hdlge, hgce (NGLDM) | ||
| hglze (SZM) | ||
| DCE(Model 3) | kurtosis, coefficient of dispersion (FO) | |
| strength (NGTDM) | ||
| joint maximum (GLCM) | ||
| PET(Model 4) | glv, lglze (SZM) | |
| complexity (NGTDM) | ||
| inverse difference moment (GLCM) | ||
| rlv (RLM) | ||
| T2-w(Model 5) | coefficient of variation (FO) | |
| entropy (NGLDM) | ||
| run emphasis (RLM) | ||
| gln (SZM) | ||
| Combinations of radiomic features | ADCr, DCE(Model 6) | auto correlation, cluster shade (GLCM, DCE) |
| szhgle (SZM, DCE) | ||
| sre (RLM, ADC) | ||
| strength (NGTDM, DCE) | ||
| ADCr, DCE, PET(Model 7) | zln (SZM, ADC) | |
| glv (SZM, PET) | ||
| dcnNorm (NGLDM, PET) | ||
| coefficient of variation, entropy (FO, PET) | ||
| Integrated model of radiomic features and quantitative parameters | ADCr, DCE, PET + quantitative parameters(Model 8) | SUVmax |
| complexity (NGTDM, PET) | ||
| inverse difference moment (GLCM, PET) | ||
| minimum (FO, T2) | ||
| kurtosis (FO, DCE) |
| Model | Sensitivity | Specificity | PPV | NPV | Accuracy | AUROC |
|---|---|---|---|---|---|---|
| 1 (Quantitative parameters) | 87.2 (82.4–90.5) | 77.5 (72.6–82.2) | 79.7 (76.2–83.5) | 85.7 (81.2–89.2) | 82.4 (78.9–85.7) | 0.884 0.867–0.898) |
| 2 (ADC) | 75.0 (67.6–81.1) | 80.6 (75.3–84.9) | 79.7 (74.6–84.1) | 76.1 (70.7–80.8) | 77.7 (72.8–81.6) | 0.826 (0.789–0.857) |
| 3 (DCE-derived RF) | 70.2 (62.2–77.0) | 79.3 (72.6–84.9) | 77.5 (72.2–82.6) | 72.5 (67.1–77.2) | 74.7 (70.1–78.9) | 0.771 (0.720–0.814) |
| 4 (PET-derived RF) | 68.4 (59.5–77.0) | 75.9 (68.5–82.2) | 74.2 (68.1–80.0) | 70.4 (64.2–76.7) | 72.1 (66.6–77.6) | 0.789 (0.733–0.841) |
| 5 (T2-derived RF) | 69.0 (62.1–74.3) | 73.5 (68.5–78.1) | 72.5 (68.1–76.5) | 70.1 (65.0–74.7) | 71.2 (66.7–75.5) | 0.725 0.679–0.765) |
| 6 (ADC, DCE-derived RF) | 83.7 (79.7–86.5) | 67.4 (63.0–71.2) | 72.3 (69.7–75.0) | 80.3 (75.8–83.6) | 75.6 (72.8–78.2) | 0.822 (0.797–0.842) |
| 7 (ADC, DCE, PET-derived RF) | 79.7 (71.6–86.5) | 86.0 (80.8–90.4) | 85.3 (80.9–89.4) | 80.8 (75.0–86.3) | 82.8 (78.2–87.1) | 0.887 0.847–0.916) |
| 8 (Radiomics features + quantitative parameters) | 88.9 (85.1–91.9) | 74.4 (69.9–78.1) | 77.9 (74.7–81.0) | 86.9 (82.5–90.5) | 81.7 (78.2–85.0) | 0.871 (0.849–0.889) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romeo, V.; Kapetas, P.; Clauser, P.; Baltzer, P.A.T.; Rasul, S.; Gibbs, P.; Hacker, M.; Woitek, R.; Pinker, K.; Helbich, T.H. A Simultaneous Multiparametric 18F-FDG PET/MRI Radiomics Model for the Diagnosis of Triple Negative Breast Cancer. Cancers 2022, 14, 3944. https://doi.org/10.3390/cancers14163944
Romeo V, Kapetas P, Clauser P, Baltzer PAT, Rasul S, Gibbs P, Hacker M, Woitek R, Pinker K, Helbich TH. A Simultaneous Multiparametric 18F-FDG PET/MRI Radiomics Model for the Diagnosis of Triple Negative Breast Cancer. Cancers. 2022; 14(16):3944. https://doi.org/10.3390/cancers14163944
Chicago/Turabian StyleRomeo, Valeria, Panagiotis Kapetas, Paola Clauser, Pascal A. T. Baltzer, Sazan Rasul, Peter Gibbs, Marcus Hacker, Ramona Woitek, Katja Pinker, and Thomas H. Helbich. 2022. "A Simultaneous Multiparametric 18F-FDG PET/MRI Radiomics Model for the Diagnosis of Triple Negative Breast Cancer" Cancers 14, no. 16: 3944. https://doi.org/10.3390/cancers14163944
APA StyleRomeo, V., Kapetas, P., Clauser, P., Baltzer, P. A. T., Rasul, S., Gibbs, P., Hacker, M., Woitek, R., Pinker, K., & Helbich, T. H. (2022). A Simultaneous Multiparametric 18F-FDG PET/MRI Radiomics Model for the Diagnosis of Triple Negative Breast Cancer. Cancers, 14(16), 3944. https://doi.org/10.3390/cancers14163944








