Characterization of RARRES1 Expression on Circulating Tumor Cells as Unfavorable Prognostic Marker in Resected Pancreatic Ductal Adenocarcinoma Patients
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials & Methods
2.1. Cell Lines
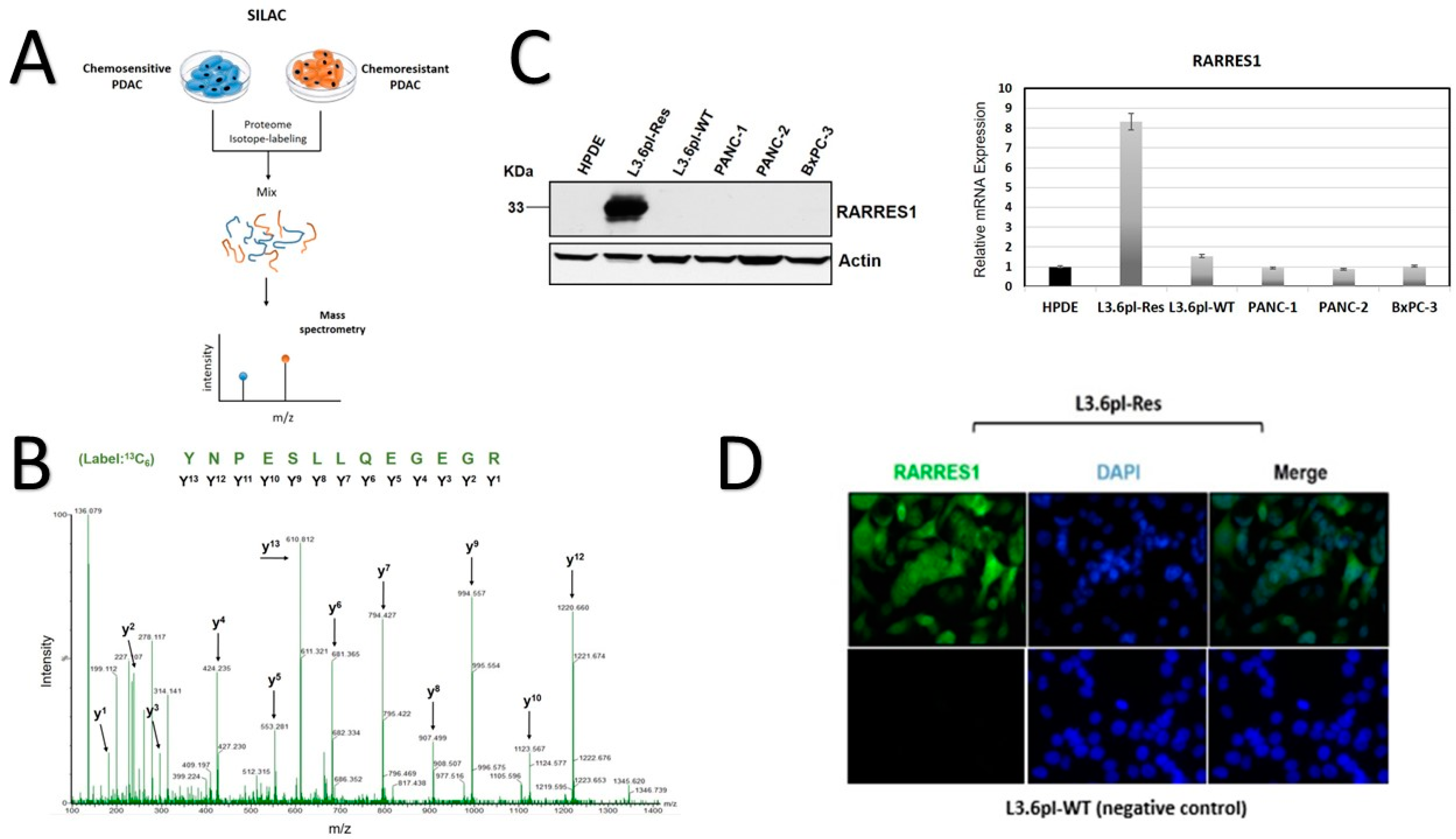
2.2. Stable Isotope Labeling with Amino Acids in Cell Culture (SILAC)
2.3. RNA Isolation and Real-Time Reverse Transcription-PCR (Real-Time RT-PCR)
2.4. Western Blot
2.5. Immunocytochemistry
2.5.1. Patient Cohort
2.5.2. Blood Collection
2.6. CTC Detection
2.7. Statistical Analysis
3. Results
3.1. SILAC-Based Identification of Abundant RARRES1 in Chemoresistant PDAC Cells
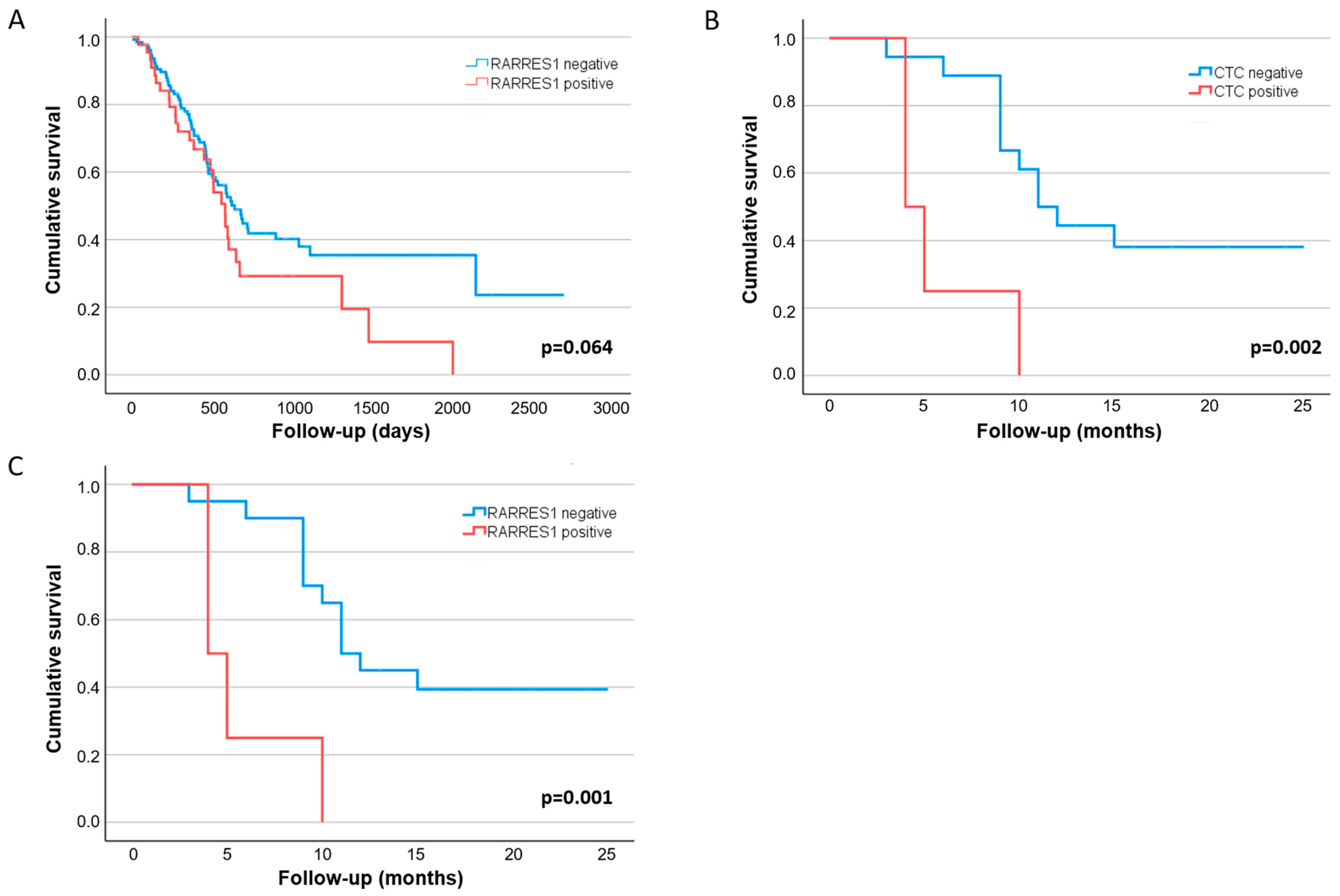
3.2. CTC Detection
3.3. Clinical Value of RARRES1 Detection on CTCs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Güngör, C.; Hofmann, B.T.; Wolters-Eisfeld, G.; Bockhorn, M. Pancreatic cancer. British J. Pharmacol. 2014, 171, 849–858. [Google Scholar] [CrossRef]
- Hosein, A.N.; Dougan, S.K.; Aguirre, A.J.; Maitra, A. Translational advances in pancreatic ductal adenocarcinoma therapy. Nat. Cancer 2022, 3, 272–286. [Google Scholar] [CrossRef]
- Hugenschmidt, H.; Labori, K.; Borgen, E.; Brunborg, C.; Schirmer, C.; Seeberg, L.; Naume, B.; Wiedswang, G. Preoperative CTC-Detection by CellSearch® Is Associated with Early Distant Metastasis and Impaired Survival in Resected Pancreatic Cancer. Cancers 2021, 13, 485. [Google Scholar] [CrossRef]
- Okubo, K.; Uenosono, Y.; Arigami, T.; Mataki, Y.; Matsushita, D.; Yanagita, S.; Kurahara, H.; Sakoda, M.; Kijima, Y.; Maemura, K.; et al. Clinical impact of circulating tumor cells and therapy response in pancreatic cancer. Eur. J. Surg. Oncol. 2017, 43, 1050–1055. [Google Scholar] [CrossRef]
- Buscail, E.; Alix-Panabières, C.; Quincy, P.; Cauvin, T.; Chauvet, A.; Degrandi, O.; Caumont, C.; Verdon, S.; Lamrissi, I.; Moranvillier, I.; et al. High Clinical Value of Liquid Biopsy to Detect Circulating Tumor Cells and Tumor Exosomes in Pancreatic Ductal Adenocarcinoma Patients Eligible for Up-Front Surgery. Cancers 2019, 11, 1656. [Google Scholar] [CrossRef]
- Yeo, D.; Bastian, A.; Strauss, H.; Saxena, P.; Grimison, P.; Rasko, J.E.J. Exploring the Clinical Utility of Pancreatic Cancer Circulating Tumor Cells. Int. J. Mol. Sci. 2022, 23, 1671. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabières, C. Liquid biopsy and minimal residual disease—Latest advances and implications for cure. Nat. Rev. Clin. Oncol. 2019, 16, 409–424. [Google Scholar] [CrossRef]
- Rhim, A.D.; Mirek, E.T.; Aiello, N.M.; Maitra, A.; Bailey, J.M.; McAllister, F.; Reichert, M.; Beatty, G.L.; Rustgi, A.K.; Vonderheide, R.H.; et al. EMT and Dissemination Precede Pancreatic Tumor Formation. Cell 2012, 148, 349–361. [Google Scholar] [CrossRef]
- Gall, T.M.; Belete, S.; Khanderia, E.; Frampton, A.E.; Jiao, L.R. Circulating Tumor Cells and Cell-Free DNA in Pancreatic Ductal Adenocarcinoma. Am. J. Pathol. 2019, 189, 71–81. [Google Scholar] [CrossRef]
- Lee, J.; Park, S.S.; Lee, Y.K.; Norton, J.A.; Jeffrey, S.S. Liquid biopsy in pancreatic ductal adenocarcinoma: Current status of circulating tumor cells and circulating tumorDNA. Mol. Oncol. 2019, 13, 1623–1650. [Google Scholar] [CrossRef] [Green Version]
- Lianidou, E.S.; Strati, A.; Markou, A. Circulating tumor cells as promising novel biomarkers in solid cancers. Crit. Rev. Clin. Lab. Sci. 2014, 51, 160–171. [Google Scholar] [CrossRef] [PubMed]
- Effenberger, K.E.; Schroeder, C.; Hanssen, A.; Wolter, S.; Eulenburg, C.; Tachezy, M.; Gebauer, F.; Izbicki, J.R.; Pantel, K.; Bockhorn, M. Improved Risk Stratification by Circulating Tumor Cell Counts in Pancreatic Cancer. Clin. Cancer Res. 2018, 24, 2844–2850. [Google Scholar] [CrossRef] [PubMed]
- Martini, V.; Timme-Bronsert, S.; Fichtner-Feigl, S.; Hoeppner, J.; Kulemann, B. Circulating Tumor Cells in Pancreatic Cancer: Current Perspectives. Cancers 2019, 11, 1659. [Google Scholar] [CrossRef]
- Ferreira, M.M.; Ramani, V.C.; Jeffrey, S.S. Circulating tumor cell technologies. Mol. Oncol. 2016, 10, 374–394. [Google Scholar] [CrossRef]
- Ma, X.-L.; Li, Y.-Y.; Zhang, J.; Huang, J.-W.; Jia, H.-Y.; Liu, L.; Li, P. Prognostic role of circulating tumor cells in patients with pancreatic cancer: A meta-analysis. Asian Pac. J. Cancer Prev. 2014, 15, 6015–6020. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Chen, W.; Zhao, Q. Prognostic value of circulating tumor cells in patients with pancreatic cancer: A meta-analysis. Tumor Biol. 2014, 35, 2473–2480. [Google Scholar] [CrossRef] [PubMed]
- Poruk, K.E.; Valero, V., III; Saunders, T.; Blackford, A.L.; Griffin, J.F.; Poling, J.; Hruban, R.H.; Anders, R.A.; Herman, J.; Zheng, L.; et al. Circulating Tumor Cell Phenotype Predicts Recurrence and Survival in Pancreatic Adenocarcinoma. Anna. Surg. 2016, 264, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Götze, J.; Nitschke, C.; Uzunoglu, F.G.; Pantel, K.; Sinn, M.; Wikman, H. Tumor-Stroma Interaction in PDAC as a New Approach for Liquid Biopsy and its Potential Clinical Implications. Front. Cell Dev. Biol. 2022, 10, 918795. [Google Scholar] [CrossRef]
- Güngör, C.; Zander, H.; Effenberger, K.E.; Vashist, Y.K.; Kalinina, T.; Izbicki, J.R.; Yekebas, E.; Bockhorn, M. Notch Signaling Activated by Replication Stress–Induced Expression of Midkine Drives Epithelial–Mesenchymal Transition and Chemoresistance in Pancreatic Cancer. Cancer Res. 2011, 71, 5009–5019. [Google Scholar] [CrossRef]
- Koch, C.; Joosse, S.A.; Schneegans, S.; Wilken, O.J.W.; Janning, M.; Loreth, D.; Müller, V.; Prieske, K.; Banys-Paluchowski, M.; Horst, L.J.; et al. Pre-Analytical and Analytical Variables of Label-Independent Enrichment and Automated Detection of Circulating Tumor Cells in Cancer Patients. Cancers (Basel) 2020, 12, 442. [Google Scholar] [CrossRef] [Green Version]
- Loreth, D.; Schuette, M.; Zinke, J.; Mohme, M.; Piffko, A.; Schneegans, S.; Stadler, J.; Janning, M.; Loges, S.; Joosse, S.; et al. CD74 and CD44 Expression on CTCs in Cancer Patients with Brain Metastasis. Int. J. Mol. Sci. 2021, 22, 6993. [Google Scholar] [CrossRef] [PubMed]
- Neves, R.P.L.; Ammerlaan, W.; Andree, K.C.; Bender, S.; Cayrefourcq, L.; Driemel, C.; Koch, C.; Luetke-Eversloh, M.V.; Oulhen, M.; Rossi, E.; et al. Proficiency Testing to Assess Technical Performance for CTC-Processing and Detection Methods in CANCER-ID. Clin. Chem. 2021, 67, 631–641. [Google Scholar] [CrossRef] [PubMed]
- RARRES1 Data Available from v21.1.proteinatlas.org. 2021. Available online: https://www.proteinatlas.org/ENSG00000118849-RARRES1/pathology/pancreatic+cancer#ihc (accessed on 20 July 2022).
- Alix-Panabières, C.; Pantel, K. Liquid Biopsy: From Discovery to Clinical Application. Cancer Discov. 2021, 11, 858–873. [Google Scholar] [CrossRef] [PubMed]
- Müller, V.; Banys-Paluchowski, M.; Friedl, T.; Fasching, P.; Schneeweiss, A.; Hartkopf, A.; Wallwiener, D.; Rack, B.; Meier-Stiegen, F.; Huober, J.; et al. Prognostic relevance of the HER2 status of circulating tumor cells in metastatic breast cancer patients screened for participation in the DETECT study program. ESMO Open 2021, 6, 100299. [Google Scholar] [CrossRef] [PubMed]
- Maimouni, S.; Issa, N.; Cheng, S.; Ouaari, C.; Cheema, A.; Kumar, D.; Byers, S. Tumor suppressor RARRES1- A novel regulator of fatty acid metabolism in epithelial cells. PLoS ONE 2018, 13, e0208756. [Google Scholar] [CrossRef] [PubMed]
- Maimouni, S.; Lee, M.-H.; Sung, Y.-M.; Hall, M.; Roy, A.; Ouaari, C.; Hwang, Y.-S.; Spivak, J.; Glasgow, E.; Swift, M.; et al. Tumor suppressor RARRES1 links tubulin deglutamylation to mitochondrial metabolism and cell survival. Oncotarget 2019, 10, 1606–1624. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; He, M.-Q.; Fan, D.-Q. RARRES1 is a novel immune-related biomarker in GBM. Am. J. Transl. Res. 2019, 11, 5655–5663. [Google Scholar]
- Yanatatsaneejit, P.; Chalermchai, T.; Kerekhanjanarong, V.; Shotelersuk, K.; Supiyaphun, P.; Mutirangura, A.; Sriuranpong, V. Promoter hypermethylation of CCNA1, RARRES1, and HRASLS3 in nasopharyngeal carcinoma. Oral Oncol. 2008, 44, 400–406. [Google Scholar] [CrossRef]
- Kloth, M.; Goering, W.; Ribarska, T.; Arsov, C.; Sørensen, K.D.; Schulz, W. The SNP rs6441224 influences transcriptional activity and prognostically relevant hypermethylation of RARRES1 in prostate cancer. Int. J. Cancer 2012, 131, E897–E904. [Google Scholar] [CrossRef]
- Wu, C.-C.; Shyu, R.-Y.; Chou, J.-M.; Jao, S.-W.; Chao, P.-C.; Kang, J.-C.; Wu, S.-T.; Huang, S.-L.; Jiang, S.-Y. RARRES1 expression is significantly related to tumour differentiation and staging in colorectal adenocarcinoma. Eur. J. Cancer 2006, 42, 557–565. [Google Scholar] [CrossRef]
- Hvichia, G.; Parveen, Z.; Wagner, C.; Janning, M.; Quidde, J.; Stein, A.; Müller, V.; Loges, S.; Neves, R.; Stoecklein, N.; et al. A novel microfluidic platform for size and deformability based separation and the subsequent molecular characterization of viable circulating tumor cells. Int. J. Cancer 2016, 138, 2894–2904. [Google Scholar] [CrossRef] [PubMed]

| Univariate | N = 33 $ | Median RFS, Months (95% CI) | p-Value | |
|---|---|---|---|---|
| Analyses | ||||
| Age | ≤67 years | 17 | 10.0 (7.2–12.8) | 0.218 |
| >67 years | 16 | 15.7 (11.0–20.5) * | ||
| Gender | male | 16 | 9.0 (3.8–14.2) | 0.170 |
| female | 17 | 15.0 (9.5–20.5) | ||
| ECOG | 0 | 20 | - | 0.296 |
| 1 | 12 | - | ||
| 2 | 1 | - | ||
| UICC stage | I-II | 26 | 16.1 (12.6–19.6) * | 0.005 |
| III | 7 | 6.0 (3.4–8.6) | ||
| R-status | R0; CRM- | 17 | 10.0 (6.0–14.0) | 0.227 |
| R0; CRM + /R1 | 16 | 16.0 (9.1–22.9) | ||
| Grading ~ | G2 | 21 | 11.0 (9.6–12.4) | 0.353 |
| G3 | 9 | 14.8 (10.3–19.4) * | ||
| Neoadjuvant treatment | no | 26 | 12.0 (6.0–18.0) | 0.089 |
| yes | 7 | 9.0 (0–19.3) | ||
| Adjuvant treatment | no | 4 | 6.0 (0.0–12.0) * | 0.009 |
| yes | 29 | 12.0 (6.7–17.3) | ||
| Clavien-Dindo | 0–2 | 20 | 16.0 (7.9–24.1) | 0.161 |
| 3–4 | 13 | 10.0 (7.7–12.3) | ||
| Ca 19-9 | <500 U/ml | 24 | 12.0 (5.9–18.1) | 0.881 |
| ≥500 U/ml | 9 | 11.0 (8.2–13.8) | ||
| CTC count per 7.5 mL detected at FUP # | <3 | 18 | 11.0 (8.2–13.8) | 0.002 |
| ≥3 | 4 | 5.8 (2.9–8.6) * | ||
| Combined analysis RARRES1 status and CTC count per 7.5 mL detected at FUP # | <3 OR RARRES1 negative | 18 | 11.0 (8.8–13.2) | 0.001 |
| ≥3 AND RARRES1 positive | 4 | 5.8 (2.9–8.6) * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nitschke, C.; Markmann, B.; Tölle, M.; Kropidlowski, J.; Belloum, Y.; Goetz, M.R.; Schlüter, H.; Kwiatkowski, M.; Sinn, M.; Izbicki, J.; et al. Characterization of RARRES1 Expression on Circulating Tumor Cells as Unfavorable Prognostic Marker in Resected Pancreatic Ductal Adenocarcinoma Patients. Cancers 2022, 14, 4405. https://doi.org/10.3390/cancers14184405
Nitschke C, Markmann B, Tölle M, Kropidlowski J, Belloum Y, Goetz MR, Schlüter H, Kwiatkowski M, Sinn M, Izbicki J, et al. Characterization of RARRES1 Expression on Circulating Tumor Cells as Unfavorable Prognostic Marker in Resected Pancreatic Ductal Adenocarcinoma Patients. Cancers. 2022; 14(18):4405. https://doi.org/10.3390/cancers14184405
Chicago/Turabian StyleNitschke, Christine, Benedikt Markmann, Marie Tölle, Jolanthe Kropidlowski, Yassine Belloum, Mara R. Goetz, Hartmut Schlüter, Marcel Kwiatkowski, Marianne Sinn, Jakob Izbicki, and et al. 2022. "Characterization of RARRES1 Expression on Circulating Tumor Cells as Unfavorable Prognostic Marker in Resected Pancreatic Ductal Adenocarcinoma Patients" Cancers 14, no. 18: 4405. https://doi.org/10.3390/cancers14184405
APA StyleNitschke, C., Markmann, B., Tölle, M., Kropidlowski, J., Belloum, Y., Goetz, M. R., Schlüter, H., Kwiatkowski, M., Sinn, M., Izbicki, J., Pantel, K., Güngör, C., Uzunoglu, F. G., & Wikman, H. (2022). Characterization of RARRES1 Expression on Circulating Tumor Cells as Unfavorable Prognostic Marker in Resected Pancreatic Ductal Adenocarcinoma Patients. Cancers, 14(18), 4405. https://doi.org/10.3390/cancers14184405









