Circulating Cell-Free DNA in Renal Cell Carcinoma: The New Era of Precision Medicine
Abstract
:Simple Summary
Abstract
1. Introduction
2. Data Acquisition
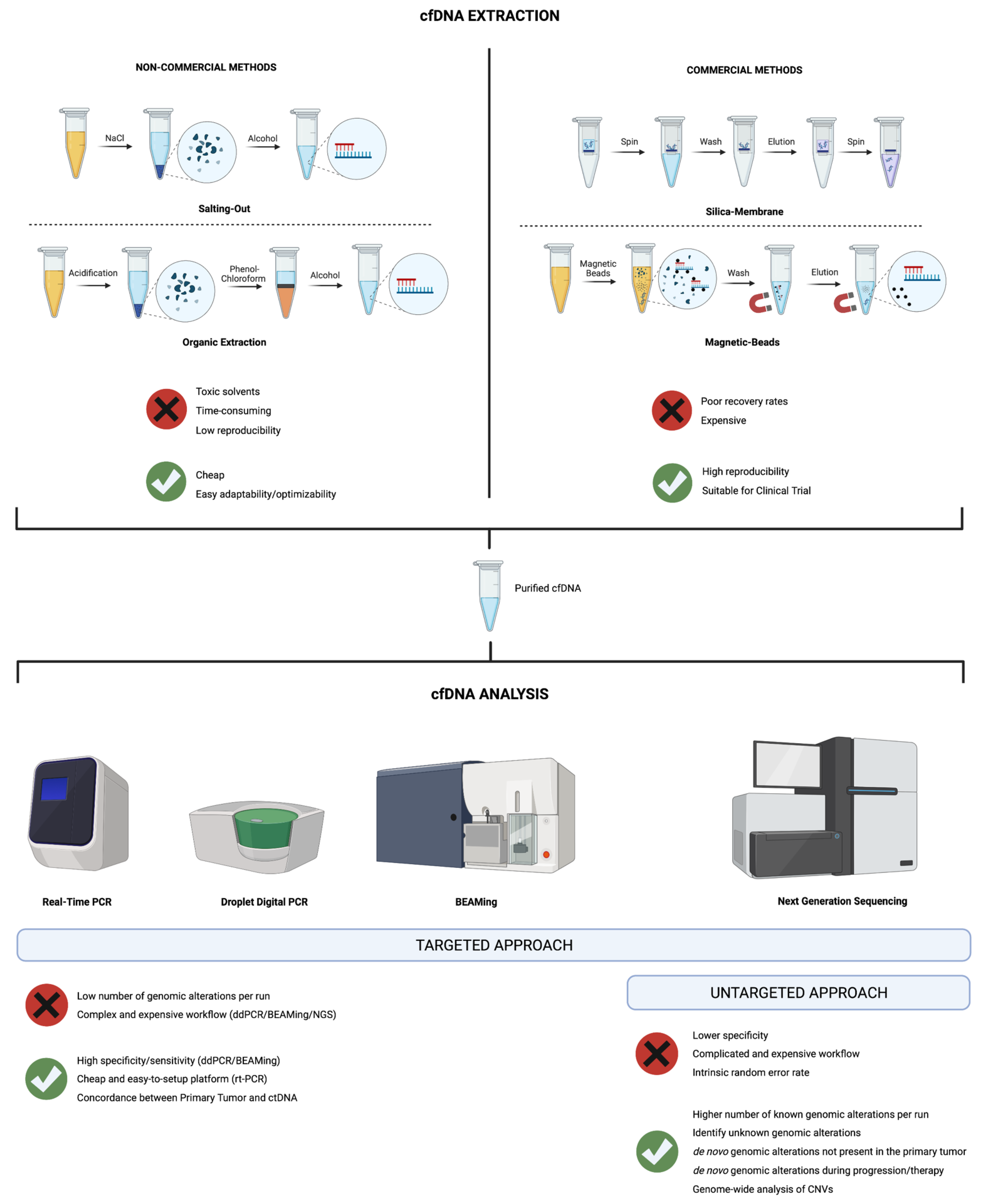
3. Isolation and Analysis of cfDNA
4. Applications of cfDNA Analysis in RCC
4.1. Diagnostic Role
4.1.1. cfDNA Levels in RCC, Healthy Controls and Non-Cancer Disease
4.1.2. cfDNA Integrity as a Tool for Differentiating RCC and Non-Cancer Controls
4.1.3. cfDNA Genetic Alterations
| Author, Year, [Ref.] | Microsatellite Markers | Patients (n) | Controls (n) | Sensitivity (%) | Specificity (%) |
|---|---|---|---|---|---|
| Goessl, 1998, [79] | D3S1307(3p), D3SI560(3p), D3SI289(3p), D3SI300(3p) | 40 RCC | 10 healthy individuals | 63 (at least one MA) 35 (more than one MA) | 100 |
| Eisenberger, 1999, [37] | D1S251 (1pq), HTPO(2p), D3S1317(3p), D3S587(3p), D3S1560(3p), D3S1289(3p), D3S1286 (3p), D3S1038(3p), D4S243(4pq), FGA(4)(4q), CSF(5q), ACTBP2(5p), D8S348(8q), D8S307(8p), D9S747(9p), D9S242(9p), IFNa(9p), D9S162(9p), D11S488(11q), THO(11p), vWA(12p), D13S802(13q), MJD(14q), D17S695(17p), D17S654(17p), D18S51(18q), MBP(18q), D21S1245(21q). | 25 RCC 1 AML 1 MN 3 OCT | 8 individuals with nephrolithiasis 8 healthy individuals | 60 (at least one MA) | 100 |
| von Knobloch, 2002, [80] | D3S1560(3p), D3S2450(3p), D3S3666(3p), D3S2408(3p), D3S1259(3p), D5S1720(5p), D5S1480(5p), D5S476(5p), D5S818(5p), D7S1796(7p), D7S1807(7p), D8S261(8p), D8S560(8p), D9S925(9p), D13S153(13p), D17S799(17p), D17S1306 (17p), D17S783(17p), D17S1298(17p), D17S807(17p) | 53 RCC 1 renal B cell lymphoma 6 TCC | 20 healthy individuals | 74 (using 9 MA) 87 (using 20 MA) | 85 |
| Perego, 2008, [59] | D3S1566(3p), D3S1285(3p), D3S1300(3p), D3S1289(3p), D3S1597(3p) | 48 RCC 1 TCC 5 OCT | 41 healthy individuals | 55.6 (at least one MA) * | NR |
4.1.4. cfDNA Epigenetic Alterations: A New Promising Method to Detect RCC
4.2. Post-Operative Recurrence and Prognostic Role
4.3. Predictive Role
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA A Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Gudbjartsson, T.; Thoroddsen, A.; Petursdottir, V.; Hardarson, S.; Magnusson, J.; Einarsson, G.V. Effect of incidental detection for survival of patients with renal cell carcinoma: Results of population-based study of 701 patients. Urology 2005, 66, 1186–1191. [Google Scholar] [CrossRef] [PubMed]
- Berger, A.; Brandina, R.; Atalla, M.A.; Herati, A.S.; Kamoi, K.; Aron, M.; Haber, G.-P.; Stein, R.J.; Desai, M.M.; Kavoussi, L.R.; et al. Laparoscopic Radical Nephrectomy for Renal Cell Carcinoma: Oncological Outcomes at 10 Years or More. J. Urol. 2009, 182, 2172–2176. [Google Scholar] [CrossRef] [PubMed]
- Eggener, S.E.; Yossepowitch, O.; Pettus, J.A.; Snyder, M.E.; Motzer, R.; Russo, P. Renal Cell Carcinoma Recurrence After Nephrectomy for Localized Disease: Predicting Survival from Time of Recurrence. J. Clin. Oncol. 2006, 24, 3101–3106. [Google Scholar] [CrossRef] [PubMed]
- Konety, B.R. Postoperative surveillance protocol for patients with localized and locally advanced renal cell carcinoma based on a validated prognostic nomogram and risk group stratification system. Urol. Oncol. Semin. Orig. Investig. 2006, 24, 87–88. [Google Scholar] [CrossRef]
- Bosma, N.A.; Warkentin, M.T.; Gan, C.L.; Karim, S.; Heng, D.Y.; Brenner, D.R.; Lee-Ying, R.M. Efficacy and Safety of First-line Systemic Therapy for Metastatic Renal Cell Carcinoma: A Systematic Review and Network Meta-analysis. Eur. Urol. Open Sci. 2022, 37, 14–26. [Google Scholar] [CrossRef] [PubMed]
- Mollica, V.; Santoni, M.; Matrana, M.R.; Basso, U.; De Giorgi, U.; Rizzo, A.; Maruzzo, M.; Marchetti, A.; Rosellini, M.; Bleve, S.; et al. Concomitant Proton Pump Inhibitors and Outcome of Patients Treated with Nivolumab Alone or Plus Ipilimumab for Advanced Renal Cell Carcinoma. Target. Oncol. 2022, 17, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Santoni, M.; Massari, F.; Bracarda, S.; Grande, E.; Matrana, M.R.; Rizzo, M.; De Giorgi, U.; Basso, U.; Aurilio, G.; Incorvaia, L.; et al. Cabozantinib in Patients with Advanced Renal Cell Carcinoma Primary Refractory to First-line Immunocombinations or Tyrosine Kinase Inhibitors. Eur. Urol. Focus 2022, 49, 2405–2415. [Google Scholar] [CrossRef]
- Acosta, A.M.; Sholl, L.M.; Fanelli, G.N.; Gordetsky, J.B.; Baniak, N.; Barletta, J.A.; Lindeman, N.I.; Hirsch, M.S. Intestinal metaplasia of the urinary tract harbors potentially oncogenic genetic variants. Mod. Pathol. 2020, 34, 457–468. [Google Scholar] [CrossRef] [PubMed]
- Coati, I.; Lotz, G.; Fanelli, G.N.; Brignola, S.; Lanza, C.; Cappellesso, R.; Pellino, A.; Pucciarelli, S.; Spolverato, G.; Guzzardo, V.; et al. Claudin-18 expression in oesophagogastric adenocarcinomas: A tissue microarray study of 523 molecularly profiled cases. Br. J. Cancer 2019, 121, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Fanelli, G.N.; Scarpitta, R.; Cinacchi, P.; Fuochi, B.; Szumera-Ciećkiewicz, A.; De Ieso, K.; Ferrari, P.; Fontana, A.; Miccoli, M.; Naccarato, A.G.; et al. Immunohistochemistry for Thymidine Kinase-1 (TK1): A Potential Tool for the Prognostic Stratification of Breast Cancer Patients. J. Clin. Med. 2021, 10, 5416. [Google Scholar] [CrossRef] [PubMed]
- Fassan, M.; Facchin, S.; Munari, G.; Fanelli, G.N.; Lorenzon, G.; Savarino, E. Noncoding RNAs as drivers of the phenotypic plasticity of oesophageal mucosa. World J. Gastroenterol. 2017, 23, 7653–7656. [Google Scholar] [CrossRef]
- Forooshani, M.K.; Scarpitta, R.; Fanelli, G.N.; Miccoli, M.; Naccarato, A.G.; Scatena, C. Is It Time to Consider the Androgen Receptor as a Therapeutic Target in Breast Cancer? Anti-Cancer Agents Med. Chem. 2022, 22, 775–786. [Google Scholar] [CrossRef]
- Pasqualetti, F.; Gonnelli, A.; Orlandi, P.; Palladino, E.; Giannini, N.; Gadducci, G.; Mattioni, R.; Montrone, S.; Calistri, E.; Mazzanti, C.M.; et al. Association of XRCC3 rs1799794 polymorphism with survival of glioblastoma multiforme patients treated with combined radio-chemotherapy. Investig. New Drugs 2021, 39, 1159–1165. [Google Scholar] [CrossRef]
- Nuzzo, P.V.; Buzzatti, G.; Ricci, F.; Rubagotti, A.; Argellati, F.; Zinoli, L.; Boccardo, F. Periostin: A Novel Prognostic and Therapeutic Target For Genitourinary Cancer? Clin. Genitourin. Cancer 2014, 12, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Nuzzo, P.V.; Rubagotti, A.; Zinoli, L.; Salvi, S.; Boccardo, F.M. The prognostic value of stromal and epithelial periostin expression in human breast cancer: Correlation with clinical pathological features and mortality outcome. BMC Cancer 2016, 16, 95. [Google Scholar] [CrossRef] [PubMed]
- Saraggi, D.; Galuppini, F.; Fanelli, G.N.; Remo, A.; Urso, E.D.; Bao, Q.R.; Bacchin, D.; Guzzardo, V.; Luchini, C.; Braconi, C.; et al. MiR-21 up-regulation in ampullary adenocarcinoma and its pre-invasive lesions. Pathol. Res. Pr. 2018, 214, 835–839. [Google Scholar] [CrossRef]
- Fanelli, G.N.; Fassan, M.; Moro, F.D.; Soligo, M.; Munari, G.; Zattoni, F.; Gardiman, M.P.; Prayer-Galetti, T. Thyroid-like follicular carcinoma of the kidney: The mutational profiling reveals a BRAF wild type status. Pathol. Res. Pr. 2019, 215, 152532. [Google Scholar] [CrossRef] [PubMed]
- Fusco, N.; Ragazzi, M.; Sajjadi, E.; Venetis, K.; Piciotti, R.; Morganti, S.; Santandrea, G.; Fanelli, G.N.; Despini, L.; Invernizzi, M.; et al. Assessment of estrogen receptor low positive status in breast cancer: Implications for pathologists and oncologists. Histol. Histopathol. 2021, 36, 1235–1245. [Google Scholar] [CrossRef]
- Penney, K.L.; Tyekucheva, S.; Rosenthal, J.; El Fandy, H.; Carelli, R.; Borgstein, S.; Zadra, G.; Fanelli, G.N.; Stefanizzi, L.; Giunchi, F.; et al. Metabolomics of Prostate Cancer Gleason Score in Tumor Tissue and Serum. Mol. Cancer Res. 2021, 19, 475–484. [Google Scholar] [CrossRef]
- Scatena, C.; Fanelli, G.; Fanelli, G.N.; Menicagli, M.; Aretini, P.; Ortenzi, V.; Civitelli, S.P.; Innocenti, L.; Sotgia, F.; Lisanti, M.P.; et al. New insights in the expression of stromal caveolin 1 in breast cancer spread to axillary lymph nodes. Sci. Rep. 2021, 11, 2755. [Google Scholar] [CrossRef] [PubMed]
- Leon, S.A.; Shapiro, B.; Sklaroff, D.M.; Yaros, M.J. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977, 37, 646–650. [Google Scholar] [PubMed]
- Song, P.; Wu, L.R.; Yan, Y.H.; Zhang, J.X.; Chu, T.; Kwong, L.N.; Patel, A.A.; Zhang, D.Y. Limitations and opportunities of technologies for the analysis of cell-free DNA in cancer diagnostics. Nat. Biomed. Eng. 2022, 6, 232–245. [Google Scholar] [CrossRef] [PubMed]
- Cucchiara, F.; Scarpitta, R.; Crucitta, S.; Scatena, C.; Arici, R.; Naccarato, A.G.; Fogli, S.; Danesi, R.; Del Re, M. Diagnosis and treatment monitoring in breast cancer: How liquid biopsy can support patient management. Pharmacogenomics 2022, 23, 119–134. [Google Scholar] [CrossRef]
- Fanelli, G.N.; Naccarato, A.G.; Scatena, C. Recent Advances in Cancer Plasticity: Cellular Mechanisms, Surveillance Strategies, and Therapeutic Optimization. Front. Oncol. 2020, 10, 569. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, P.; Scatena, C.; Ghilli, M.; Bargagna, I.; Lorenzini, G.; Nicolini, A. Molecular Mechanisms, Biomarkers and Emerging Therapies for Chemotherapy Resistant TNBC. Int. J. Mol. Sci. 2022, 23, 1665. [Google Scholar] [CrossRef] [PubMed]
- Berchuck, J.E.; Baca, S.C.; McClure, H.M.; Korthauer, K.; Tsai, H.K.; Nuzzo, P.V.; Kelleher, K.M.; He, M.; Steinharter, J.A.; Zacharia, S.; et al. Detecting Neuroendocrine Prostate Cancer Through Tissue-Informed Cell-Free DNA Methylation Analysis. Clin. Cancer Res. 2022, 28, 928–938. [Google Scholar] [CrossRef]
- Lasseter, K.; Nassar, A.H.; Hamieh, L.; Berchuck, J.E.; Nuzzo, P.V.; Korthauer, K.; Shinagare, A.B.; Ogorek, B.; McKay, R.; Thorner, A.R.; et al. Plasma cell-free DNA variant analysis compared with methylated DNA analysis in renal cell carcinoma. Genet. Med. 2020, 22, 1366–1373. [Google Scholar] [CrossRef]
- Malapelle, U.; Pisapia, P.; Addeo, A.; Arrieta, O.; Bellosillo, B.; Cardona, A.F.; Cristofanilli, M.; De Miguel-Perez, D.; Denninghoff, V.; Durán, I.; et al. Liquid biopsy from research to clinical practice: Focus on non-small cell lung cancer. Expert Rev. Mol. Diagn. 2021, 21, 1165–1178. [Google Scholar] [CrossRef]
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Klutstein, M.; Nejman, D.; Greenfield, R.; Cedar, H. DNA Methylation in Cancer and Aging. Cancer Res. 2016, 76, 3446–3450. [Google Scholar] [CrossRef] [PubMed]
- Esteller, M. Epigenetics in cancer. N. Engl. J. Med. 2008, 358, 1148–1159. [Google Scholar] [CrossRef]
- Greenberg, M.V.C.; Bourc’His, D. The diverse roles of DNA methylation in mammalian development and disease. Nat. Rev. Mol. Cell Biol. 2019, 20, 590–607. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.Y.; Singhania, R.; Fehringer, G.; Chakravarthy, A.; Roehrl, M.H.A.; Chadwick, D.; Zuzarte, P.C.; Borgida, A.; Wang, T.T.; Li, T.; et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes. Nature 2018, 563, 579–583. [Google Scholar] [CrossRef]
- Slieker, R.C.; Bos, S.D.; Goeman, J.J.; Bovée, J.V.; Talens, R.P.; van der Breggen, R.; Suchiman, H.E.D.; Lameijer, E.-W.; Putter, H.; van den Akker, E.B.; et al. Identification and systematic annotation of tissue-specific differentially methylated regions using the Illumina 450k array. Epigenetics Chromatin 2013, 6, 26. [Google Scholar] [CrossRef]
- Kundaje, A.; Meuleman, W.; Ernst, J.; Bilenky, M.; Yen, A.; Heravi-Moussavi, A.; Kheradpour, P.; Zhang, Z.; Wang, J.; Ziller, M.J.; et al. Integrative analysis of 111 reference human epigenomes. Nature 2015, 518, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Eisenberger, C.F.; Schoenberg, M.; Enger, C.; Hortopan, S.; Shah, S.; Chow, N.-H.; Marshall, F.F.; Sidransky, D. Diagnosis of Renal Cancer by Molecular Urinalysis. JNCI J. Natl. Cancer Inst. 1999, 91, 2028–2032. [Google Scholar] [CrossRef] [PubMed]
- Hoque, M.O.; Begum, S.; Topaloglu, O.; Jeronimo, C.; Mambo, E.; Westra, W.H.; Califano, J.A.; Sidransky, D. Quantitative Detection of Promoter Hypermethylation of Multiple Genes in the Tumor, Urine, and Serum DNA of Patients with Renal Cancer. Cancer Res. 2004, 64, 5511–5517. [Google Scholar] [CrossRef]
- Thierry, A.R.; Mouliere, F.; El Messaoudi, S.; Mollevi, C.; Lopez-Crapez, E.; Rolet, F.; Gillet, B.; Gongora, C.; Dechelotte, P.; Robert, B.; et al. Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat. Med. 2014, 20, 430–435. [Google Scholar] [CrossRef] [PubMed]
- Freidin, M.B.; Freydin, M.; Leung, M.; Fernandez, A.M.; Nicholson, A.G.; Lim, E. Circulating Tumor DNA Outperforms Circulating Tumor Cells for KRAS Mutation Detection in Thoracic Malignancies. Clin. Chem. 2015, 61, 1299–1304. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hid-dessen, A.L.; Legler, T.C.; et al. High-Throughput Droplet Digital PCR System for Absolute Quantitation of DNA Copy Number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef] [PubMed]
- Diehl, F.; Li, M.; He, Y.; Kinzler, K.W.; Vogelstein, B.; Dressman, D. BEAMing: Single-molecule PCR on microparticles in water-in-oil emulsions. Nat. Methods 2006, 3, 551–559. [Google Scholar] [CrossRef] [PubMed]
- Beaver, J.A.; Jelovac, D.; Balukrishna, S.; Cochran, R.L.; Croessmann, S.; Zabransky, D.J.; Wong, H.Y.; Toro, P.V.; Cidado, J.; Blair, B.G.; et al. Detection of Cancer DNA in Plasma of Patients with Early-Stage Breast Cancer. Clin. Cancer Res. 2014, 20, 2643–2650. [Google Scholar] [CrossRef]
- Higgins, M.J.; Jelovac, D.; Barnathan, E.; Blair, B.; Slater, S.; Powers, P.; Zorzi, J.; Jeter, S.C.; Oliver, G.R.; Fetting, J.; et al. Detection of tumor PIK3CA status in metastatic breast cancer using peripheral blood. Clin. Cancer Res. 2012, 18, 3462–3469. [Google Scholar] [CrossRef]
- Oxnard, G.R.; Thress, K.S.; Alden, R.S.; Lawrance, R.; Paweletz, C.P.; Cantarini, M.; Yang, J.C.-H.; Barrett, J.C.; Jänne, P.A. Association Between Plasma Genotyping and Outcomes of Treatment with Osimertinib (AZD9291) in Advanced Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2016, 34, 3375–3382. [Google Scholar] [CrossRef]
- Taniguchi, K.; Uchida, J.; Nishino, K.; Kumagai, T.; Okuyama, T.; Okami, J.; Higashiyama, M.; Kodama, K.; Imamura, F.; Kato, K. Quantitative Detection of EGFR Mutations in Circulating Tumor DNA Derived from Lung Adenocarcinomas. Clin. Cancer Res. 2011, 17, 7808–7815. [Google Scholar] [CrossRef] [PubMed]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef]
- Siravegna, G.; Mussolin, B.; Buscarino, M.; Corti, G.; Cassingena, A.; Crisafulli, G.; Ponzetti, A.; Cremolini, C.; Amatu, A.; Lauricella, C.; et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat. Med. 2015, 21, 795–801. [Google Scholar] [CrossRef] [PubMed]
- Glenn, T.C. Field guide to next-generation DNA sequencers. Mol. Ecol. Resour. 2011, 11, 759–769. [Google Scholar] [CrossRef] [PubMed]
- Leary, R.J.; Sausen, M.; Kinde, I.; Papadopoulos, N.; Carpten, J.D.; Craig, D.; O’Shaughnessy, J.; Kinzler, K.W.; Parmigiani, G.; Vogelstein, B.; et al. Detection of Chromosomal Alterations in the Circulation of Cancer Patients with Whole-Genome Sequencing. Sci. Transl. Med. 2012, 4, 162ra154. [Google Scholar] [CrossRef] [Green Version]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, S.B.; Narayan, A.; Kole, A.J.; Decker, R.H.; Teysir, J.; Carriero, N.J.; Lee, A.; Nemati, R.; Nath, S.K.; Mane, S.M.; et al. Early Assessment of Lung Cancer Immunotherapy Response via Circulating Tumor DNA. Clin. Cancer Res. 2018, 24, 1872–1880. [Google Scholar] [CrossRef]
- Phallen, J.; Sausen, M.; Adleff, V.; Leal, A.; Hruban, C.; White, J.; Anagnostou, V.; Fiksel, J.; Cristiano, S.; Papp, E.; et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 2017, 9, eaan2415. [Google Scholar] [CrossRef] [PubMed]
- Aravanis, A.M.; Lee, M.; Klausner, R.D. Next-Generation Sequencing of Circulating Tumor DNA for Early Cancer Detection. Cell 2017, 168, 571–574. [Google Scholar] [CrossRef]
- Israel, G.M.; Hindman, N.; Bosniak, M.A. Evaluation of Cystic Renal Masses: Comparison of CT and MR Imaging by Using the Bosniak Classification System. Radiology 2004, 231, 365–371. [Google Scholar] [CrossRef]
- Zill, O.A.; Banks, K.C.; Fairclough, S.R.; Mortimer, S.A.; Vowles, J.V.; Mokhtari, R.; Gandara, D.R.; Mack, P.C.; Odegaard, J.I.; Nagy, R.J.; et al. The Landscape of Actionable Genomic Alterations in Cell-Free Circulating Tumor DNA from 21,807 Advanced Cancer Patients. Clin. Cancer Res. 2018, 24, 3528–3538. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.; Zhu, L.; Jiang, Z.; Cheng, K. Monitoring of Plasma Cell-Free DNA in Predicting Postoperative Recurrence of Clear Cell Renal Cell Carcinoma. Urol. Int. 2013, 91, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra224. [Google Scholar] [CrossRef] [PubMed]
- Perego, R.A.; Corizzato, M.; Brambilla, P.; Ferrero, S.; Bianchi, C.; Fasoli, E.; Signorini, S.; Torsello, B.; Invernizzi, L.; Bombelli, S.; et al. Concentration and microsatellite status of plasma DNA for monitoring patients with renal carcinoma. Eur. J. Cancer 2008, 44, 1039–1047. [Google Scholar] [CrossRef] [PubMed]
- De Martino, M.; Klatte, T.; Haitel, A.; Marberger, M. Serum cell-free DNA in renal cell carcinoma. Cancer 2012, 118, 82–90. [Google Scholar] [CrossRef]
- Feng, G.; Ye, X.; Fang, F.; Pu, C.; Huang, H.; Li, G. Quantification of Plasma Cell-Free DNA 1 in Predicting Therapeutic Efficacy of Sorafenib on Metastatic Clear Cell Renal Cell Carcinoma. Dis. Markers 2013, 34, 105–111. [Google Scholar] [CrossRef]
- Skrypkina, I.; Tsyba, L.; Onyshchenko, K.; Morderer, D.; Kashparova, O.; Nikolaienko, O.; Panasenko, G.; Vozianov, S.; Romanenko, A.; Rynditch, A. Concentration and Methylation of Cell-Free DNA from Blood Plasma as Diagnostic Markers of Renal Cancer. Dis. Markers 2016, 2016, 3693096. [Google Scholar] [CrossRef] [PubMed]
- Henriksen, T.V.; Reinert, T.; Christensen, E.; Sethi, H.; Birkenkamp-Demtröder, K.; Gögenur, M.; Gögenur, I.; Zimmermann, B.G.; Dyrskjøt, L.; Andersen, C.L.; et al. The effect of surgical trauma on circulating free DNA levels in cancer patients—implications for studies of circulating tumor DNA. Mol. Oncol. 2020, 14, 1670–1679. [Google Scholar] [CrossRef] [PubMed]
- Van Der Meer, A.J.; Kroeze, A.; Hoogendijk, A.J.; Soussan, A.A.; Van Der Schoot, C.E.; Wuillemin, W.A.; Voermans, C.; Van Der Poll, T.; Zeerleder, S. Systemic inflammation induces release of cell-free DNA from hematopoietic and parenchymal cells in mice and humans. Blood Adv. 2019, 3, 724–728. [Google Scholar] [CrossRef] [PubMed]
- Mondelo-Macía, P.; Castro-Santos, P.; Castillo-García, A.; Muinelo-Romay, L.; Diaz-Peña, R. Circulating Free DNA and Its Emerging Role in Autoimmune Diseases. J. Pers. Med. 2021, 11, 151. [Google Scholar] [CrossRef] [PubMed]
- Hauser, S.; Zahalka, T.; Ellinger, J.; Fechner, G.; Heukamp, L.; Von Ruecker, A.; Müller, S.C.; Bastian, P.J. Cell-free circulating DNA: Diagnostic value in patients with renal cell cancer. Anticancer Res. 2010, 30, 2785–2789. [Google Scholar] [PubMed]
- Gang, F.; Guorong, L.; An, Z.; Anne, G.-P.; Christian, G.; Jacques, T. Prediction of Clear Cell Renal Cell Carcinoma by Integrity of Cell-free DNA in Serum. Urology 2010, 75, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Busch, J.; Jung, M.; Rabenhorst, S.; Ralla, B.; Kilic, E.; Mergemeier, S.; Budach, N.; Fendler, A.; Jung, K. Diagnostic and prognostic potential of circulating cell-free genomic and mitochondrial DNA fragments in clear cell renal cell carcinoma patients. Clin. Chim. Acta 2016, 452, 109–119. [Google Scholar] [CrossRef]
- Pal, S.K.; Sonpavde, G.; Agarwal, N.; Vogelzang, N.J.; Srinivas, S.; Haas, N.B.; Signoretti, S.; McGregor, B.A.; Jones, J.; Lanman, R.B.; et al. Evolution of Circulating Tumor DNA Profile from First-line to Subsequent Therapy in Metastatic Renal Cell Carcinoma. Eur. Urol. 2017, 72, 557–564. [Google Scholar] [CrossRef]
- Maia, M.C.; Bergerot, P.G.; Dizman, N.; Hsu, J.; Jones, J.; Lanman, R.B.; Banks, K.C.; Pal, S.K. Association of Circulating Tumor DNA (ctDNA) Detection in Metastatic Renal Cell Carcinoma (mRCC) with Tumor Burden. Kidney Cancer 2017, 1, 65–70. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, Y.; Uemura, M.; Nakano, K.; Hayashi, Y.; Wang, C.; Ishizuya, Y.; Kinouchi, T.; Hayashi, T.; Matsuzaki, K.; Jingushi, K.; et al. Increased level and fragmentation of plasma circulating cell-free DNA are diagnostic and prognostic markers for renal cell carcinoma. Oncotarget 2018, 9, 20467–20475. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, Y.; Xu, B.; Li, F.; Wang, Y.; Li, M.; Du, R.; Zhou, Y.; Salgia, M.; Yang, L.; et al. Circulating tumor DNA analysis of metastatic renal cell carcinoma. Mol. Clin. Oncol. 2020, 14, 16. [Google Scholar] [CrossRef] [PubMed]
- Corrò, C.; Hejhal, T.; Poyet, C.; Sulser, T.; Hermanns, T.; Winder, T.; Prager, G.; Wild, P.J.; Frew, I.; Moch, H.; et al. Detecting circulating tumor DNA in renal cancer: An open challenge. Exp. Mol. Pathol. 2017, 102, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.G.; Moser, T.; Mouliere, F.; Field-Rayner, J.; Eldridge, M.; Riediger, A.L.; Chandrananda, D.; Heider, K.; Wan, J.; Warren, A.Y.; et al. Comprehensive characterization of cell-free tumor DNA in plasma and urine of patients with renal tumors. Genome Med. 2020, 12, 23. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.C.M.; Heider, K.; Gale, D.; Murphy, S.; Fisher, E.; Mouliere, F.; Ruiz-Valdepenas, A.; Santonja, A.; Morris, J.; Chandrananda, D.; et al. ctDNA monitoring using patient-specific sequencing and integration of variant reads. Sci. Transl. Med. 2020, 12. [Google Scholar] [CrossRef] [PubMed]
- Hahn, A.W.; Gill, D.M.; Maughan, B.; Agarwal, A.; Arjyal, L.; Gupta, S.; Streeter, J.; Bailey, E.; Pal, S.K.; Agarwal, N. Correlation of genomic alterations assessed by next-generation sequencing (NGS) of tumor tissue DNA and circulating tumor DNA (ctDNA) in metastatic renal cell carcinoma (mRCC): Potential clinical implications. Oncotarget 2017, 8, 33614–33620. [Google Scholar] [CrossRef]
- Bacon, J.V.; Annala, M.; Soleimani, M.; Lavoie, J.-M.; So, A.; Gleave, M.E.; Fazli, L.; Wang, G.; Chi, K.N.; Kollmannsberger, C.K.; et al. Plasma Circulating Tumor DNA and Clonal Hematopoiesis in Metastatic Renal Cell Carcinoma. Clin. Genitourin. Cancer 2020, 18, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Mouliere, F.; Chandrananda, D.; Piskorz, A.M.; Moore, E.K.; Morris, J.; Ahlborn, L.B.; Mair, R.; Goranova, T.; Marass, F.; Heider, K.; et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci. Transl. Med. 2018, 10, eaat4921. [Google Scholar] [CrossRef] [PubMed]
- Goessl, C.; Heicappell, R.; Münker, R.; Anker, P.; Stroun, M.; Krause, H.; Müller, M.; Miller, K. Microsatellite analysis of plasma DNA from patients with clear cell renal carcinoma. Cancer Res. 1998, 58, 4728–4732. [Google Scholar] [PubMed]
- von Knobloch, R.; Hegele, A.; Brandt, H.; Varga, Z.; Wille, S.; Kälble, T.; Heidenreich, A.; Hofmann, R. High frequency of serum DNA alterations in renal cell carcinoma detected by fluorescent microsatellite analysis. Int. J. Cancer 2002, 98, 889–894. [Google Scholar] [CrossRef] [PubMed]
- Lasseigne, B.N.; Brooks, J.D. The Role of DNA Methylation in Renal Cell Carcinoma. Mol. Diagn. Ther. 2018, 22, 431–442. [Google Scholar] [CrossRef]
- Hauser, S.; Zahalka, T.; Fechner, G.; Müller, S.C.; Ellinger, J. Serum DNA hypermethylation in patients with kidney cancer: Results of a prospective study. Anticancer Res. 2013, 33, 4651–4656. [Google Scholar] [PubMed]
- Liu, M.C.; Oxnard, G.R.; Klein, E.A.; Swanton, C.; Seiden, M.V. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 2020, 31, 745–759. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.Y.; Burgener, J.M.; Bratman, S.V.; De Carvalho, D.D. Preparation of cfMeDIP-seq libraries for methylome profiling of plasma cell-free DNA. Nat. Protoc. 2019, 14, 2749–2780. [Google Scholar] [CrossRef] [PubMed]
- Nuzzo, P.V.; Berchuck, J.E.; Korthauer, K.; Spisak, S.; Nassar, A.H.; Alaiwi, S.A.; Chakravarthy, A.; Shen, S.Y.; Bakouny, Z.; Boccardo, F.; et al. Detection of renal cell carcinoma using plasma and urine cell-free DNA methylomes. Nat. Med. 2020, 26, 1041–1043. [Google Scholar] [CrossRef]
- Zuccato, J.A.; Patil, V.; Mansouri, S.; Liu, J.C.; Nassiri, F.; Mamatjan, Y.; Chakravarthy, A.; Karimi, S.; Almeida, J.P.; Bernat, A.-L.; et al. DNA methylation-based prognostic subtypes of chordoma tumors in tissue and plasma. Neuro-oncology 2022, 24, 442–454. [Google Scholar] [CrossRef] [PubMed]
- Nassiri, F.; Chakravarthy, A.; Feng, S.; Shen, S.Y.; Nejad, R.; Zuccato, J.A.; Voisin, M.R.; Patil, V.; Horbinski, C.; Aldape, K.; et al. Detection and discrimination of intracranial tumors using plasma cell-free DNA methylomes. Nat. Med. 2020, 26, 1044–1047. [Google Scholar] [CrossRef] [PubMed]
- Gonzalgo, M.L.; Eisenberger, C.F.; Lee, S.M.; Trock, B.J.; Marshall, F.F.; Hortopan, S.; Sidransky, D.; Schoenberg, M.P. Prognostic significance of preoperative molecular serum analysis in renal cancer. Clin. Cancer Res. 2002, 8, 1878–1881. [Google Scholar] [PubMed]
- Rouvinov, K.; Mermershtain, W.; Dresler, H.; Ariad, S.; Riff, R.; Shani-Shrem, N.; Keizman, D.; Douvdevani, A. Circulating Cell-Free DNA Levels in Patients with Metastatic Renal Cell Carcinoma. Oncol. Res. Treat. 2017, 40, 707–710. [Google Scholar] [CrossRef]
| Author, Year, [Ref.] | Gene | Methylated /RCC Patients (n), (%) | Methylated /Controls (n), (%) | Sensitivity (%) | Specificity (%) | Cutoff Value | AUC | Sample Source, Amount | Detection Method |
|---|---|---|---|---|---|---|---|---|---|
| Hoque, 2004, [38] | APC | 1/18 (5.5) | 1/30 (3.3) | 5.5 | 96.7 | 4.5 | NR | Serum, NR | PCR |
| ARF | 1/18 (5.5) | 1/30 (3.3) | 5.5 | 96.7 | 0 | NR | |||
| CDH1 | 6/18 (33.3) | 2/30 (6.6) | 33.3 | 93.4 | 0.3 | NR | |||
| GSTP1 | 1/18 (5.5) | 0/30 (0) | 5.5 | 100 | 0 | NR | |||
| MGMT | 0/18 (0) | 1/30 (3.3) | 0 | 96.7 | 0 | NR | |||
| p16 | 4/18 (22.2) | 0/30 (0) | 22.2 | 100 | 0 | NR | |||
| RAR-82 | 1/18 (5.5) | 0/30 (0) | 5.5 | 100 | 0.1 | NR | |||
| RASSF1A | 2/18 (11.1) | 1/30 (3.3) | 11.1 | 96.7 | 0.1 | NR | |||
| TIMP3 | 3/18 (16.6) | 0/30 (0) | 17 | 100 | 1 | NR | |||
| De Martino, 2011, [60] | RASSF1A | 72/157 (45.9) | 3/43 (7) | 45.9 | 93 | 0 | 0.694 | Serum, 1 mL | qPCR |
| PTGS2 | 60/157 (38.2) | 15/43 (34.9) | 38.2 | 65.1 | 0 | 0.517 | |||
| P16 | 73/157 (46.5) | 19/43 (44.2) | 46.5 | 55.8 | 0 | 0.512 | |||
| VHL | 79/157 (50.3) | 4/43 (8.3) | 50.3 | 90.7 | 0 | 0.705 | |||
| Hauser, 2013, [82] | APC | 19/35(54.3) | 5/54 (9.3) | 54.3 | 90.7 | 0.37 | 0.72 | Serum, 1 mL | PCR |
| GSTP1 | 6/35(17.1) | 1/54(1.9) | 17.1 | 98.1 | 0.75 | 0.57 | |||
| p14(ARF) | 5/35(14.3) | 0/54(0) | 14.3 | 100 | 0.26 | 0.57 | |||
| P16 | 9/35(25.7) | 9/54(16.7) | 25.7 | 83.3 | 0 | NR | |||
| PTGS2 | 8/35(22.9) | 2/54(3.7) | 22.9 | 96.3 | 0.47 | 0.59 | |||
| RAR-B | 14/35(40) | 8/54(14.8) | 40.0 | 85.2 | 0.19 | 0.61 | |||
| RASSF1A | 8/35(22.9) | 1/54(1.9) | 22.9 | 98.2 | 0.09 | 0.60 | |||
| TIMP3 | 20/35(57.1) | 21/54(38.9) | 57.1 | 61.1 | 0 | NR | |||
| Skrypkina, 2016, [62] | APC | 14/27 (51.9) | 1/15(6.7) | 51.9 | 93.3 | 0 | NR | Plasma, 2 mL | PCR |
| FHIT | 15/27 (55.6) | 0/15 (0) | 55.6 | 100 | 0 | NR | |||
| ITGA9 | 0/27(0) | 0/15 (0) | 0 | 100 | 0 | NR | |||
| LRRC3B | 20/27 (74.1) | 5/15 (33.3) | 74.1 | 66.7 | 0 | NR | |||
| RASSF1 | 17/27 (63.0) | 1/15 (6.7) | 62.9 | 93.3 | 0 | NR | |||
| VHL | 0/27 (0) | 0/15 (0) | 0 | 100 | 0 | NR |
| DIAGNOSTIC ROLE OF cfDNA IN RCC | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| cfDNA Feature Analyzed | Author, Year, [Ref.] | Study Type | Purpose | Patients (n) | Controls | Sample Source, Amount | Timing Sample | cfDNA Isolation Method | Detection Method | Results |
| cfDNA levels | Perego, 2008, [59] | Prospective | Diagnostic Prognostic | 48 RCC (stages I–IV), 1 TCC, 5 OCT | 41 healthy individuals | Plasma, 1 mL | Pre-operative Post-operative | QIAamp DNA Mini kit (Qiagen) | qPCR | RCC patients had a higher plasma cfDNA concentration compared to the healthy controls. Plasma DNA concentration decreased after nephrectomy. During follow-up, plasma DNA increased in 12 patients without evidence of neoplasia: 3 patients successively relapsed. Pre-operative plasma DNA of 9 patients harbored LOH in 5 cases (55.6%). Augmented plasma DNA of 7 patients displayed LOH in 3 cases (42.9%) at follow-up, and in 1 case preceded the recurrence of the disease. |
| De Martino, 2012, [60] | Prospective | Diagnostic Prognostic | 157 RCC (stage I–IV) | 43 benign renal tumors | Serum, 1 mL | Pre-operative | QIAampUltrasens Virus kit (Qiagen) | PCR | Total cfDNA levels and CpG island methylation of RASSF1A and VHL were highly diagnostic for RCC. cfDNA levels were associated with poorer DFS. RASSF1A and VHL methylation was not associated with DFS. | |
| Feng, 2013, [61] | Prospective | Diagnostic Predictive | 18 RCC (stage IV) | 10 healthy individuals | Plasma, 0.4 mL | Six timepoints during treatment with Sorafenib: before treatment, 4–8–12–16–24 weeks | QIAamp DNA Blood Mini Kit (Qiagen) | qPCR | Pretreatment cfDNA levels were increased in RCC vs. healthy individuals and were associated with stage, grade, and the number of metastases. cfDNA levels from weeks 8 to 24 of treatment were higher in those with disease progression than in those with stable disease or partial response. Levels of cfDNA at 8 weeks were predictive of progression. | |
| Wan, 2013, [57] | Prospective | Diagnostic Prognostic | 92 RCC (stage I–IV) | 44 healthy individuals | Plasma, 0.4 mL | Pre-operative Post-operative | QIAamp DNA Blood Mini Kit (Qiagen) | qPCR | Pretreatment levels of plasma cfDNA in pts with mRCC were significantly higher than in those with localized RCC or healthy individuals. cfDNA levels were associated with Fuhrman grade, TNM stage, and tumor size. Of pts with localized RCC, those with recurrence had a significantly higher plasma cfDNA level than those without. The pts with a high plasma cfDNA level had a significantly higher recurrence rate than those with a low plasma cfDNA level before and after nephrectomy. | |
| Skrypkina, 2016, [62] | Prospective | Diagnostic | 27 RCC (stage I–IV) | 15 healthy individuals | Plasma, 2 mL | Pre-operative | Proba Na kit (DNA-Technology) | qPCR | Levels of cfDNA levels were significantly higher in RCC pts than in controls. Hypermethylation of CpG islands of LRRC3B, APC, FHIT, and RASSF1 genes was detected in RCC and was not correlated with clinicopathologic features. | |
| cfDNA integrity | Hauser, 2010, [66] | Prospective | Diagnostic Prognostic | 35 RCC (stage I–IV) | 54 healthy individuals | Serum, 1 mL | Pre-operative | ChargeSwitchgDNA Kit (Invitrogen) | qPCR | cfDNA integrity (ACTB-384/ACTB-106 ratio) analysis distinguished between RCC and healthy controls. High level of ACTB384 compared to ACTB106 was found in RCC patients. No significant correlation of cfDNA levels and DNA integrity with pT-stage, grading or histological subtype was observed. |
| Gang, 2010, [67] | Prospective | Diagnostic Prognostic | 78 RCC (stage I–III) | 42 healthy individuals | Serum, 0.4 mL | Pre-operative Post-operative | QIAamp DNA Blood Mini Kit (Qiagen) | PCR | cfDNA integrity distinguished between RCC and healthy controls and was correlated with tumor stage and size. | |
| Lu, 2016, [68] | Retrospective | Diagnostic Prognostic | 229 RCC (stage I-IV) | 40 healthy individuals | Plasma, 1 mL | Pre-operative Pre-therapy | QIAamp Circulating Nucleic Acid Kit (Qiagen) | PCR | cfDNA integrity did not differ in metastatic, non-metastatic and controls, but decreased from controls to metastatic patients. | |
| cfDNA genetic alterations | Goessl, 1998, [79] | Prospective | Diagnostic | 40 RCC (stages I–IV) | 10 healthy individuals | Plasma, 1 mL | Pre-operative | Qiamp Blood Kit (Qiagen) | PCR | Analysis of LOH and microsatellite instability of four chromosome 3p microsatellites: LOH was found in at least 1 locus in 63% of pts, and 35% exhibited LOH at more than one locus. Microsatellite instability of plasma cfDNA was detected in 3% of the patient. No alterations were found in the controls. |
| Eisenberger, 1999, [37] | Prospective | Diagnostic | 25 RCC (stages I–II–III), 1 AML, 1 MN, 3 OCT | 8 individuals with nephrolithiasis 8 healthy individuals | Serum, NR Urine, NR | Pre-operative | Digestion methods with proteinase K (Boehringer Mannheim GmbH, Mannheim, Germany) in the presence of sodium dodecyl sulfate, followed by phenol–chloroform extraction and ethanol precipitation | PCR | Analysis of LOH and microsatellite instability of 28 microsatellites markers analysis: 60% of RCC had one or more microsatellite cfDNA alterations in the serum. No alterations were found in the controls. | |
| von Knobloch, 2002, [80] | Prospective | Diagnostic | 53 RCC (stages I–II–III), 1 renal B cell lymphoma, 6 TCC | 20 healthy individuals | Serum, 2–4 mL | Pre-operative | Qiamp Midi-Kit (Qiagen) | PCR | Analysis of microsatellite instability of 20 microsatellites markers analysis: serum cfDNA alterations was detected in 74% of cases using 9 markers and 15% of controls using 10 markers. | |
| Perego, 2008, [59] | Prospective | Diagnostic Prognostic | 48 RCC (stages I–IV), 1 TCC, 5 OCT | 41 healthy individuals | Plasma, 1 mL | Pre-operative Post-operative | QIAamp DNA Mini kit (Qiagen) | PCR | RCC pts had a higher plasma cfDNA concentration compared to the healthy controls. Plasma DNA concentration decreased after nephrectomy. During follow-up, plasma DNA increased in 12 patients without evidence of neoplasia and 3 patients successively relapsed. Pre-operative plasma DNA of 9 patients harbored LOH in 5 cases (55.6%). Augmented plasma DNA of 7 patients displayed LOH in 3 cases (42.9%) at follow-up, and in 1 case preceded the recurrence of disease. | |
| Bettegowda, 2014, [58] | Prospective | Diagnostic | 5 RCC (stage IV) | - | Plasma, 2 mL | - | QIAamp Circulating Nucleic Acid Kit (Qiagen) | PCR | ctDNA from RCC showed the lowest levels compared to other cancers. | |
| Pal, 2017, [69] | Prospective | Diagnostic Predictive | 220 RCC (stage IV) | - | Plasma, 1.5–2 mL | Pre-therapy Post-therapy | QIAamp Circulating Nucleic Acid Kit (Qiagen) | Targeted sequencing (73 genes) | GAs were detected in 78.6% of patients. GAs in TP53 and NF1 increased with subsequent therapies. | |
| Maia, 2017, [70] | Retrospective | Diagnostic Predictive | 34 RCC (stage IV) | - | Plasma, 1.5–2 mL | Pre-therapy Post-therapy | QIAamp Circulating Nucleic Acid Kit (Qiagen) | Targeted sequencing (73 genes) | GAs were detected in 53% pf patients. cfDNA showed to be is a surrogate of tumor burden burden. No associations were found between IMDC risk, histology or treatment type and presence/absence of cfDNA | |
| Hahn, 2017, [76] | Prospective | Diagnostic | 19 RCC (stage IV) | - | Plasma, 1.5–2 mL | Pre-operative | QIAamp Circulating Nucleic Acid Kit (Qiagen) | Targeted sequencing (73 genes) | The median GAs rate in cfDNA was detected 2.2% of patients. Median mutation rate was similar between cfDNA and tumor tissue whereas concordance rate was 8.6% | |
| Corrò, 2017, [73] | Prospective | Diagnostic | 9 RCC (stage I–III) | - | Plasma, 1 mL Serum, 1 mL | Pre-operative | QIAamp Circulating free DNA Kit (Qiagen) | PCR | The VHL mutation in plasma or serum was detected in 1one patients | |
| Mouliere, 2018, [78] | Prospective | Diagnostic | 33 RCC (stage not available) | 65 healthy controls | Plasma, 2 mL | Pre-operative | QIAamp Circulating Nucleic Acid Kit (Qiagen) | Low-pass whole-genome sequencing | Enrichment of ctDNA in fragment sizes between 90 and 150 bp improved RCC detection | |
| Yamamoto, 2019, [71] | Prospective | Diagnostic Prognostic | 53 RCC (stage III–IV) | - | Plasma, 1–3 mL | Pre-therapy Post-therapy | QIAamp Circulating Nucleic Acid Kit (Qiagen) | ddPCR and targeted sequencing (48 genes) | GAs were detected in 30% of patients. ctDNA status and cfDNA fragmentation were associated with PFS and OS. Patients with detectable ctDNA had poor responses to therapy | |
| Lasseter, 2020, [28] | Retrospective | Diagnostic | 40 RCC (stage IV) | 34 healthy controls | Plasma, 1 mL | Pre-therapy Post-therapy | QIAamp Circulating Nucleic Acid Kit (Qiagen) | Targeted sequencing (27 genes) and cfMeDIP-seq | Genetic variants were found in 21% of the patients. cfMeDIP-Seq performed in 34 RCC patientsdetected all RCC cases (sensitivity 100%, specificity 88%) | |
| Zhang, 2020, [72] | Prospective | Diagnostic | 50 RCC (stage IV) | - | Plasma, NA | During therapy | QIAamp Circulating Nucleic Acid Kit (Qiagen) | Targeted sequencing (120 genes) | GAs were identified in all 50 patients. The number of GAs was significantly associated with the number of lines of therapy | |
| Bacon, 2020, [77] | Prospective | Diagnostic Prognostic | 55 RCC (stage IV) | - | Plasma, 1.5–2 mL | Pre-therapy | QIAamp Circulating Nucleic Acid Kit (Qiagen) | Targeted sequencing (981 cancer genes) | ctDNA was detected in 33% of patients and the average VAF was 3.9% Patients with detectable ctDNA had shorter PFS and OS | |
| Smith, 2020, [74] | Prospective | Diagnostic | 29 RCC (stage I–IV) | - | Plasma, 2 mL | Pre-operative Post-operative | QIAamp Circulating Nucleic Acid Kit (Qiagen) | Targeted sequencing (297 genes) | cfDNA levels in RCC were significantly lower that in other analyzed cancers of similar size and stage. Targeted sequecing methods detected 27.5% of RCC patients. Post-operative cfDNA was correlated with clinical response to treatment. | |
| Wan, 2020, [75] | Prospective | Diagnostic | 24 RCC (stage I–II) | 45 healthy controls | Plasma, 1–2 mL | Pre-operative Post-operative | QIAamp Circulating Nucleic Acid Kit (Qiagen) | Targeted sequencing | cfDNA from RCC had the lowest levels compared to other analyzed cancers. AUC of the method was 0.66 | |
| cfDNA epigenetic alterations | Hoque, 2004, [38] | Prospective | Diagnostic | 18 RCC (stages I–IV) | 30 healthy individuals | Serum, 1 mL Urine, NR | Pre-operative | Digestion methods with proteinase K (Boehringer Mannheim GmbH, Mannheim, Germany) in the presence of sodium dodecyl sulfate, followed by phenol–chloroform extraction and ethanol precipitation | qPCR | Aberrant methylation of nine gene promoters’ analysis: 67% of RCC pts and 1% of controls were methylation positive for at least one gene tested |
| De Martino, 2012, [60] | Prospective | Diagnostic Prognostic | 157 RCC (stage I–IV) | 43 benign renal tumors | Serum, 1 mL | Pre-operative | QIAampUltrasens Virus kit (Qiagen) | PCR | cfDNA levels and CpG island methylation of RASSF1A and VHL were highly diagnostic for RCC. cfDNA levels were associated with poorer DFS. RASSF1A and VHL methylation was not associated with DFS. | |
| Hauser, 2013, [82] | Prospective | Diagnostic | 35 RCC (stage I–IV) | 54 healthy individuals | Serum, 1 mL | Pre-operative | ChargeSwitchgDNA Kit (Invitrogen) | PCR | cfDNA methylation analysis in eight selected genes (APC, GSTP1, p14(ARF), p16, RAR-B, RASSF1A, TIMP3, PTGS2): in 30 of 35 pts with RCC, at least one gene was methylated. All genes, except p16 and TIMP3, were significantly methylated in RCC pts compared to healthy individuals. APC was correlated with advanced tumor stage | |
| Skrypkina, 2016, [62] | Prospective | Diagnostic | 27 RCC (stage I–IV) | 15 healthy individuals | Plasma, 2 mL | Pre-operative | Proba Na kit (DNA-Technology) | qPCR | Levels of cfDNA were significantly higher in RCC patients than in controls. Hypermethylation of CpG islands of LRRC3B, APC, FHIT, and RASSF1 genes was detected in RCC and was not correlated with clinicopathologic features. | |
| Liu, 2020, [83] | Prospective | Diagnostic | 56 RCC in the training cohort 25 RCC in the validation cohort | 1521 healthy individuals in the training cohort 610 healthy individuals in the validation cohort | Plasma, up to 10 mL | Pre-operative | QIAamp Circulating Nucleic Acid Kit (Qiagen) | WGBS (103,456 regions identified) | WGBS from multiple cancer types was used to build a classifier to identify the cfDNA and the TOO. cfDNA from RCC has the lowest detection, but TOO was classified correctly from all RCC patients. | |
| Lasseter, 2020, [28] | Retrospective | Diagnostic | 40 RCC (stage IV) | 34 healthy controls | Plasma, 1 mL | Pre-therapy Post-therapy | QIAamp Circulating Nucleic Acid Kit (Qiagen) | Tumor sequencing (27 genes) and cfMeDIP-seq | Genetic variants were found in 21% of the patients. cfMeDIP-Seq perfomed in 34 RCC patients detected all RCC cases (sensitivity 100%, specificity 88%) | |
| Nuzzo, 2020, [85] | Retrospective | Diagnostic | 99 RCC (stage I–IV) | 28 healthy controls | Plasma, 1 mL | Pre-operative | QIAamp Circulating Nucleic Acid Kit (Qiagen) | cfMeDIP-seq | Based on a classifier built on top 300 differentially methylated regions, cfMeDIP-seq detected cfDNA RCC in 97% of the patients | |
| PROGNOSTIC ROLE OF cfDNA IN RCC | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Author, Year | Study Type | Purpose | Patients (n) | Controls | Sample Source, Amount | Timing Sample | cfDNA Isolation Method | Detection Method | Results |
| Gonzalgo, 2002, [88] | Retrospective | Prognostic | 25 RCC (stages I–II–III), 1 AML, 1 MN, 3 OCT | _ | Serum, NR Urine, NR | Pre-operative | Digestion methods with proteinase K (Boehringer Mannheim GmbH, Mannheim, Germany) in the presence of sodium dodecyl sulfate, followed by phenol–chloroform extraction and ethanol precipitation | PCR | Analysis of 28 microsatellites markers in RCC patients who had recurrent disease 2 years after nephrectomy: recurrence disease was detected in 7% of patients with no detectable pre-operative serum LOH, 17% with 1 detectable serum LOH and 100% with 2 or 3 detectable serum LOH. |
| Perego, 2008, [59] | Prospective | Diagnostic Prognostic | 48 RCC (stages I–IV), 1 TCC, 5 OCT | 41 healthy individuals | Plasma, 1 mL | Pre-operative Post-operative | QIAamp DNA Mini kit (Qiagen) | qPCR | RCC pts had a higher plasma cfDNA concentration compared to the healthy controls. Plasma DNA concentration decreased after nephrectomy. During follow-up, plasma DNA increased in 12 patients without evidence of neoplasia: 3 patients successively relapsed. Pre-operative plasma DNA of 9 patients harbored LOH in 5 cases (55.6%). Augmented plasma DNA of 7 patients displayed LOH in 3 cases (42.9%) at follow-up, and in 1 case preceded the recurrence of disease. |
| De Martino, 2012, [60] | Prospective | Diagnostic Prognostic | 157 RCC (stage I–IV) | 43 benign renal tumors | Serum, 1 mL | Pre-operative | QIAampUltrasens Virus kit (Qiagen) | PCR | Total cfDNA levels and CpG island methylation of RASSF1A and VHL were highly diagnostic for RCC. cfDNA levels were associated with poorer DFS. RASSF1A and VHL methylation was not associated with DFS. |
| Wan, 2013, [57] | Prospective | Prognostic | 92 RCC (stage I–IV) | 44 healthy individuals | Plasma, 0.4 mL | Pre-operative Post-operative | QIAamp DNA Blood Mini Kit (Qiagen) | qPCR | Pretreatment levels of plasma cfDNA in pts with mRCC were significantly higher than in those with localized RCC or nealthy individuals. cfDNA levels were associated with Fuhrman grade, TNM stage, and tumor size. Of pts with localized RCC, those with recurrence had a significantly higher plasma cfDNA level than those without. The pts with a high plasma cfDNA level had a significantly higher recurrence rate than those with a low plasma cfDNA level before and after nephrectomy. |
| Yamamoto, 2019, [71] | Prospective | Diagnostic Prognostic | 53 RCC (stage III–IV) | - | Plasma, 1–3 mL | Pre-therapy Post-therapy | QIAamp Circulating Nucleic Acid Kit (Qiagen) | ddPCR and targeted sequencing (48 genes) | GAs were detected in 30% of patients. cfDNA status and cfDNA fragmentation were associated with PFS and OS. Patients with detectable ctDNA had poor responses to therapy |
| Bacon, 2020, [77] | Prospective | Diagnostic Prognostic | 55 RCC (stage IV) | - | Plasma, 1.5–2 mL | Pre-therapy | QIAamp Circulating Nucleic Acid Kit (Qiagen) | Targeted sequencing (981 cancer genes) | cfDNA was detected in 33% of patients and the average VAF was 3.9% Patients with detectable cfDNA had shorter PFS and OS |
| Smith, 2020, [74] | Prospective | Diagnostic | 29 RCC (stage I–IV) | - | Plasma, 2 mL | Pre-operative Post-operative | QIAamp Circulating Nucleic Acid Kit (Qiagen) | Targeted sequencing (297 variants) | cfDNA levels in RCC were significantly lower than in other analyzed cancers of similar size and stage. Targeted sequecing methods detected 27.5% of RCC patients. Post-operative cfDNA was coreelated with clinical response to treatment. |
| PREDICTIVE ROLE OF cfDNA IN RCC | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Author, Year | Study Type | Purpose | Patients (n) | Controls | Sample Source, Amount | Timing Sample | cfDNA Isolation Method | Detection Method | Results |
| Feng, 2013, [61] | Prospective | Diagnostic Predictive | 18 RCC (stage IV) | 10 healthy individuals | Plasma, 0.4 mL | Six timepoints during treatment with Sorafenib: before treatment, 4–8–12–16–24 weeks | QIAamp DNA Blood Mini Kit (Qiagen) | qPCR | Pretreatment cfDNA levels were increased in RCC vs. healthy individual and were associated with stage, grade, and number of metastases. cfDNA levels from weeks 8 to 24 of treatment were higher in those with disease progression than in those with stable disease or partial response. Levels of cfDNA at 8 weeks were predictive of progression. |
| Rouvinov, 2017, [89] | Prospective | Predictive | 23 RCC (stage IV) | - | Serum, 1 mL | Six timepoints during treatment with targeted therapy: before treatment, 4–8–12–16–24 weeks | QIAamp Blood Kit (Qiagene) | qPCR | Patients with normal pretreatment cfDNA level had a better PFS versus patients with increased levels. In multivariate analysis, cfDNA levels was associated with PFS. |
| Pal, 2017, [69] | Prospective | Predictive | 220 RCC (stage IV) | - | Plasma, 1.5–2 mL | Pre-therapy Post-therapy | QIAamp Circulating Nucleic Acid Kit (Qiagen) | Targeted sequencing (73 genes) | GAs were detected in 78.6% of patients. The number of GAs was not correlated to line of therapy, but GAs in TP53 and NF1 increased with subsequent therapies. |
| Maia, 2017, [70] | Retrospective | Predictive | 34 RCC (stage IV) | - | Plasma, 1.5–2 mL | Pre-therapy Post-therapy | QIAamp Circulating Nucleic Acid Kit (Qiagen) | Targeted sequencing (73 genes) | GAs were detected in 53% pf patients. cfDNA is a surrogate of tumor burner. No associations were found between ctDNA and IMDC risk, histology and response to treatment. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Francini, E.; Fanelli, G.N.; Pederzoli, F.; Spisak, S.; Minonne, E.; Raffo, M.; Pakula, H.; Tisza, V.; Scatena, C.; Naccarato, A.G.; et al. Circulating Cell-Free DNA in Renal Cell Carcinoma: The New Era of Precision Medicine. Cancers 2022, 14, 4359. https://doi.org/10.3390/cancers14184359
Francini E, Fanelli GN, Pederzoli F, Spisak S, Minonne E, Raffo M, Pakula H, Tisza V, Scatena C, Naccarato AG, et al. Circulating Cell-Free DNA in Renal Cell Carcinoma: The New Era of Precision Medicine. Cancers. 2022; 14(18):4359. https://doi.org/10.3390/cancers14184359
Chicago/Turabian StyleFrancini, Edoardo, Giuseppe Nicolò Fanelli, Filippo Pederzoli, Sandor Spisak, Erika Minonne, Massimiliano Raffo, Hubert Pakula, Viktoria Tisza, Cristian Scatena, Antonio Giuseppe Naccarato, and et al. 2022. "Circulating Cell-Free DNA in Renal Cell Carcinoma: The New Era of Precision Medicine" Cancers 14, no. 18: 4359. https://doi.org/10.3390/cancers14184359
APA StyleFrancini, E., Fanelli, G. N., Pederzoli, F., Spisak, S., Minonne, E., Raffo, M., Pakula, H., Tisza, V., Scatena, C., Naccarato, A. G., Loda, M., & Nuzzo, P. V. (2022). Circulating Cell-Free DNA in Renal Cell Carcinoma: The New Era of Precision Medicine. Cancers, 14(18), 4359. https://doi.org/10.3390/cancers14184359









