Inhibition of Autophagy Promotes the Anti-Tumor Effect of Metformin in Oral Squamous Cell Carcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. CCK-8 Assay
2.3. Colony Formation Assay
2.4. Scratch Assay
2.5. Transwell Invasion Assay
2.6. Apoptosis Assay
2.7. Cell Cycle Assay
2.8. ROS Assay
2.9. MRFP-GFP-LC3 Analysis
2.10. Transmission Electron Microscopy (TEM)
2.11. Western Blot
2.12. Tumor Xenograft Studies
2.13. Statistical Analysis
3. Results
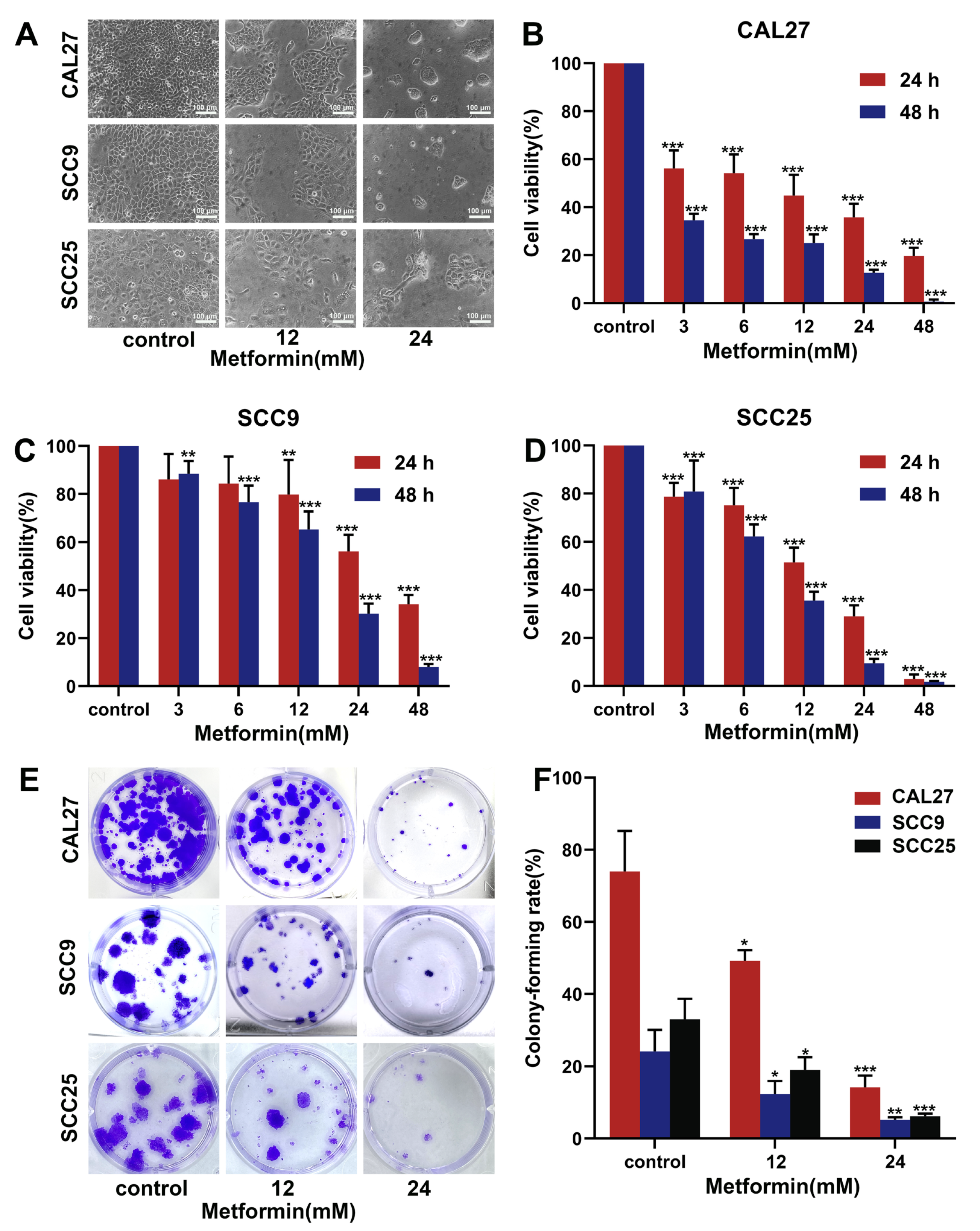
3.1. Metformin Suppresses the Proliferation of OSCC Cells
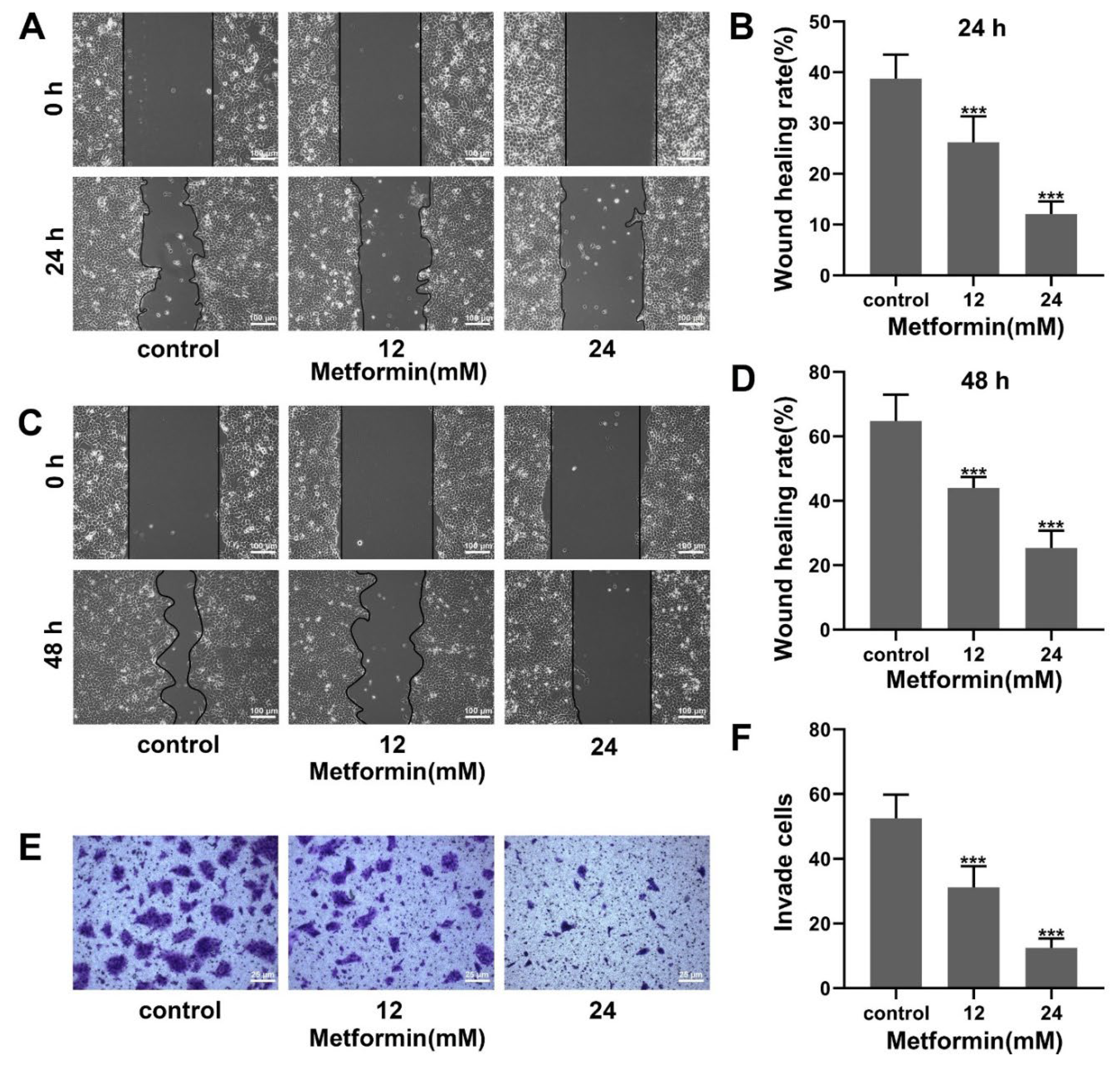
3.2. Metformin Suppresses Migration and Invasion in OSCC Cells
3.3. Metformin Induces G1 Phase Cell-Cycle Block in OSCC Cells
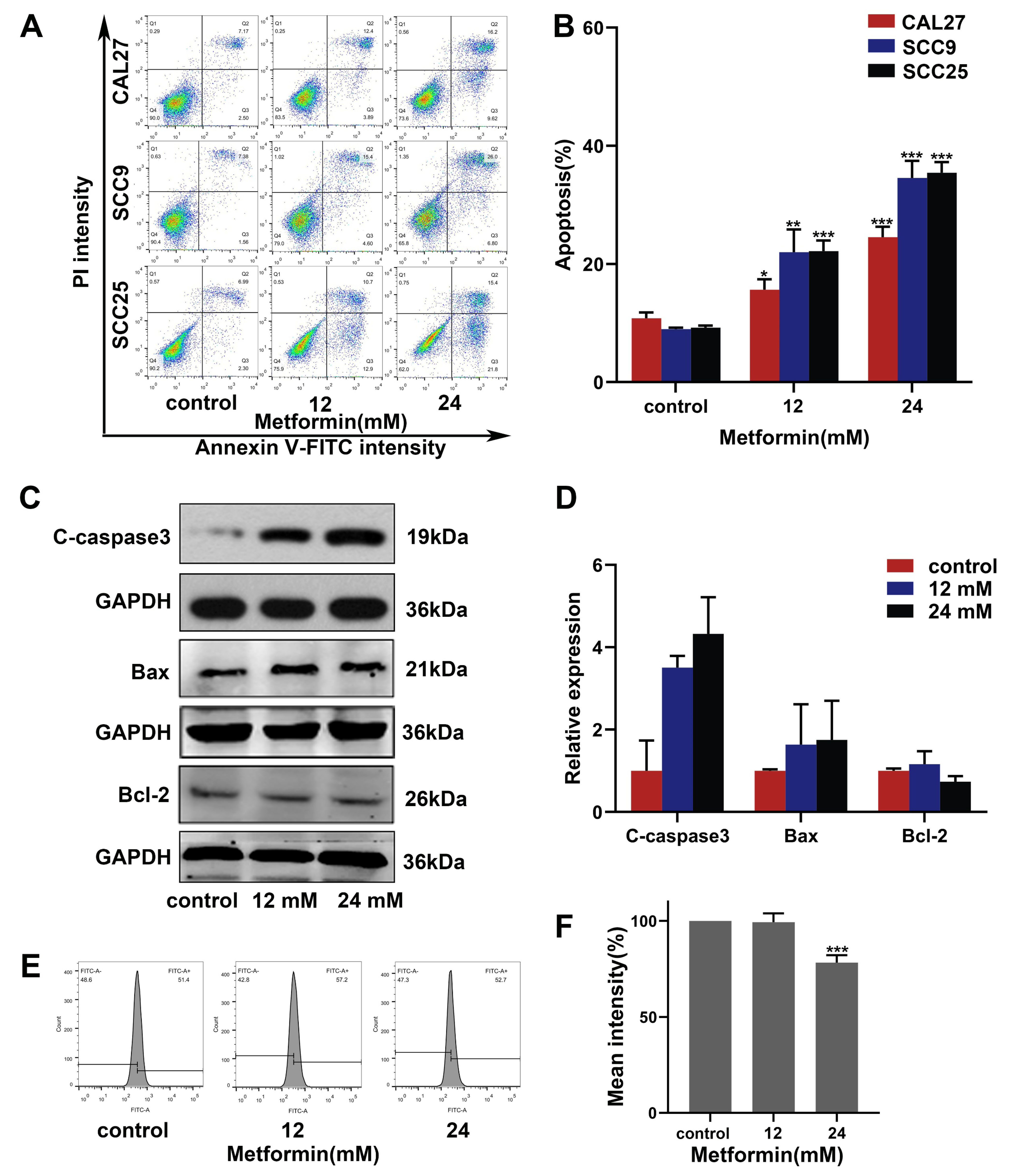
3.4. Metformin Promotes Apoptosis in OSCC Cells via a Non-ROS-Dependent Pathway
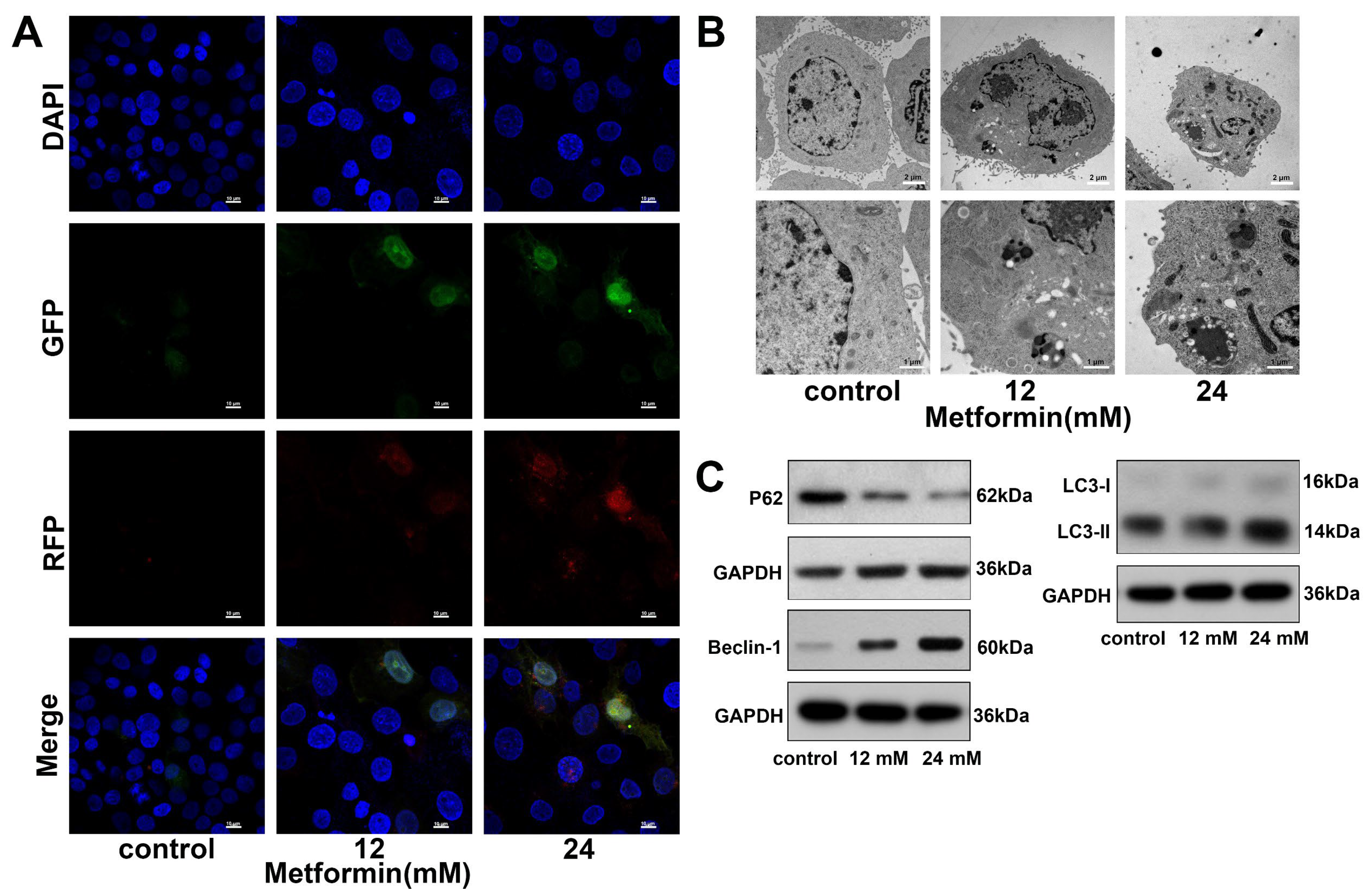
3.5. Metformin Promotes Autophagy in OSCC Cells
3.6. Inhibiting Autophagy with HCQ Increases the Apoptosis of OSCC Cells Induced by Metformin
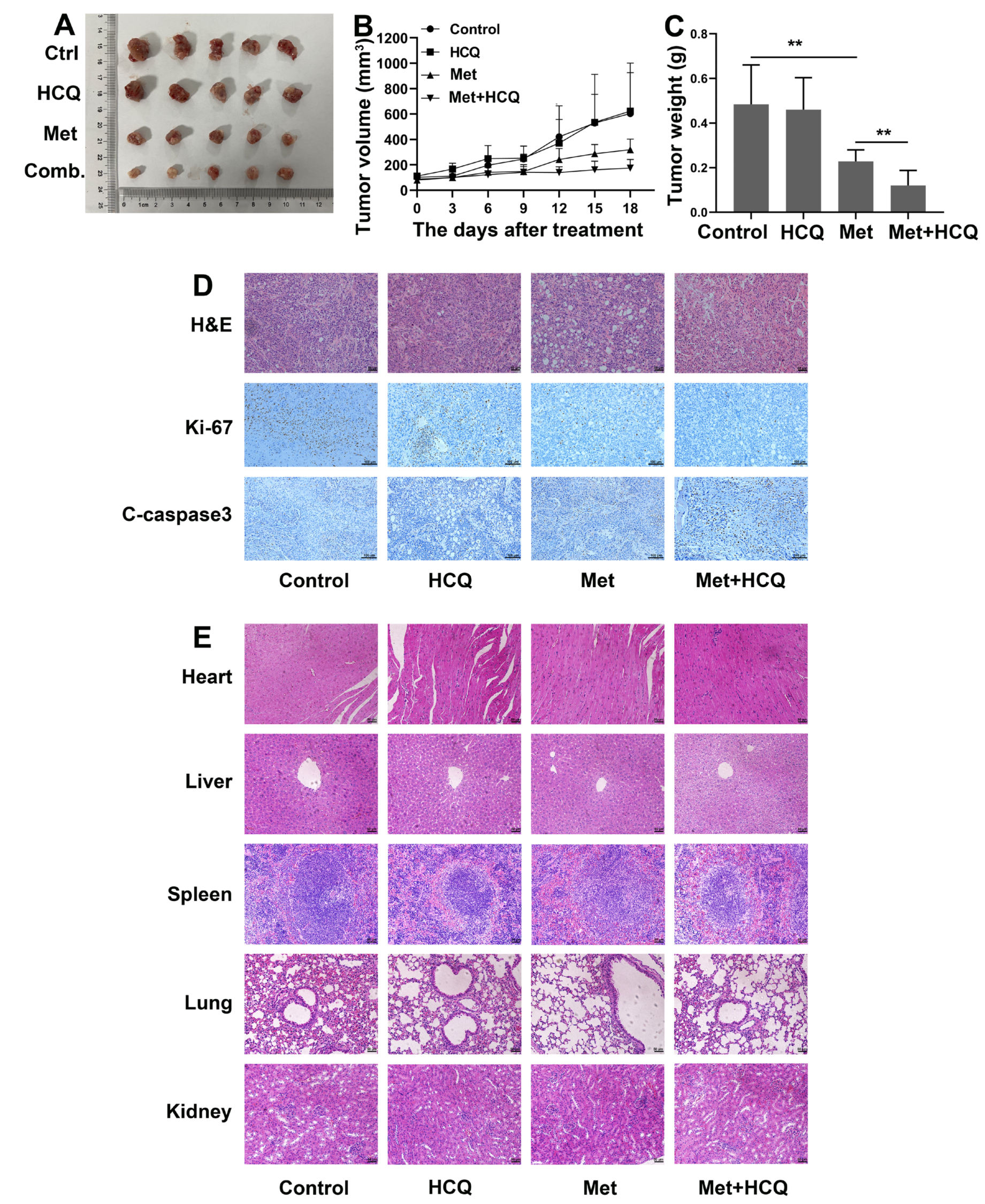
3.7. Metformin and HCQ Synergistically Suppress OSCC Growth In Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sorroche, B.; Talukdar, F.; Lima, S.; Melendez, M.; de Carvalho, A.; de Almeida, G.; De Marchi, P.; Lopes, M.; Pinto, L.R.; Carvalho, A.; et al. DNA methylation markers from negative surgical margins can predict recurrence of oral squamous cell carcinoma. Cancers 2021, 13, 2915. [Google Scholar] [CrossRef] [PubMed]
- Togni, L.; Mascitti, M.; Sartini, D.; Campagna, R.; Pozzi, V.; Salvolini, E.; Offidani, A.; Santarelli, A.; Emanuelli, M. Nicotinamide N-Methyltransferase in head and neck tumors: A comprehensive review. Biomolecules 2021, 11, 1594. [Google Scholar] [CrossRef]
- Dan, H.; Liu, S.; Liu, J.; Liu, D.; Yin, F.; Wei, Z.; Wang, J.; Zhou, Y.; Jiang, L.; Ji, N.; et al. RACK1 promotes cancer progression by increasing the M2/M1 macrophage ratio via the NF-kappaB pathway in oral squamous cell carcinoma. Mol. Oncol. 2020, 14, 795–807. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Chen, C.; Chen, X.; Fei, Y.; Jiang, L.; Wang, G. Vitamin C promotes apoptosis and cell cycle arrest in oral squamous cell carcinoma. Front. Oncol. 2020, 10, 976. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Massey, S.; Story, D.; Li, L. Metformin: An old drug with new applications. Int. J. Mol. Sci. 2018, 19, 2863. [Google Scholar] [CrossRef]
- Bailey, C.J.; Turner, R.C. Metformin. N. Engl. J. Med. 1996, 334, 574–579. [Google Scholar] [CrossRef]
- Lu, T.; Li, M.; Zhao, M.; Huang, Y.; Bi, G.; Liang, J.; Chen, Z.; Zheng, Y.; Xi, J.; Lin, Z.; et al. Metformin inhibits human non-small cell lung cancer by regulating AMPK–CEBPB–PDL1 signaling pathway. Cancer Immunol. Immunother. 2021, 71, 1733–1746. [Google Scholar] [CrossRef]
- Li, C.-Q.; Liu, Z.-Q.; Liu, S.-S.; Zhang, G.-T.; Jiang, L.; Chen, C.; Luo, D.-Q. Transcriptome analysis of liver cancer cell Huh-7 treated with metformin. Front. Pharmacol. 2022, 13, 822023. [Google Scholar] [CrossRef]
- Lord, S.R.; Cheng, W.-C.; Liu, D.; Gaude, E.; Haider, S.; Metcalf, T.; Patel, N.; Teoh, E.J.; Gleeson, F.; Bradley, K.; et al. Integrated pharmacodynamic analysis identifies two metabolic adaption pathways to metformin in breast cancer. Cell Metab. 2018, 28, 679–688 e4. [Google Scholar] [CrossRef] [Green Version]
- Moro, M.; Caiola, E.; Ganzinelli, M.; Zulato, E.; Rulli, E.; Marabese, M.; Centonze, G.; Busico, A.; Pastorino, U.; de Braud, F.G.; et al. Metformin enhances cisplatin-induced apoptosis and prevents resistance to cisplatin in co-mutated KRAS/LKB1 NSCLC. J. Thorac. Oncol. 2018, 13, 1692–1704. [Google Scholar] [CrossRef]
- Bowker, S.L.; Majumdar, S.R.; Veugelers, P.; Johnson, J.A. Increased cancer-related mortality for patients with type 2 diabetes who use sulfonylureas or insulin: Response to farooki and schneider. Diabetes Care 2006, 29, 1990–1991. [Google Scholar] [CrossRef]
- Sui, X.; Xu, Y.; Wang, X.; Han, W.; Pan, H.; Xiao, M. Metformin: A novel but controversial drug in cancer prevention and treatment. Mol. Pharm. 2015, 12, 3783–3791. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Feng, X.; Tian, H.; Fu, X.; Gu, W.; Wen, Y. Metformin inhibits mTOR and c-Myc by decreasing YAP protein expression in OSCC cells. Oncol. Rep. 2021, 45, 1249–1260. [Google Scholar] [CrossRef]
- Chen, X.; He, W.-T.; Hu, L.; Li, J.; Fang, Y.; Wang, X.; Xu, X.; Wang, Z.; Huang, K.; Han, J. Pyroptosis is driven by non-selective gasdermin-D pore and its morphology is different from MLKL channel-mediated necroptosis. Cell Res. 2016, 26, 1007–1020. [Google Scholar] [CrossRef]
- Chen, S.-Y.; Chiu, L.-Y.; Ma, M.-C.; Wang, J.-S.; Chien, C.-L.; Lin, W.-W. zVAD-induced autophagic cell death requires c-Src-dependent ERK and JNK activation and reactive oxygen species generation. Autophagy 2011, 7, 217–228. [Google Scholar] [CrossRef]
- He, Y.; Zhang, M.; Wu, Y.; Jiang, H.; Fu, H.; Cai, Y.; Xu, Z.; Liu, C.; Chen, B.; Yang, T. Aberrant activation of Notch-1 signaling inhibits podocyte restoration after islet transplantation in a rat model of diabetic nephropathy. Cell Death Dis. 2018, 9, 950. [Google Scholar] [CrossRef]
- Su, M.; Mei, Y.; Sinha, S. Role of the crosstalk between autophagy and apoptosis in cancer. J. Oncol. 2013, 2013, 102735. [Google Scholar] [CrossRef]
- Ghavami, S.; Eshragi, M.; Ande, S.R.; Chazin, W.J.; Klonisch, T.; Halayko, A.J.; Mcneill, K.D.; Hashemi, M.; Kerkhoff, C.; Los, M. S100A8/A9 induces autophagy and apoptosis via ROS-mediated cross-talk between mitochondria and lysosomes that involves BNIP3. Cell Res. 2010, 20, 314–331. [Google Scholar] [CrossRef]
- Zhang, Z.; Yan, J.; Bowman, A.B.; Bryan, M.R.; Singh, R.; Aschner, M. Dysregulation of TFEB contributes to manganese-induced autophagic failure and mitochondrial dysfunction in astrocytes. Autophagy 2020, 16, 1506–1523. [Google Scholar] [CrossRef] [PubMed]
- Wirth, M.; Zhang, W.; Razi, M.; Nyoni, L.; Joshi, D.; O’Reilly, N.; Johansen, T.; Tooze, S.A.; Mouilleron, S. Molecular determinants regulating selective binding of autophagy adapters and receptors to ATG8 proteins. Nat. Commun. 2019, 10, 2055. [Google Scholar] [CrossRef] [PubMed]
- Amaravadi, R.K.; Kimmelman, A.C.; Debnath, J. Targeting autophagy in cancer: Recent advances and future directions. Cancer Discov. 2019, 9, 1167–1181. [Google Scholar] [CrossRef]
- Eisenberg-Lerner, A.; Bialik, S.; Simon, H.-U.; Kimchi, A. Life and death partners: Apoptosis, autophagy and the cross-talk between them. Cell Death Differ. 2009, 16, 966–975. [Google Scholar] [CrossRef] [PubMed]
- Rangwala, R.; Chang, Y.C.; Hu, J.; Algazy, K.M.; Evans, T.L.; Fecher, L.A.; Schuchter, L.M.; Torigian, D.A.; Panosian, J.T.; Troxel, A.B.; et al. Combined MTOR and autophagy inhibition. Phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy 2014, 10, 1391–1402. [Google Scholar] [CrossRef] [PubMed]
- Amaravadi, R.K.; Lippincott-Schwartz, J.; Yin, X.-M.; Weiss, W.A.; Takebe, N.; Timmer, W.; DiPaola, R.S.; Lotze, M.T.; White, E. Principles and current strategies for targeting autophagy for cancer treatment. Clin. Cancer Res. 2011, 17, 654–666. [Google Scholar] [CrossRef]
- Dourado, M.R.; Korvala, J.; Åström, P.; De Oliveira, C.E.; Cervigne, N.K.; Mofatto, L.S.; Bastos, D.C.; Messetti, A.C.P.; Graner, E.; Leme, A.F.P.; et al. Extracellular vesicles derived from cancer-associated fibroblasts induce the migration and invasion of oral squamous cell carcinoma. J. Extracell. Vesicles 2019, 8, 1578525. [Google Scholar] [CrossRef]
- Chen, J.; Yang, J.; Li, H.; Yang, Z.; Zhang, X.; Li, X.; Wang, J.; Chen, S.; Song, M. Single-cell transcriptomics reveal the intratumoral landscape of infiltrated T-cell subpopulations in oral squamous cell carcinoma. Mol. Oncol. 2021, 15, 866–886. [Google Scholar] [CrossRef]
- Peres, M.A.; Macpherson, L.M.; Weyant, R.J.; Daly, B.; Venturelli, R.; Mathur, M.R.; Listl, S.; Celeste, R.K.; Guarnizo-Herreño, C.C.; Kearns, C.; et al. Oral diseases: A global public health challenge. Lancet 2019, 394, 249–260. [Google Scholar] [CrossRef]
- Zhao, X.; Zhang, C.; Le, Z.; Zeng, S.; Pan, C.; Shi, J.; Wang, J.; Zhao, X. Telomerase reverse transcriptase interference synergistically promotes tumor necrosis factor-related apoptosis-inducing ligand-induced oral squamous cell carcinoma apoptosis and suppresses proliferation in vitro and in vivo. Int. J. Mol. Med. 2018, 42, 1283–1294. [Google Scholar] [CrossRef]
- Evans, J.M.M.; Donnelly, L.A.; Emslie-Smith, A.M.; Alessi, D.R.; Morris, A.D. Metformin and reduced risk of cancer in diabetic patients. Br. Med. J. 2005, 330, 1304–1305. [Google Scholar] [CrossRef] [Green Version]
- Zheng, L.; Yang, W.; Wu, F.; Wang, C.; Yu, L.; Tang, L.; Qiu, B.; Li, Y.; Guo, L.; Wu, M.; et al. Prognostic significance of AMPK activation and therapeutic effects of metformin in hepatocellular carcinoma. Clin. Cancer Res. 2013, 19, 5372–5380. [Google Scholar] [CrossRef]
- He, Y.; Gan, M.; Wang, Y.; Huang, T.; Wang, J.; Han, T.; Yu, B. EGFR-ERK induced activation of GRHL1 promotes cell cycle progression by up-regulating cell cycle related genes in lung cancer. Cell Death Dis. 2021, 12, 430. [Google Scholar] [CrossRef]
- Pan, W.; Cheng, Y.; Zhang, H.; Liu, B.; Mo, X.; Li, T.; Li, L.; Cheng, X.; Zhang, L.; Ji, J.; et al. CSBF/C10orf99, a novel potential cytokine, inhibits colon cancer cell growth through inducing G1 arrest. Sci. Rep. 2014, 4, 6812. [Google Scholar] [CrossRef]
- Ouellet, S.; Vigneault, F.; Lessard, M.; Leclerc, S.; Drouin, R.; Guérin, S.L. Transcriptional regulation of the cyclin-dependent kinase inhibitor 1A (p21) gene by NFI in proliferating human cells. Nucleic Acids Res. 2006, 34, 6472–6487. [Google Scholar] [CrossRef]
- Yang, X.; Ye, T.; Liu, H.; Lv, P.; Duan, C.; Wu, X.; Jiang, K.; Lu, H.; Xia, D.; Peng, E.; et al. Expression profiles, biological functions and clinical significance of circRNAs in bladder cancer. Mol. Cancer 2021, 20, 4. [Google Scholar] [CrossRef]
- Kim, H.-E.; Du, F.; Fang, M.; Wang, X. Formation of apoptosome is initiated by cytochrome c -induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc. Natl. Acad. Sci. USA 2005, 102, 17545–17550. [Google Scholar] [CrossRef]
- Hardwick, J.M.; Soane, L. Multiple functions of BCL-2 family proteins. Cold Spring Harb. Perspect. Biol. 2013, 5, a008722. [Google Scholar] [CrossRef]
- D’Autréaux, B.; Toledano, M.B. ROS as signalling molecules: Mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 2007, 8, 813–824. [Google Scholar] [CrossRef]
- Chen, H.; Sun, B.; Sun, H.; Xu, L.; Wu, G.; Tu, Z.; Cheng, X.; Fan, X.; Mai, Z.; Tang, Q.; et al. Bak instead of Bax plays a key role in metformin-induced apoptosis s in HCT116 cells. Cell Death Discov. 2021, 7, 363. [Google Scholar] [CrossRef]
- Sharma, P.; Kumar, S. Metformin inhibits human breast cancer cell growth by promoting apoptosis via a ROS-independent pathway involving mitochondrial dysfunction: Pivotal role of superoxide dismutase (SOD). Cell. Oncol. 2018, 41, 637–650. [Google Scholar] [CrossRef]
- Chen, C.; Lu, Y.; Siu, H.M.; Guan, J.; Zhu, L.; Zhang, S.; Yue, J.; Zhang, L. Identification of novel vacuolin-1 analogues as autophagy inhibitors by virtual drug screening and chemical synthesis. Molecules 2017, 22, 891. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.H.; Jackson, S.; Seaman, M.; Brown, K.; Kempkes, B.; Hibshoosh, H.; Levine, B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999, 402, 672–676. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.J.; Ye, L.; Huang, W.F.; Guo, L.J.; Xu, Z.G.; Wu, H.L.; Yang, C.; Liu, H.F. p62 links the autophagy pathway and the ubiqutin–proteasome system upon ubiquitinated protein degradation. Cell. Mol. Biol. Lett. 2016, 21, 29. [Google Scholar] [CrossRef] [PubMed]
- Levy, J.M.M.; Towers, C.G.; Thorburn, A. Targeting autophagy in cancer. Nat. Cancer 2017, 17, 528–542. [Google Scholar] [CrossRef] [PubMed]
- Prerna, K.; Dubey, V.K. Beclin1-mediated interplay between autophagy and apoptosis: New understanding. Int. J. Biol. Macromol. 2022, 204, 258–273. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, W.; Chen, C.; Zhou, J.; Chen, X.; Cai, K.; Shen, M.; Chen, X.; Jiang, L.; Wang, G. Inhibition of Autophagy Promotes the Anti-Tumor Effect of Metformin in Oral Squamous Cell Carcinoma. Cancers 2022, 14, 4185. https://doi.org/10.3390/cancers14174185
Zhao W, Chen C, Zhou J, Chen X, Cai K, Shen M, Chen X, Jiang L, Wang G. Inhibition of Autophagy Promotes the Anti-Tumor Effect of Metformin in Oral Squamous Cell Carcinoma. Cancers. 2022; 14(17):4185. https://doi.org/10.3390/cancers14174185
Chicago/Turabian StyleZhao, Wei, Chen Chen, Jianjun Zhou, Xiaoqing Chen, Kuan Cai, Miaomiao Shen, Xuan Chen, Lei Jiang, and Guodong Wang. 2022. "Inhibition of Autophagy Promotes the Anti-Tumor Effect of Metformin in Oral Squamous Cell Carcinoma" Cancers 14, no. 17: 4185. https://doi.org/10.3390/cancers14174185
APA StyleZhao, W., Chen, C., Zhou, J., Chen, X., Cai, K., Shen, M., Chen, X., Jiang, L., & Wang, G. (2022). Inhibition of Autophagy Promotes the Anti-Tumor Effect of Metformin in Oral Squamous Cell Carcinoma. Cancers, 14(17), 4185. https://doi.org/10.3390/cancers14174185





