Identification of Lethal Inhibitors and Inhibitor Combinations for Mono-Driver versus Multi-Driver Triple-Negative Breast Cancer Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines, Media, and Drugs
2.2. Cell Culture and Viability Assays
2.3. Curve Fitting by the Hill Equation and the Biphasic Equation
2.4. Drug Synergy Analysis and Combination Index Calculation
2.5. Time Course Experiments
2.6. Cell Treatments and Western Blots
2.7. DU-4475 Apoptosis and Necrosis Assay
3. Results
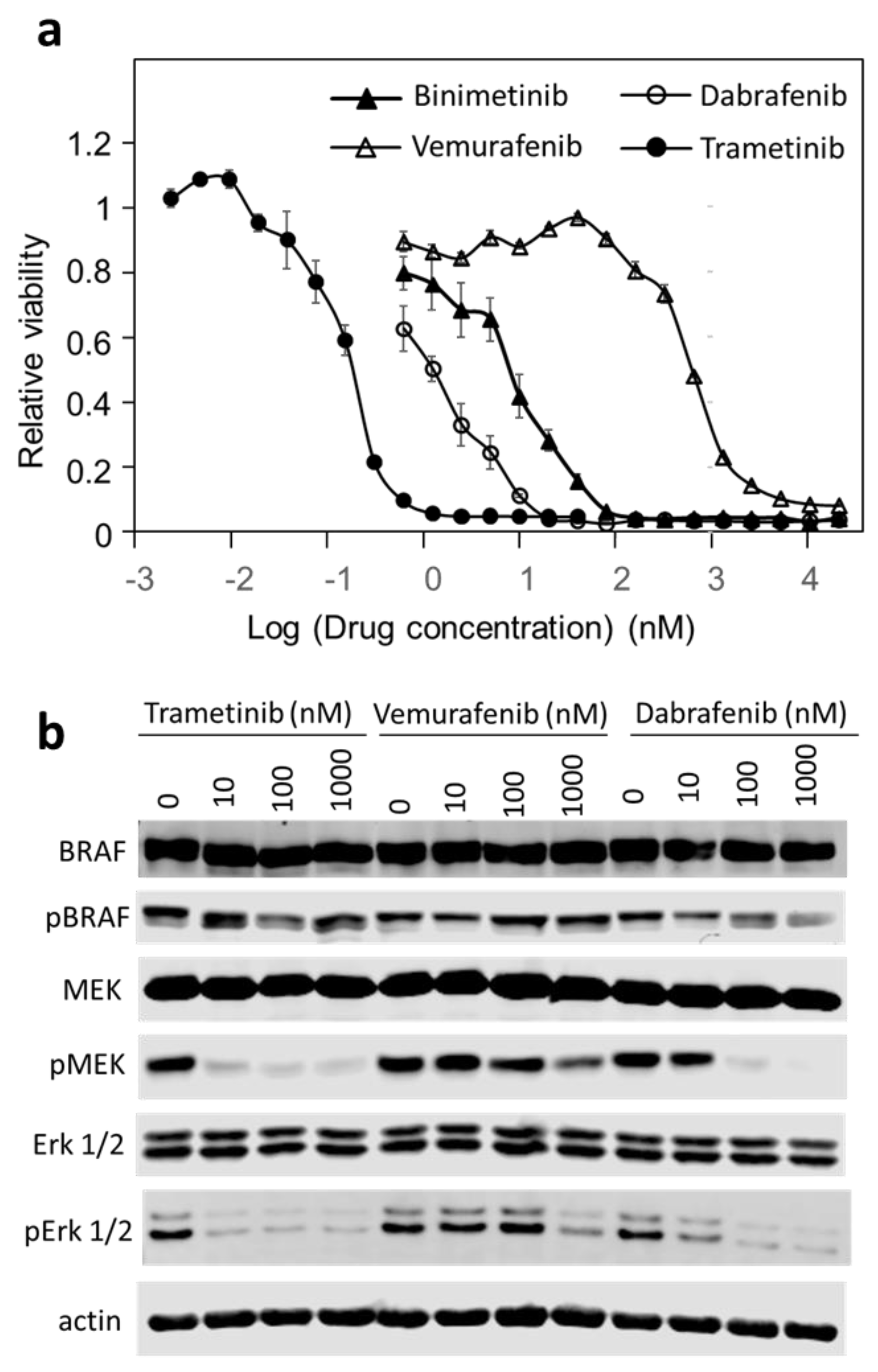
3.1. DU-4475 Cell Line Is Exceptionally Sensitive to BRAF and Mek Kinase Inhibitors
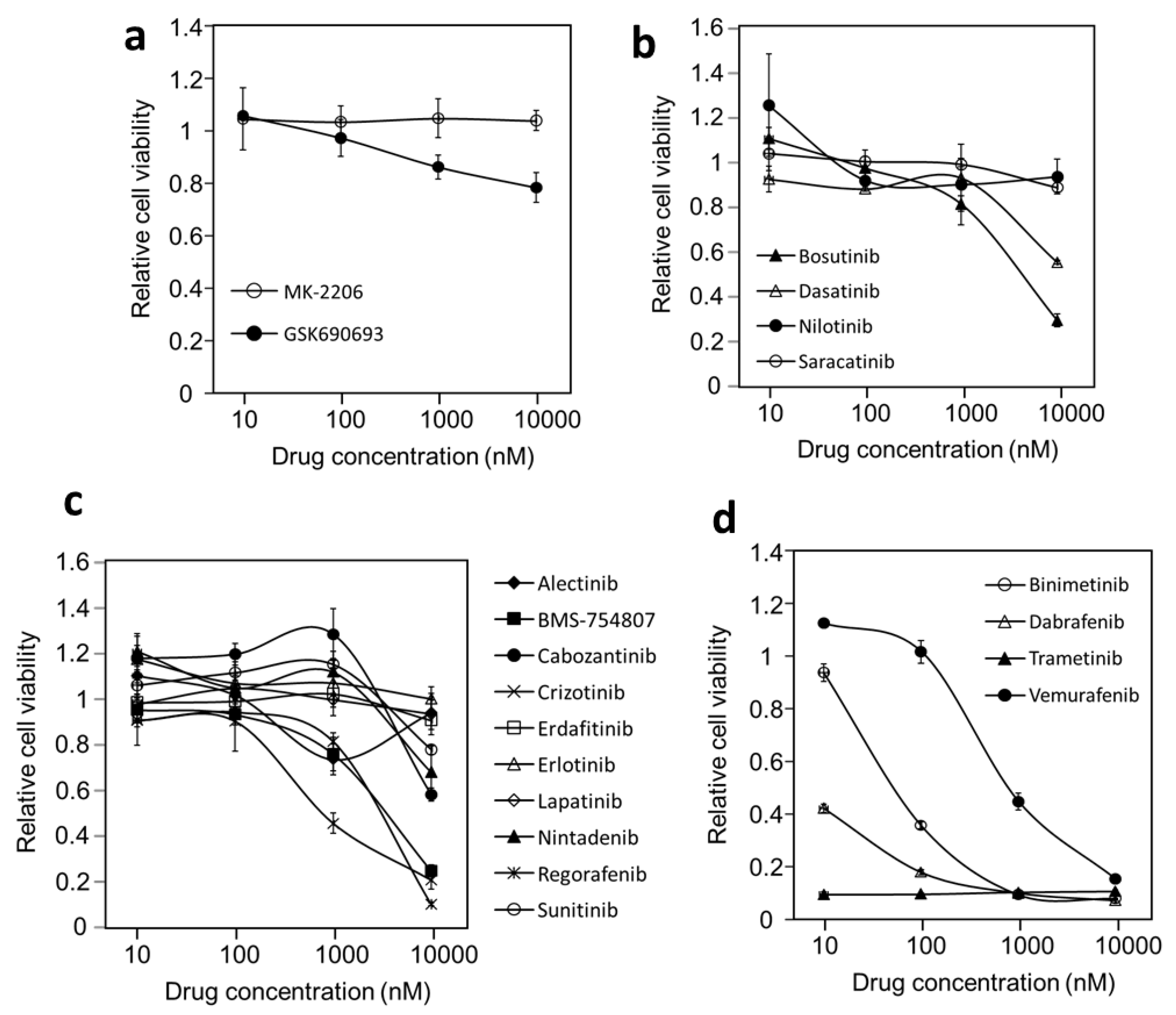
3.2. Probing Oncogenic Protein Kinase Drivers in DU-4475 Cell Line Viability
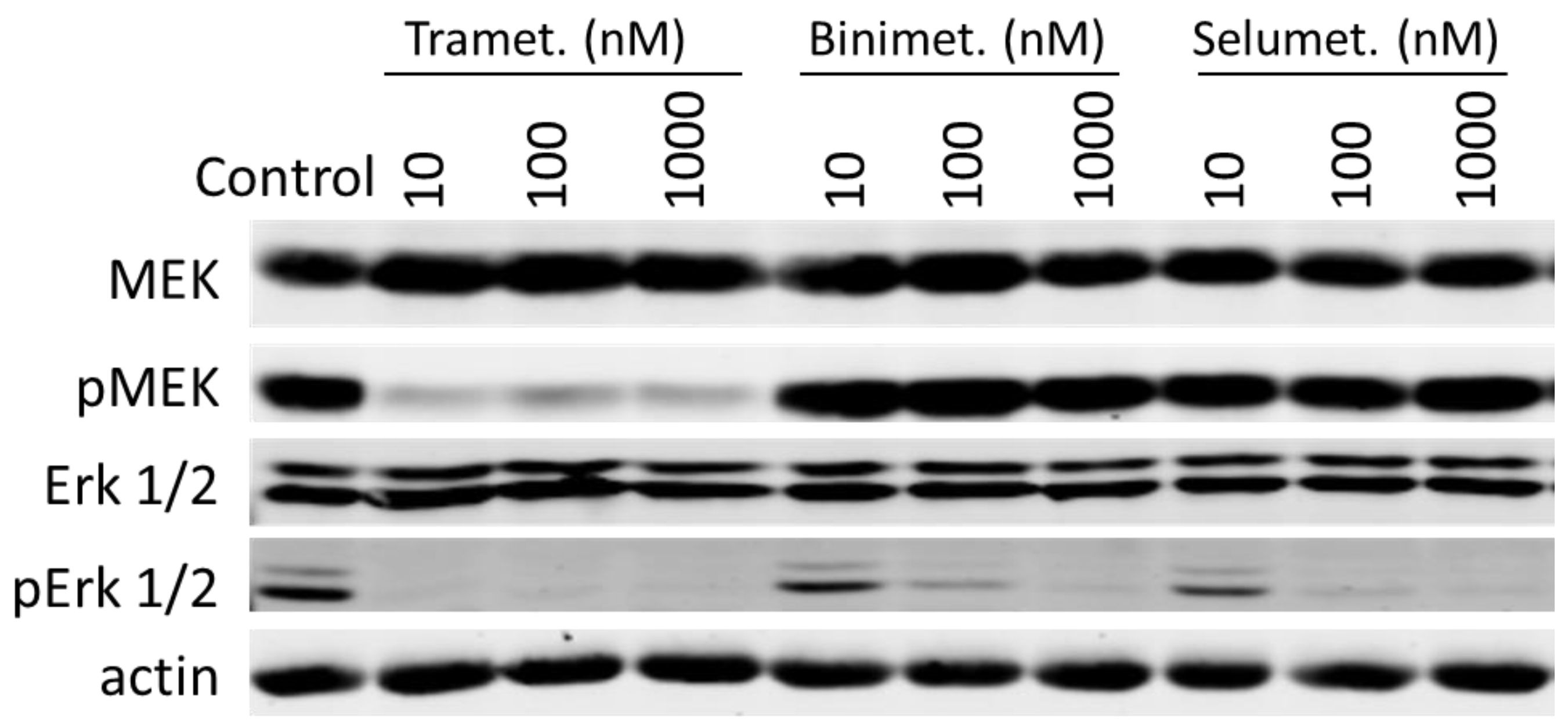
3.3. Trametinib and Dabrafenib Fully Block Mek and Erk Activation in DU-4475
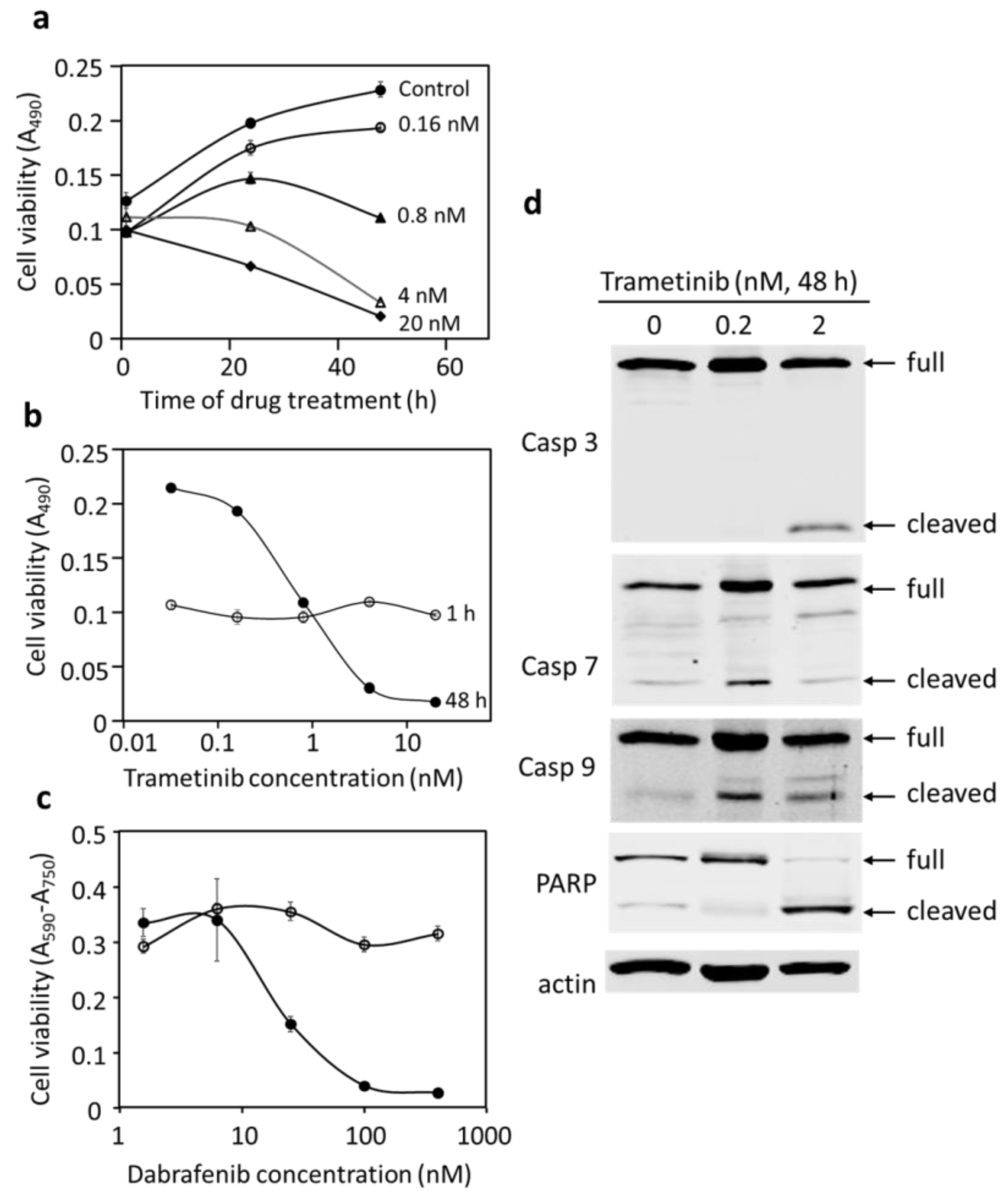
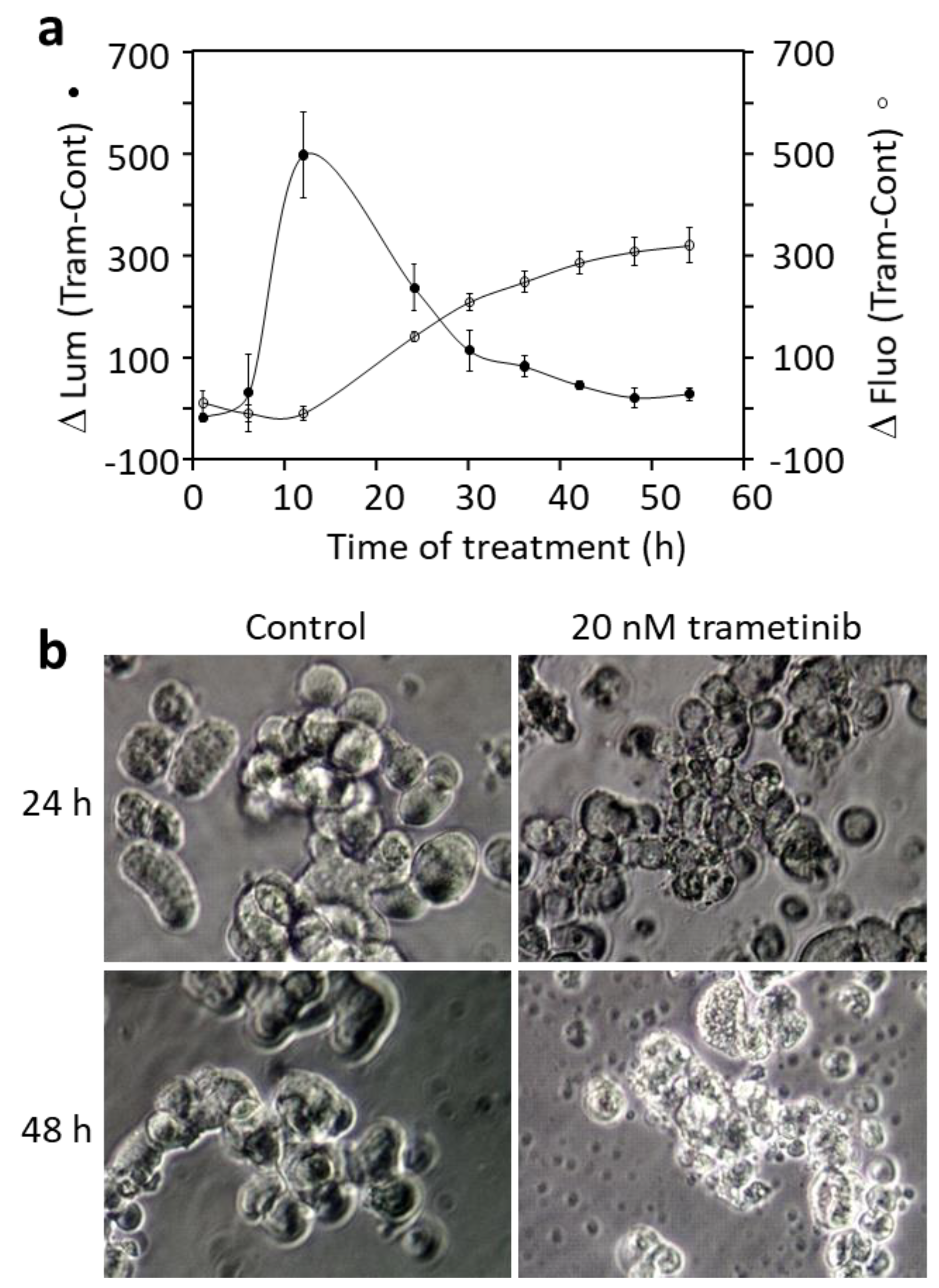
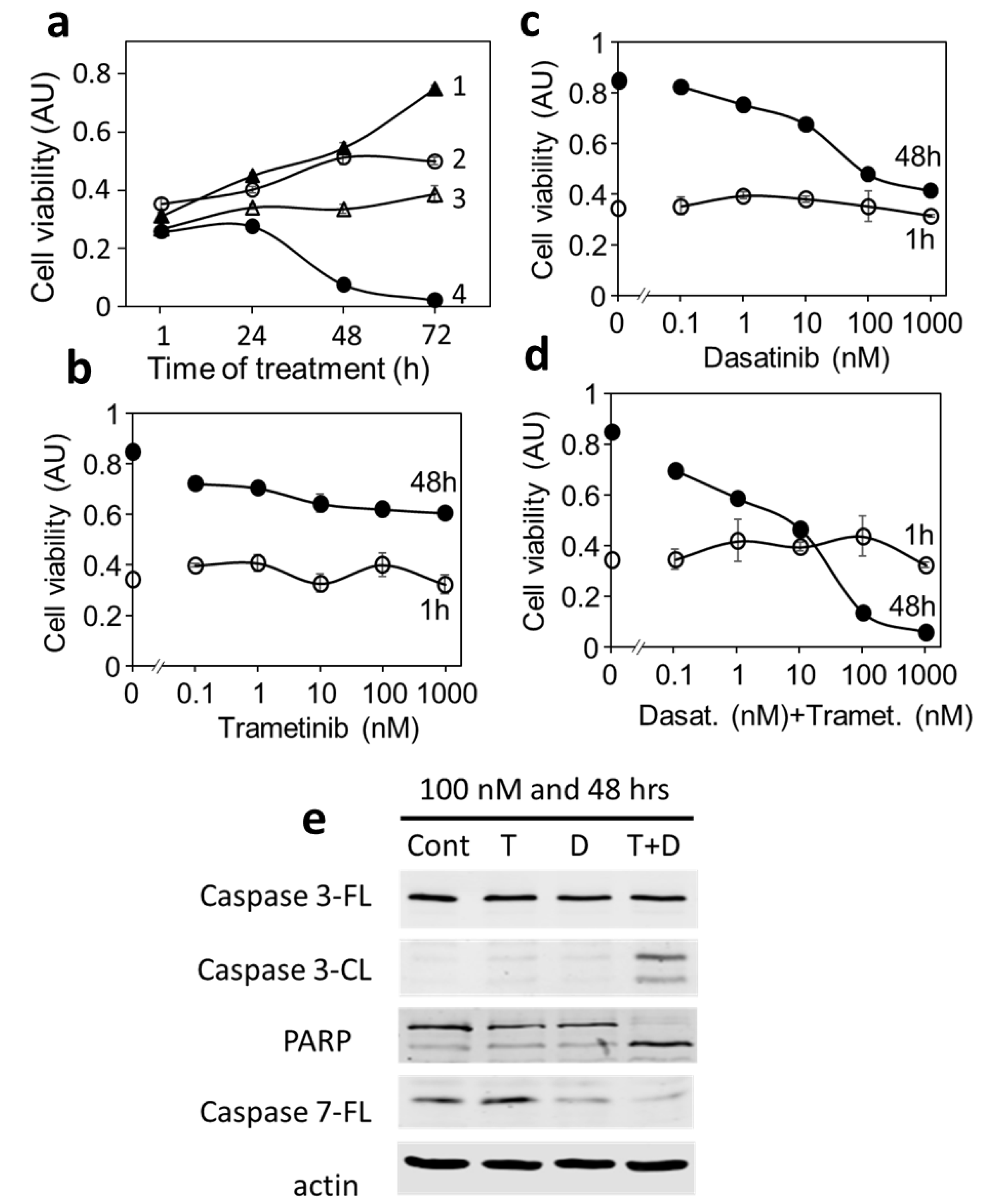
3.4. Blocking BRAF or Mek Inhibits Proliferation and Causes Cell Death in DU-4475
3.5. MDA-MB-231 Is a Multi-Driver TNBC Cell Line Dependent on Both the MAP Kinase Pathway and Src Kinase for Proliferation
3.6. Blocking Each Driver Partially Inhibits Cell Proliferation, While Blocking Both Drivers Induces Apoptosis in MDA-MB-231
4. Discussion
4.1. DU-4475 as a Mono-Driver Cancer Cell Model
4.2. BRAF V600E as a Therapeutic Target in TNBC
4.3. Blocking Both Oncogenic Drivers in MDA-MB-231 Causes Synthetic Lethality
4.4. Single-Drug Lethality versus Synthetic Lethality
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- DeSantis, C.E.; Ma, J.; Gaudet, M.M.; Newman, L.A.; Miller, K.D.; Goding Sauer, A.; Jemal, A.; Siegel, R.L. Breast cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 438–451. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Network, C.G.A. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciriello, G.; Gatza, M.L.; Beck, A.H.; Wilkerson, M.D.; Rhie, S.K.; Pastore, A.; Zhang, H.; McLellan, M.; Yau, C.; Kandoth, C.; et al. Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 2015, 163, 506–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foulkes, W.D.; Smith, I.E.; Reis-Filho, J.S. Triple-negative breast cancer. N. Engl. J. Med. 2010, 363, 1938–1948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bianchini, G.; Balko, J.M.; Mayer, I.A.; Sanders, M.E.; Gianni, L. Triple-negative breast cancer: Challenges and opportunities of a heterogeneous disease. Nat. Rev. Clin. Oncol. 2016, 13, 674–690. [Google Scholar] [CrossRef]
- Boyle, P. Triple-negative breast cancer: Epidemiological considerations and recommendations. Ann. Oncol. 2012, 23 (Suppl. S6), vi7–vi12. [Google Scholar] [CrossRef]
- Gupta, G.K.; Collier, A.L.; Lee, D.; Hoefer, R.A.; Zheleva, V.; van Reesema, L.L.S.; Tang-Tan, A.M.; Guye, M.L.; Chang, D.Z.; Winston, J.S.; et al. Perspectives on Triple-Negative Breast Cancer: Current Treatment Strategies, Unmet Needs, and Potential Targets for Future Therapies. Cancers 2020, 12, 2392. [Google Scholar] [CrossRef]
- Pal, S.K.; Childs, B.H.; Pegram, M. Triple negative breast cancer: Unmet medical needs. Breast Cancer Res. Treat. 2011, 125, 627–636. [Google Scholar] [CrossRef] [Green Version]
- Schroeder, M.C.; Rastogi, P.; Geyer, C.E.; Miller, L.D.; Thomas, A. Early and Locally Advanced Metaplastic Breast Cancer: Presentation and Survival by Receptor Status in Surveillance, Epidemiology, and End Results (SEER) 2010–2014. Oncologist 2018, 23, 481–488. [Google Scholar] [CrossRef] [Green Version]
- Howlader, N.; Cronin, K.A.; Kurian, A.W.; Andridge, R. Differences in Breast Cancer Survival by Molecular Subtypes in the United States. Cancer Epidemiol. Biomark. Prev. 2018, 27, 619–626. [Google Scholar] [CrossRef] [Green Version]
- Thomas, A.; Rhoads, A.; Pinkerton, E.; Schroeder, M.C.; Conway, K.M.; Hundley, W.G.; McNally, L.R.; Oleson, J.; Lynch, C.F.; Romitti, P.A. Incidence and Survival Among Young Women with Stage I-III Breast Cancer: SEER 2000-2015. JNCI Cancer Spectr. 2019, 3, pkz040. [Google Scholar] [CrossRef]
- Carey, L.; Winer, E.; Viale, G.; Cameron, D.; Gianni, L. Triple-negative breast cancer: Disease entity or title of convenience? Nat. Rev. Clin. Oncol. 2010, 7, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Newman, L.A.; Kaljee, L.M. Health Disparities and Triple-Negative Breast Cancer in African American Women: A Review. JAMA Surg. 2017, 152, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, J.; Ginsburg, O.; Rochon, P.A.; Sun, P.; Narod, S.A. Differences in breast cancer stage at diagnosis and cancer-specific survival by race and ethnicity in the United States. JAMA 2015, 313, 165–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, C.C.; Bartholomew, L.K.; Carpentier, M.Y.; Bluethmann, S.M.; Vernon, S.W. Adherence to adjuvant hormonal therapy among breast cancer survivors in clinical practice: A systematic review. Breast Cancer Res. Treat. 2012, 134, 459–478. [Google Scholar] [CrossRef] [Green Version]
- Puhalla, S.; Bhattacharya, S.; Davidson, N.E. Hormonal therapy in breast cancer: A model disease for the personalization of cancer care. Mol. Oncol. 2012, 6, 222–236. [Google Scholar] [CrossRef] [Green Version]
- Figueroa-Magalhães, M.C.; Jelovac, D.; Connolly, R.; Wolff, A.C. Treatment of HER2-positive breast cancer. Breast 2014, 23, 128–136. [Google Scholar] [CrossRef]
- Waks, A.G.; Winer, E.P. Breast Cancer Treatment: A Review. JAMA 2019, 321, 288–300. [Google Scholar] [CrossRef]
- Goetz, M.P.; Gradishar, W.J.; Anderson, B.O.; Abraham, J.; Aft, R.; Allison, K.H.; Blair, S.L.; Burstein, H.J.; Dang, C.; Elias, A.D.; et al. NCCN Guidelines Insights: Breast Cancer, Version 3.2018. J. Natl. Compr. Cancer Netw. 2019, 17, 118–126. [Google Scholar] [CrossRef] [Green Version]
- Andreopoulou, E.; Schweber, S.J.; Sparano, J.A.; McDaid, H.M. Therapies for triple negative breast cancer. Expert Opin. Pharmacother. 2015, 16, 983–998. [Google Scholar] [CrossRef] [PubMed]
- Isakoff, S.J.; Mayer, E.L.; He, L.; Traina, T.A.; Carey, L.A.; Krag, K.J.; Rugo, H.S.; Liu, M.C.; Stearns, V.; Come, S.E.; et al. TBCRC009: A Multicenter Phase II Clinical Trial of Platinum Monotherapy with Biomarker Assessment in Metastatic Triple-Negative Breast Cancer. J. Clin. Oncol. 2015, 33, 1902–1909. [Google Scholar] [CrossRef] [PubMed]
- American Cancer Society. Triple-Negative Breast Cancer. Available online: https://www.cancer.org/cancer/breast-cancer/about/types-of-breast-cancer/triple-negative.html. (accessed on 10 September 2021).
- Berger, E.R.; Park, T.; Saridakis, A.; Golshan, M.; Greenup, R.A.; Ahuja, N. Immunotherapy Treatment for Triple Negative Breast Cancer. Pharmaceuticals 2021, 14, 763. [Google Scholar] [CrossRef] [PubMed]
- Marquart, J.; Chen, E.Y.; Prasad, V. Estimation of the Percentage of US Patients with Cancer Who Benefit from Genome-Driven Oncology. JAMA Oncol. 2018, 4, 1093–1098. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Druker, B.J. Perspectives on the development of a molecularly targeted agent. Cancer Cell 2002, 1, 31–36. [Google Scholar] [CrossRef] [Green Version]
- Baudino, T.A. Targeted Cancer Therapy: The Next Generation of Cancer Treatment. Curr. Drug Discov. Technol. 2015, 12, 3–20. [Google Scholar] [CrossRef]
- Jakhetiya, A.; Garg, P.K.; Prakash, G.; Sharma, J.; Pandey, R.; Pandey, D. Targeted therapy of gastrointestinal stromal tumours. World J. Gastrointest. Surg. 2016, 8, 345–352. [Google Scholar] [CrossRef] [Green Version]
- Lorentzen, H.F. Targeted therapy for malignant melanoma. Curr. Opin. Pharmacol. 2019, 46, 116–121. [Google Scholar] [CrossRef]
- Lynch, T.J.; Bell, D.W.; Sordella, R.; Gurubhagavatula, S.; Okimoto, R.A.; Brannigan, B.W.; Harris, P.L.; Haserlat, S.M.; Supko, J.G.; Haluska, F.G.; et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2004, 350, 2129–2139. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R. Properties of FDA-approved small molecule protein kinase inhibitors: A 2020 update. Pharmacol. Res. 2020, 152, 104609. [Google Scholar] [CrossRef] [PubMed]
- Baselga, J.; Im, S.A.; Iwata, H.; Cortés, J.; De Laurentiis, M.; Jiang, Z.; Arteaga, C.L.; Jonat, W.; Clemons, M.; Ito, Y.; et al. Buparlisib plus fulvestrant versus placebo plus fulvestrant in postmenopausal, hormone receptor-positive, HER2-negative, advanced breast cancer (BELLE-2): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2017, 18, 904–916. [Google Scholar] [CrossRef]
- Di Leo, A.; Johnston, S.; Lee, K.S.; Ciruelos, E.; Lønning, P.E.; Janni, W.; O’Regan, R.; Mouret-Reynier, M.A.; Kalev, D.; Egle, D.; et al. Buparlisib plus fulvestrant in postmenopausal women with hormone-receptor-positive, HER2-negative, advanced breast cancer progressing on or after mTOR inhibition (BELLE-3): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2018, 19, 87–100. [Google Scholar] [CrossRef]
- André, F.; Ciruelos, E.; Rubovszky, G.; Campone, M.; Loibl, S.; Rugo, H.S.; Iwata, H.; Conte, P.; Mayer, I.A.; Kaufman, B.; et al. Alpelisib for PIK3CA-Mutated, Hormone Receptor-Positive Advanced Breast Cancer. N. Engl. J. Med. 2019, 380, 1929–1940. [Google Scholar] [CrossRef]
- Martín, M.; Chan, A.; Dirix, L.; O’Shaughnessy, J.; Hegg, R.; Manikhas, A.; Shtivelband, M.; Krivorotko, P.; Batista López, N.; Campone, M.; et al. A randomized adaptive phase II/III study of buparlisib, a pan-class I PI3K inhibitor, combined with paclitaxel for the treatment of HER2- advanced breast cancer (BELLE-4). Ann. Oncol. 2017, 28, 313–320. [Google Scholar] [CrossRef]
- Banerji, U.; Dean, E.J.; Pérez-Fidalgo, J.A.; Batist, G.; Bedard, P.L.; You, B.; Westin, S.N.; Kabos, P.; Garrett, M.D.; Tall, M.; et al. A Phase I Open-Label Study to Identify a Dosing Regimen of the Pan-AKT Inhibitor AZD5363 for Evaluation in Solid Tumors and in PIK3CA-Mutated Breast and Gynecologic Cancers. Clin. Cancer Res. 2018, 24, 2050–2059. [Google Scholar] [CrossRef] [Green Version]
- Saura, C.; Roda, D.; Roselló, S.; Oliveira, M.; Macarulla, T.; Pérez-Fidalgo, J.A.; Morales-Barrera, R.; Sanchis-García, J.M.; Musib, L.; Budha, N.; et al. A First-in-Human Phase I Study of the ATP-Competitive AKT Inhibitor Ipatasertib Demonstrates Robust and Safe Targeting of AKT in Patients with Solid Tumors. Cancer Discov. 2017, 7, 102–113. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.B.; Dent, R.; Im, S.A.; Espié, M.; Blau, S.; Tan, A.R.; Isakoff, S.J.; Oliveira, M.; Saura, C.; Wongchenko, M.J.; et al. Ipatasertib plus paclitaxel versus placebo plus paclitaxel as first-line therapy for metastatic triple-negative breast cancer (LOTUS): A multicentre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol. 2017, 18, 1360–1372. [Google Scholar] [CrossRef]
- Hashimoto, K.; Tsuda, H.; Koizumi, F.; Shimizu, C.; Yonemori, K.; Ando, M.; Kodaira, M.; Yunokawa, M.; Fujiwara, Y.; Tamura, K. Activated PI3K/AKT and MAPK pathways are potential good prognostic markers in node-positive, triple-negative breast cancer. Ann. Oncol. 2014, 25, 1973–1979. [Google Scholar] [CrossRef]
- Giltnane, J.M.; Balko, J.M. Rationale for targeting the Ras/MAPK pathway in triple-negative breast cancer. Discov. Med. 2014, 17, 275–283. [Google Scholar] [PubMed]
- Bartholomeusz, C.; Xie, X.; Pitner, M.K.; Kondo, K.; Dadbin, A.; Lee, J.; Saso, H.; Smith, P.D.; Dalby, K.N.; Ueno, N.T. MEK Inhibitor Selumetinib (AZD6244; ARRY-142886) Prevents Lung Metastasis in a Triple-Negative Breast Cancer Xenograft Model. Mol. Cancer Ther. 2015, 14, 2773–2781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagaria, T.S.; Shi, C.; Leduc, C.; Hoskin, V.; Sikdar, S.; Sangrar, W.; Greer, P.A. Combined targeting of Raf and Mek synergistically inhibits tumorigenesis in triple negative breast cancer model systems. Oncotarget 2017, 8, 80804–80819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zawistowski, J.S.; Bevill, S.M.; Goulet, D.R.; Stuhlmiller, T.J.; Beltran, A.S.; Olivares-Quintero, J.F.; Singh, D.; Sciaky, N.; Parker, J.S.; Rashid, N.U.; et al. Enhancer Remodeling during Adaptive Bypass to MEK Inhibition Is Attenuated by Pharmacologic Targeting of the P-TEFb Complex. Cancer Discov. 2017, 7, 302–321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakai, K.; Hung, M.C.; Yamaguchi, H. A perspective on anti-EGFR therapies targeting triple-negative breast cancer. Am. J. Cancer Res. 2016, 6, 1609–1623. [Google Scholar]
- Jafarian, A.H.; Kooshkiforooshani, M.; Farzad, F.; Mohamadian Roshan, N. The Relationship between Fibroblastic Growth Factor Receptor-1 (FGFR1) Gene Amplification in Triple Negative Breast Carcinomas and Clinicopathological Prognostic Factors. Iran J. Pathol. 2019, 14, 299–304. [Google Scholar] [CrossRef]
- Perez-Garcia, J.; Muñoz-Couselo, E.; Soberino, J.; Racca, F.; Cortes, J. Targeting FGFR pathway in breast cancer. Breast 2018, 37, 126–133. [Google Scholar] [CrossRef] [Green Version]
- André, F.; Cortés, J. Rationale for targeting fibroblast growth factor receptor signaling in breast cancer. Breast Cancer Res. Treat. 2015, 150, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Adams, B.D.; Wali, V.B.; Cheng, C.J.; Inukai, S.; Booth, C.J.; Agarwal, S.; Rimm, D.L.; Győrffy, B.; Santarpia, L.; Pusztai, L.; et al. miR-34a Silences c-SRC to Attenuate Tumor Growth in Triple-Negative Breast Cancer. Cancer Res. 2016, 76, 927–939. [Google Scholar] [CrossRef] [Green Version]
- Tzeng, Y.T.; Liu, P.F.; Li, J.Y.; Liu, L.F.; Kuo, S.Y.; Hsieh, C.W.; Lee, C.H.; Wu, C.H.; Hsiao, M.; Chang, H.T.; et al. Kinome-Wide siRNA Screening Identifies Src-Enhanced Resistance of Chemotherapeutic Drugs in Triple-Negative Breast Cancer Cells. Front. Pharmacol. 2018, 9, 1285. [Google Scholar] [CrossRef] [Green Version]
- Tryfonopoulos, D.; Walsh, S.; Collins, D.M.; Flanagan, L.; Quinn, C.; Corkery, B.; McDermott, E.W.; Evoy, D.; Pierce, A.; O’Donovan, N.; et al. Src: A potential target for the treatment of triple-negative breast cancer. Ann. Oncol. 2011, 22, 2234–2240. [Google Scholar] [CrossRef]
- Shen, J.; Li, L.; Howlett, N.G.; Cohen, P.S.; Sun, G. Application of a Biphasic Mathematical Model of Cancer Cell Drug Response for Formulating Potent and Synergistic Targeted Drug Combinations to Triple Negative Breast Cancer Cells. Cancers 2020, 12, 1087. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Li, L.; Yang, T.; Cohen, P.S.; Sun, G. Biphasic Mathematical Model of Cell-Drug Interaction That Separates Target-Specific and Off-Target Inhibition and Suggests Potent Targeted Drug Combinations for Multi-Driver Colorectal Cancer Cells. Cancers 2020, 12, 436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chou, T.C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010, 70, 440–446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rashband, W.S. ImageJ 1.53K; U. S. National Institutes of Health: Bethesda, MD, USA, 1997–2018. Available online: https://imagej.nih.gov/ij/ (accessed on 10 September 2021).
- Kupcho, K.; Shultz, J.; Hurst, R.; Hartnett, J.; Zhou, W.; Machleidt, T.; Grailer, J.; Worzella, T.; Riss, T.; Lazar, D.; et al. A real-time, bioluminescent annexin V assay for the assessment of apoptosis. Apoptosis 2019, 24, 184–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.; Soares, J.; Greninger, P.; Edelman, E.J.; Lightfoot, H.; Forbes, S.; Bindal, N.; Beare, D.; Smith, J.A.; Thompson, I.R.; et al. Genomics of Drug Sensitivity in Cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013, 41, D955–D961. [Google Scholar] [CrossRef] [Green Version]
- Langlois, A.J.; Holder, W.D., Jr.; Iglehart, J.D.; Nelson-Rees, W.A.; Wells, S.A., Jr.; Bolognesi, D.P. Morphological and biochemical properties of a new human breast cancer cell line. Cancer Res. 1979, 39, 2604–2613. [Google Scholar]
- Riaz, M.; van Jaarsveld, M.T.; Hollestelle, A.; Prager-van der Smissen, W.J.; Heine, A.A.; Boersma, A.W.; Liu, J.; Helmijr, J.; Ozturk, B.; Smid, M.; et al. miRNA expression profiling of 51 human breast cancer cell lines reveals subtype and driver mutation-specific miRNAs. Breast Cancer Res. 2013, 15, R33. [Google Scholar] [CrossRef] [Green Version]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef] [Green Version]
- Thota, R.; Johnson, D.B.; Sosman, J.A. Trametinib in the treatment of melanoma. Expert Opin. Biol. Ther. 2015, 15, 735–747. [Google Scholar] [CrossRef]
- Yoshida, T.; Kakegawa, J.; Yamaguchi, T.; Hantani, Y.; Okajima, N.; Sakai, T.; Watanabe, Y.; Nakamura, M. Identification and characterization of a novel chemotype MEK inhibitor able to alter the phosphorylation state of MEK1/2. Oncotarget 2012, 3, 1533–1545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, Z.M.; Real, A.M.; Marsiglia, W.M.; Chow, A.; Duffy, M.E.; Yerabolu, J.R.; Scopton, A.P.; Dar, A.C. Structural basis for the action of the drug trametinib at KSR-bound MEK. Nature 2020, 588, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Del Pino, G.L.; Li, K.; Park, E.; Schmoker, A.M.; Ha, B.H.; Eck, M.J. Allosteric MEK inhibitors act on BRAF/MEK complexes to block MEK activation. Proc. Natl. Acad. Sci. USA 2021, 118, e2107207118. [Google Scholar] [CrossRef] [PubMed]
- Maiello, M.R.; D’Alessio, A.; Bevilacqua, S.; Gallo, M.; Normanno, N.; De Luca, A. EGFR and MEK Blockade in Triple Negative Breast Cancer Cells. J. Cell Biochem. 2015, 116, 2778–2785. [Google Scholar] [CrossRef]
- Zhou, Y.; Lin, S.; Tseng, K.F.; Han, K.; Wang, Y.; Gan, Z.H.; Min, D.L.; Hu, H.Y. Selumetinib suppresses cell proliferation, migration and trigger apoptosis, G1 arrest in triple-negative breast cancer cells. BMC Cancer 2016, 16, 818. [Google Scholar] [CrossRef] [Green Version]
- Chou, T.C.; Talalay, P. Quantitative analysis of dose-effect relationships: The combined effects of multiple drugs or enzyme inhibitors. Adv. Enzym. Regul. 1984, 22, 27–55. [Google Scholar] [CrossRef]
- Pircher, M.; Winder, T.; Trojan, A. Response to Vemurafenib in Metastatic Triple-Negative Breast Cancer Harbouring a BRAF V600E Mutation: A Case Report and Electronically Captured Patient-Reported Outcome. Case Rep. Oncol. 2021, 14, 616–621. [Google Scholar] [CrossRef]
- Wang, L.; Lu, Q.; Jiang, K.; Hong, R.; Wang, S.; Xu, F. BRAF V600E Mutation in Triple-Negative Breast Cancer: A Case Report and Literature Review. Oncol. Res. Treat. 2022, 45, 54–61. [Google Scholar] [CrossRef]
- Seo, T.; Noguchi, E.; Yoshida, M.; Mori, T.; Tanioka, M.; Sudo, K.; Shimomura, A.; Yonemori, K.; Fujiwara, Y.; Tamura, K. Response to Dabrafenib and Trametinib of a Patient with Metaplastic Breast Carcinoma Harboring a BRAF V600E Mutation. Case Rep. Oncol. Med. 2020, 2020, 2518383. [Google Scholar] [CrossRef] [Green Version]
- Madsen, R.R.; Knox, R.G.; Pearce, W.; Lopez, S.; Mahler-Araujo, B.; McGranahan, N.; Vanhaesebroeck, B.; Semple, R.K. Oncogenic PIK3CA promotes cellular stemness in an allele dose-dependent manner. Proc. Natl. Acad. Sci. USA 2019, 116, 8380–8389. [Google Scholar] [CrossRef] [Green Version]
- Nijman, S.M. Synthetic lethality: General principles, utility and detection using genetic screens in human cells. FEBS Lett. 2011, 585, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Topatana, W.; Juengpanich, S.; Li, S.; Cao, J.; Hu, J.; Lee, J.; Suliyanto, K.; Ma, D.; Zhang, B.; Chen, M.; et al. Advances in synthetic lethality for cancer therapy: Cellular mechanism and clinical translation. J. Hematol. Oncol. 2020, 13, 118. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.J.; Ashworth, A. PARP inhibitors: Synthetic lethality in the clinic. Science 2017, 355, 1152–1158. [Google Scholar] [CrossRef] [PubMed]

| Cell Line | Most Potent PKI | Target | IC50 (nM) | Z-Score |
|---|---|---|---|---|
| BT-20 | AKT inhibitor III | Akt | 3505 | −1 |
| BT-549 | GW441756 | NTRK1 | 3036 | −1.3 |
| DU-4475 | Dabrafenib | BRAF | 6.3 | −3.5 |
| DU-4475 | Trametinib | Mek | 0.5 | −2.1 |
| HCC1143 | Jak3_7406 | Jak3 | 6312 | −1.3 |
| HCC1187 | GW441756 | NTRK1 | 89.7 | −4.7 |
| HCC1395 | Panopanib | CSF1R, KIT, PDGFR | 2432 | −1.7 |
| HCC1599 | Jak1_3715 | Jak 1 | 2185 | −3.9 |
| HCC1806 | GSK1904529A | IGF1R, IR | 3466 | −1.4 |
| HCC1937 | WZ3105 | Src, Rock2, NTRK2, FLT3 | 273 | −0.8 |
| HCC38 | AKT inhibitor III | Akt 1, 2, 3 | 2085 | −1.6 |
| HCC70 | AKT inhibitor III | Akt 1, 2, 3 | 775 | −2.6 |
| HCC70 | MK-2206 | Akt 1, 2 | 1245 | −2.2 |
| Hs-578-T | AT7868 | Akt | 1160 | −2 |
| MDA-MB-157 | Motesanib | VEGFR, RET, KIT, PDGFR | 6243 | −0.8 |
| MDA-MB-231 | Alectinib | Alk | 4639 | −2 |
| MDA-MB-436 | GW441756 | NTRK1 | 5057 | −0.8 |
| MDA-MB-453 | FGFR_0939 | FGFR4 | 643 | −2.6 |
| MDA-MB-468 | Amuvatinib | Kit, PDGFRA, FLT3 | 1267 | −2.2 |
| Inhibitor | Main Target Kinase | Signaling Pathway |
|---|---|---|
| Alectinib | Alk, Ret | Receptor PTKs |
| BMS-754807 | Insulin receptor, IGF-1R | Receptor PTKs |
| Cabozantinib | Tet, VEGFR | Receptor PTKs |
| Crizotinib | Alk, Ros1, Met | Receptor PTKs |
| Erdafitinib | FGFR | Receptor PTKs |
| Erlotinib | EGFR | Receptor PTKs |
| Lapatinib | EGFR | Receptor PTKs |
| Nintadenib | VEGFR, PDGFR, FGFR | Receptor PTKs |
| Regorafenib | VEGFR | Receptor PTKs |
| Sunitinib | PDGFR, VEGFR | Receptor PTKs |
| MK-2206 | Akt | PI 3-Kinase pathway |
| GSK690693 | Akt | PI 3-kinase pathway |
| Vemurafenib | BRAF | MAP kinase pathway |
| Dabrafenib | BRAF | MAP kinase pathway |
| Trametinib | Mek | MAP kinase pathway |
| Binimetinib | Mek | MAP kinase pathway |
| Bosutinib | Src, Abl | Cytoplasmic PTKs |
| Dasatinib | Src, Abl | Cytoplasmic PTKs |
| Saracatinib | Src, Abl | Cytoplasmic PTKs |
| Nilotinib | Abl | Cytoplasmic PTKs |
| Inhibitor | Target Kinase | IC50 (nM) | Imax (%) | n |
|---|---|---|---|---|
| Dabrafenib | BRAF | 2.4 ± 0.5 | 98.0 ± 0.25 | 1.35 ± 0.2 |
| Vemurafenib | BRAF | 507 ± 16 | 95.0 ± 0.9 | 1.29 ± 0.07 |
| Trametinib | Mek | 0.28 ± 0.03 | 96.2 ± 0.35 | 1.76 ± 0.23 |
| Binimetinib | Mek | 7.3 ± 1.5 | 96.4 ± 0.8 | 1.50 ± 0.34 |
| Inhibitor | Hill Analysis | Biphasic Analysis | |||||
|---|---|---|---|---|---|---|---|
| IC50 (nM) | n | Imax (%) | F1 (%) | Kd1 (nM) | F2 (%) | Kd2 (μM) | |
| Dasatinib | 66 ± 8.5 | 0.65 ± 0.03 | 73 ± 1.3 | 55 ± 0.8 | 27 ± 3.3 | 45 ± 0.8 | 18 ± 0.5 |
| Trametinib | 13 ± 1.3 | 0.97 ± 0.03 | 26 ± 3.1 | 22 ± 1.9 | 7.7 ± 2.9 | 79 ± 1.9 | >100 |
| Binimetinib | 199 ± 5.8 | 0.92 ± 0.07 | 19 ± 0.5 | 16 ± 0.5 | 125 ± 21 | 84 ± 0.5 | >100 |
| Selumetinib | 563 ± 172 | 0.74 ± 0.11 | 43 ± 4.7 | 28 ± 3.8 | 156 ± 55 | 72 ± 3.9 | 97 ± 13 |
| Dasa + Tram | 8.2 ± 0.3 | 0.75 ± 0.03 | 98 ± 0.4 | ND | ND | ND | ND |
| Dasa + Bini | 64 ± 2.9 | 0.90 ± 0.03 | 99 ± 0.5 | ND | ND | ND | ND |
| Dasa + Selu | 78 ± 11 | 0.71 ± 0.04 | 94 ± 0.3 | ND | ND | ND | ND |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ku, G.C.; Chapdelaine, A.G.; Ayrapetov, M.K.; Sun, G. Identification of Lethal Inhibitors and Inhibitor Combinations for Mono-Driver versus Multi-Driver Triple-Negative Breast Cancer Cells. Cancers 2022, 14, 4027. https://doi.org/10.3390/cancers14164027
Ku GC, Chapdelaine AG, Ayrapetov MK, Sun G. Identification of Lethal Inhibitors and Inhibitor Combinations for Mono-Driver versus Multi-Driver Triple-Negative Breast Cancer Cells. Cancers. 2022; 14(16):4027. https://doi.org/10.3390/cancers14164027
Chicago/Turabian StyleKu, Geng Chia, Abygail G. Chapdelaine, Marina K. Ayrapetov, and Gongqin Sun. 2022. "Identification of Lethal Inhibitors and Inhibitor Combinations for Mono-Driver versus Multi-Driver Triple-Negative Breast Cancer Cells" Cancers 14, no. 16: 4027. https://doi.org/10.3390/cancers14164027
APA StyleKu, G. C., Chapdelaine, A. G., Ayrapetov, M. K., & Sun, G. (2022). Identification of Lethal Inhibitors and Inhibitor Combinations for Mono-Driver versus Multi-Driver Triple-Negative Breast Cancer Cells. Cancers, 14(16), 4027. https://doi.org/10.3390/cancers14164027




