How Reliable Are Gene Expression-Based and Immunohistochemical Biomarkers Assessed on a Core-Needle Biopsy? A Study of Paired Core-Needle Biopsies and Surgical Specimens in Early Breast Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
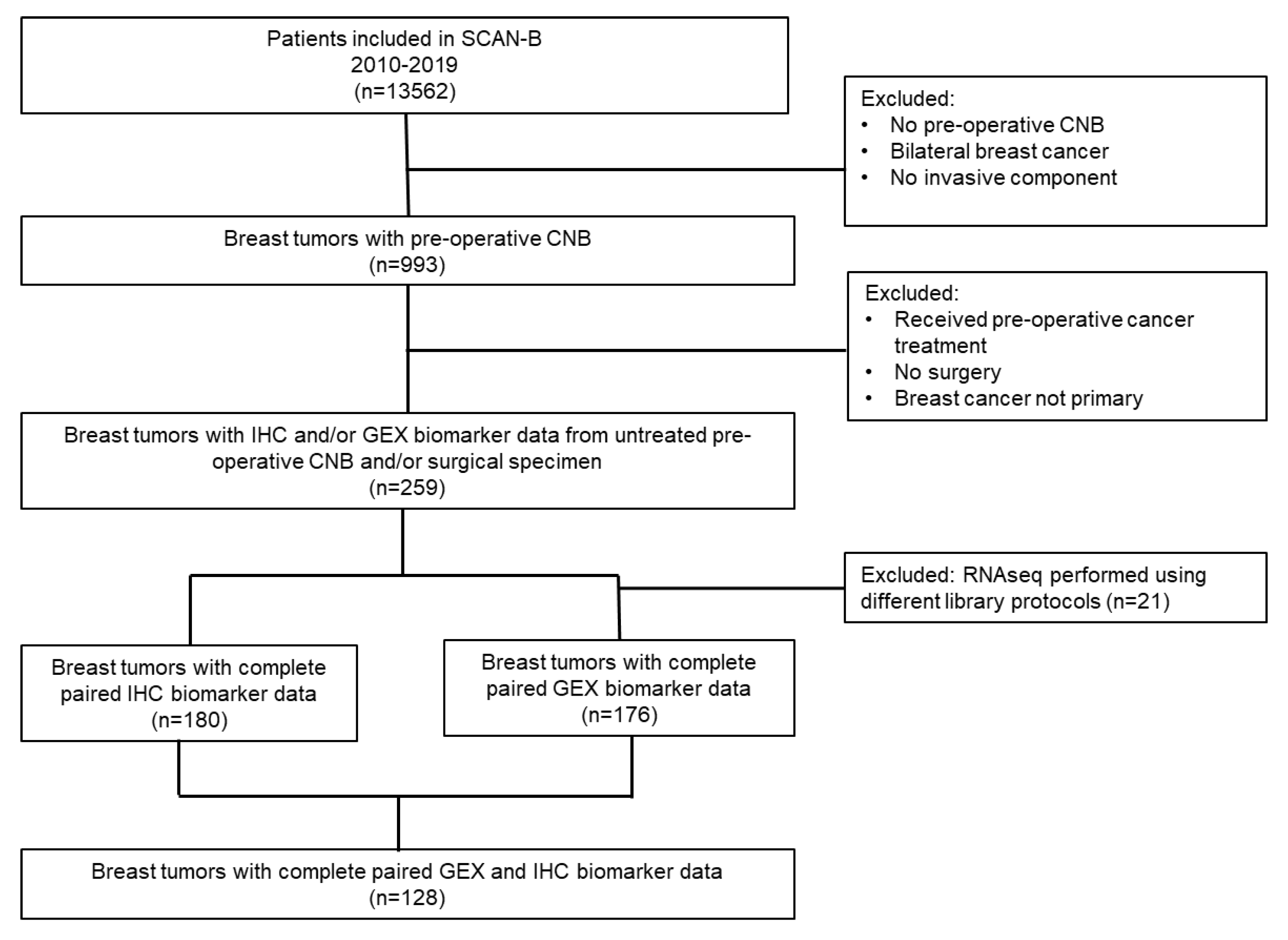
2. Materials and Methods
2.1. Study Population and Sample Collection
2.2. Biomarker Assessment Using Immunohistochemistry (IHC)
2.3. Gene Expression Analysis (GEX)
2.4. Statistical Analysis
3. Results
3.1. Distribution of Patient and Tumor Characteristics
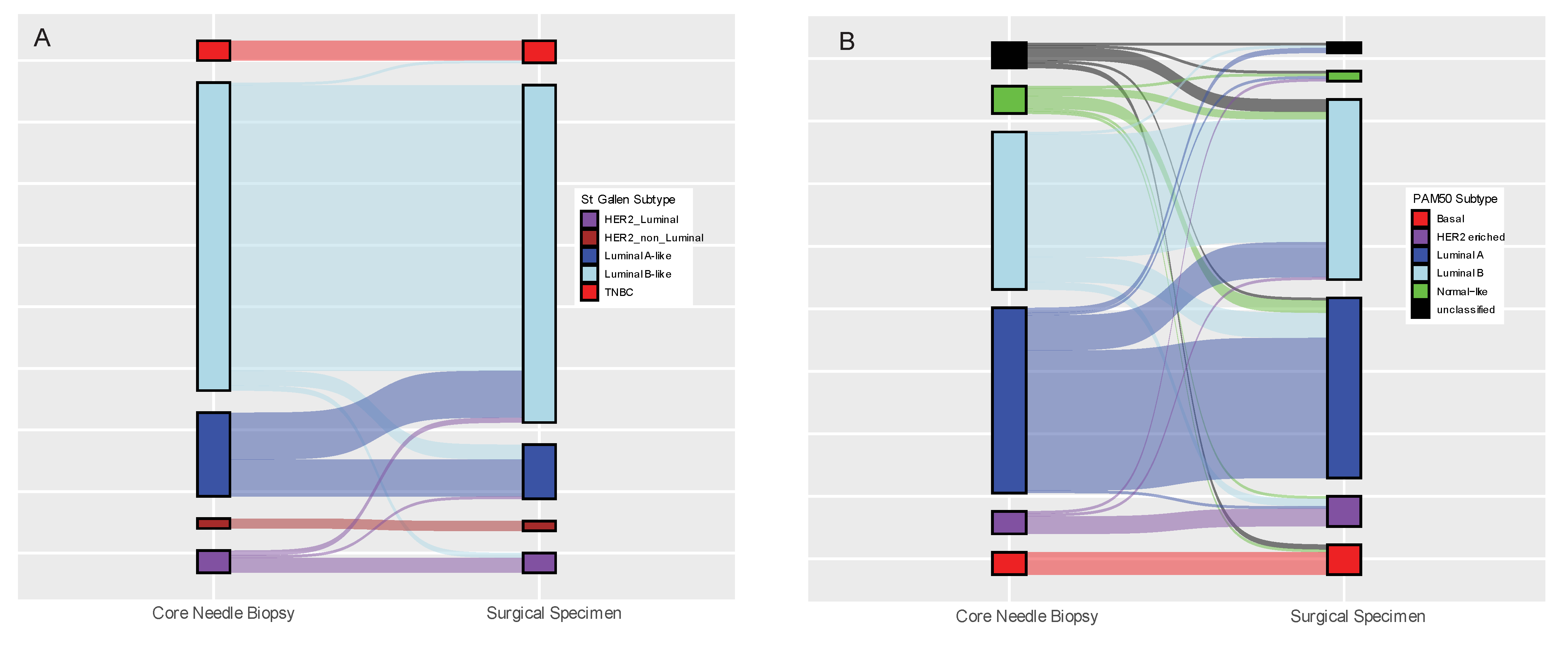
3.2. Concordance of Biomarker Status Analysed Using the Same Method (IHC or GEX) in Paired Core-Needle Biopsy and Surgical Specimen
3.3. Concordance between Biomarker Status Analyzed by GEX versus IHC for CNB or Surgical Specimens, Respectively
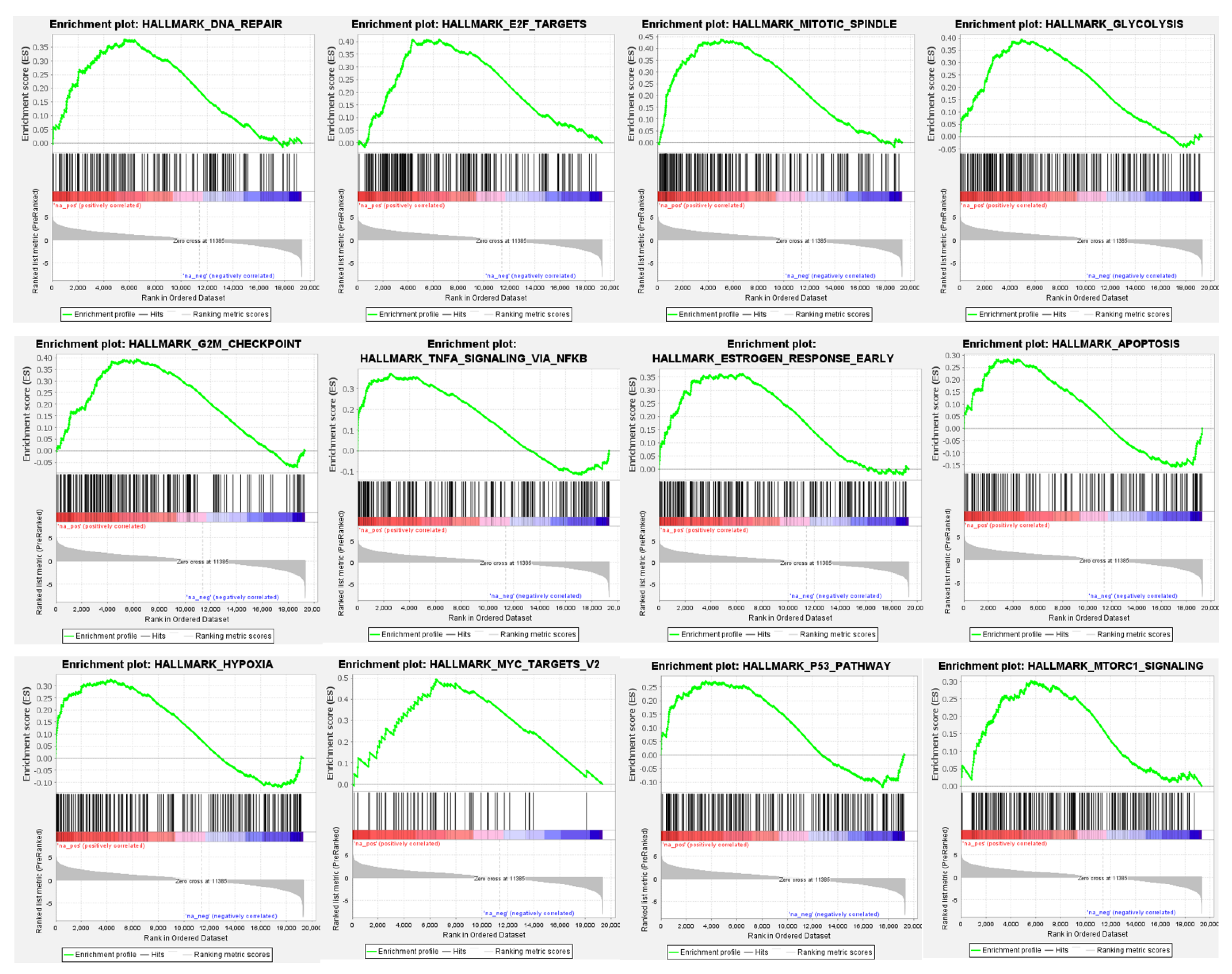
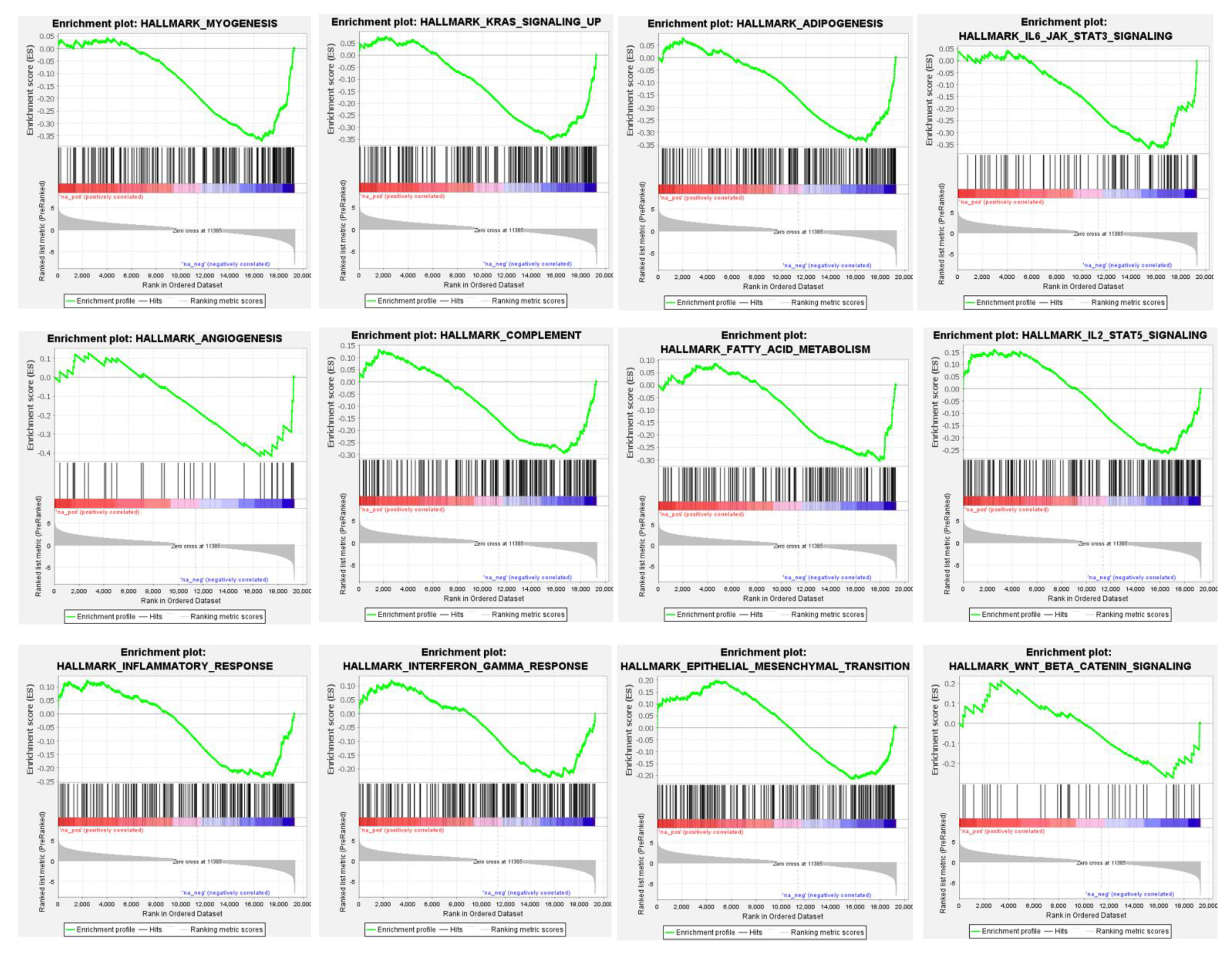
3.4. Differential Global Gene Expression Profiles of Paired CNBs and Surgical Specimens
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kooistra, B.; Wauters, C.; Strobbe, L.; Wobbes, T. Preoperative cytological and histological diagnosis of breast lesions: A critical review. Eur. J. Surg. Oncol. 2010, 36, 934–940. [Google Scholar] [CrossRef] [PubMed]
- Parker, S.H.; Burbank, F.; Jackman, R.J.; Aucreman, C.J.; Cardenosa, G.; Cink, T.M.; Coscia, J.L., Jr.; Eklund, G.W.; Evans, W.P.; Garver, P.R., III; et al. Percutaneous large-core breast biopsy: A multi-institutional study. Radiology 1994, 193, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Dahlstrom, J.E.; Jain, S.; Sutton, T.; Sutton, S. Diagnostic accuracy of stereotactic core biopsy in a mammographic breast cancer screening programme. Histopathology 1996, 28, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Verkooijen, H.M.; Peeters, P.H.; Buskens, E.; Koot, V.C.; Borel Rinkes, I.H.; Mali, W.P.; van Vroonhoven, T.J. Diagnostic accuracy of large-core needle biopsy for nonpalpable breast disease: A meta-analysis. Br. J. Cancer 2000, 82, 1017–1021. [Google Scholar] [CrossRef] [PubMed]
- Hoorntje, L.E.; Peeters, P.H.; Borel Rinkes, I.H.; Verkooijen, H.M.; Pijnappel, R.M.; Mali, W.P. Stereotactic large core needle biopsy for all nonpalpable breast lesions? Breast Cancer Res. Treat. 2002, 73, 177–182. [Google Scholar] [CrossRef]
- Verkooijen, H.M. Diagnostic accuracy of stereotactic large-core needle biopsy for nonpalpable breast disease: Results of a multicenter prospective study with 95% surgical confirmation. Int. J. Cancer 2002, 99, 853–859. [Google Scholar] [CrossRef]
- Dillon, M.F.; Hill, A.D.; Quinn, C.M.; O’Doherty, A.; McDermott, E.W.; O’Higgins, N. The accuracy of ultrasound, stereotactic, and clinical core biopsies in the diagnosis of breast cancer, with an analysis of false-negative cases. Ann. Surg. 2005, 242, 701–707. [Google Scholar] [CrossRef]
- Cancercentrum: Nationellt Kvalitetsregister för Bröstcancer (NKBC). 2020. Available online: https://statistik.incanet.se/brostcancer/ (accessed on 10 October 2021).
- Burstein, H.J.; Curigliano, G.; Loibl, S.; Dubsky, P.; Gnant, M.; Poortmans, P.; Colleoni, M.; Denkert, C.; Piccart-Gebhart, M.; Regan, M.; et al. Estimating the benefits of therapy for early-stage breast cancer: The St. Gallen International Consensus Guidelines for the primary therapy of early breast cancer 2019. Ann. Oncol. 2019, 30, 1541–1557. [Google Scholar] [CrossRef]
- Masuda, N.; Lee, S.J.; Ohtani, S.; Im, Y.H.; Lee, E.S.; Yokota, I.; Kuroi, K.; Im, S.A.; Park, B.W.; Kim, S.B.; et al. Adjuvant Capecitabine for Breast Cancer after Preoperative Chemotherapy. N. Engl. J. Med. 2017, 376, 2147–2159. [Google Scholar] [CrossRef]
- von Minckwitz, G.; Huang, C.S.; Mano, M.S.; Loibl, S.; Mamounas, E.P.; Untch, M.; Wolmark, N.; Rastogi, P.; Schneeweiss, A.; Redondo, A.; et al. Trastuzumab Emtansine for Residual Invasive HER2-Positive Breast Cancer. N. Engl. J. Med. 2019, 380, 617–628. [Google Scholar] [CrossRef]
- Tutt, A.N.J.; Garber, J.E.; Kaufman, B.; Viale, G.; Fumagalli, D.; Rastogi, P.; Gelber, R.D.; de Azambuja, E.; Fielding, A.; Balmaña, J.; et al. Adjuvant Olaparib for Patients with BRCA1- or BRCA2-Mutated Breast Cancer. N. Engl. J. Med. 2021, 384, 2394–2405. [Google Scholar] [CrossRef] [PubMed]
- EBCTCG. Long-term outcomes for neoadjuvant versus adjuvant chemotherapy in early breast cancer: Meta-analysis of individual patient data from ten randomised trials. Lancet Oncol. 2018, 19, 27–39. [Google Scholar] [CrossRef]
- Robertson, S.; Rönnlund, C.; de Boniface, J.; Hartman, J. Re-testing of predictive biomarkers on surgical breast cancer specimens is clinically relevant. Breast Cancer Res. Treat. 2019, 174, 795–805. [Google Scholar] [CrossRef] [PubMed]
- Greer, L.T.; Rosman, M.; Mylander, W.C.; Hooke, J.; Kovatich, A.; Sawyer, K.; Buras, R.R.; Shriver, C.D.; Tafra, L. Does breast tumor heterogeneity necessitate further immunohistochemical staining on surgical specimens? J. Am. Coll. Surg. 2013, 216, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Zell, J.A.; Tsang, W.Y.; Taylor, T.H.; Mehta, R.S.; Anton-Culver, H. Prognostic impact of human epidermal growth factor-like receptor 2 and hormone receptor status in inflammatory breast cancer (IBC): Analysis of 2,014 IBC patient cases from the California Cancer Registry. Breast Cancer Res. 2009, 11, R9. [Google Scholar] [CrossRef]
- Denkert, C.; Loibl, S.; Müller, B.M.; Eidtmann, H.; Schmitt, W.D.; Eiermann, W.; Gerber, B.; Tesch, H.; Hilfrich, J.; Huober, J.; et al. Ki67 levels as predictive and prognostic parameters in pretherapeutic breast cancer core biopsies: A translational investigation in the neoadjuvant GeparTrio trial. Ann. Oncol. 2013, 24, 2786–2793. [Google Scholar] [CrossRef]
- Harris, L.N.; Ismaila, N.; McShane, L.M.; Andre, F.; Collyar, D.E.; Gonzalez-Angulo, A.M.; Hammond, E.H.; Kuderer, N.M.; Liu, M.C.; Mennel, R.G.; et al. Use of Biomarkers to Guide Decisions on Adjuvant Systemic Therapy for Women with Early-Stage Invasive Breast Cancer: American Society of Clinical Oncology Clinical Practice Guideline. J. Clin. Oncol. 2016, 34, 1134–1150. [Google Scholar] [CrossRef]
- Roepman, P.; Horlings, H.M.; Krijgsman, O.; Kok, M.; Bueno-de-Mesquita, J.M.; Bender, R.; Linn, S.C.; Glas, A.M.; van de Vijver, M.J. Microarray-based determination of estrogen receptor, progesterone receptor, and HER2 receptor status in breast cancer. Clin. Cancer Res. 2009, 15, 7003–7011. [Google Scholar] [CrossRef]
- Wilson, T.R.; Xiao, Y.; Spoerke, J.M.; Fridlyand, J.; Koeppen, H.; Fuentes, E.; Huw, L.Y.; Abbas, I.; Gower, A.; Schleifman, E.B.; et al. Development of a robust RNA-based classifier to accurately determine ER, PR, and HER2 status in breast cancer clinical samples. Breast Cancer Res. Treat. 2014, 148, 315–325. [Google Scholar] [CrossRef]
- Rantalainen, M.; Klevebring, D.; Lindberg, J.; Ivansson, E.; Rosin, G.; Kis, L.; Celebioglu, F.; Fredriksson, I.; Czene, K.; Frisell, J.; et al. Sequencing-based breast cancer diagnostics as an alternative to routine biomarkers. Sci. Rep. 2016, 6, 38037. [Google Scholar] [CrossRef]
- Bastani, M.; Vos, L.; Asgarian, N.; Deschenes, J.; Graham, K.; Mackey, J.; Greiner, R. A machine learned classifier that uses gene expression data to accurately predict estrogen receptor status. PLoS ONE 2013, 8, e82144. [Google Scholar] [CrossRef]
- Brueffer, C.; Vallon-Christersson, J.; Grabau, D.; Ehinger, A.; Häkkinen, J.; Hegardt, C.; Malina, J.; Chen, Y.; Bendahl, P.O.; Manjer, J.; et al. Clinical Value of RNA Sequencing-Based Classifiers for Prediction of the Five Conventional Breast Cancer Biomarkers: A Report from the Population-Based Multicenter Sweden Cancerome Analysis Network-Breast Initiative. JCO Precis. Oncol. 2018, 2, PO.17.00135. [Google Scholar] [CrossRef]
- Cardoso, F.; van’t Veer, L.J.; Bogaerts, J.; Slaets, L.; Viale, G.; Delaloge, S.; Pierga, J.Y.; Brain, E.; Causeret, S.; DeLorenzi, M.; et al. 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. N. Engl. J. Med. 2016, 375, 717–729. [Google Scholar] [CrossRef]
- Gnant, M.; Filipits, M.; Greil, R.; Stoeger, H.; Rudas, M.; Bago-Horvath, Z.; Mlineritsch, B.; Kwasny, W.; Knauer, M.; Singer, C.; et al. Predicting distant recurrence in receptor-positive breast cancer patients with limited clinicopathological risk: Using the PAM50 Risk of Recurrence score in 1478 postmenopausal patients of the ABCSG-8 trial treated with adjuvant endocrine therapy alone. Ann. Oncol. 2014, 25, 339–345. [Google Scholar] [CrossRef]
- Filipits, M.; Nielsen, T.O.; Rudas, M.; Greil, R.; Stöger, H.; Jakesz, R.; Bago-Horvath, Z.; Dietze, O.; Regitnig, P.; Gruber-Rossipal, C.; et al. The PAM50 risk-of-recurrence score predicts risk for late distant recurrence after endocrine therapy in postmenopausal women with endocrine-responsive early breast cancer. Clin. Cancer Res. 2014, 20, 1298–1305. [Google Scholar] [CrossRef]
- Sparano, J.A.; Gray, R.J.; Ravdin, P.M.; Makower, D.F.; Pritchard, K.I.; Albain, K.S.; Hayes, D.F.; Geyer, C.E.; Dees, E.C., Jr.; Goetz, M.P.; et al. Clinical and Genomic Risk to Guide the Use of Adjuvant Therapy for Breast Cancer. N. Engl. J. Med. 2019, 380, 2395–2405. [Google Scholar] [CrossRef]
- Paik, S.; Shak, S.; Tang, G.; Kim, C.; Baker, J.; Cronin, M.; Baehner, F.L.; Walker, M.G.; Watson, D.; Park, T.; et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 2004, 351, 2817–2826. [Google Scholar] [CrossRef]
- Parker, J.S.; Mullins, M.; Cheang, M.C.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef]
- Saal, L.H.; Vallon-Christersson, J.; Häkkinen, J.; Hegardt, C.; Grabau, D.; Winter, C.; Brueffer, C.; Tang, M.H.; Reuterswärd, C.; Schulz, R.; et al. The Sweden Cancerome Analysis Network—Breast (SCAN-B) Initiative: A large-scale multicenter infrastructure towards implementation of breast cancer genomic analyses in the clinical routine. Genome Med. 2015, 7, 20. [Google Scholar] [CrossRef]
- Rydén, L.; Loman, N.; Larsson, C.; Hegardt, C.; Vallon-Christersson, J.; Malmberg, M.; Lindman, H.; Ehinger, A.; Saal, L.H.; Borg, Å. Minimizing inequality in access to precision medicine in breast cancer by real-time population-based molecular analysis in the SCAN-B initiative. Br. J. Surg. 2018, 105, e158–e168. [Google Scholar] [CrossRef]
- Cancercentrum R: Kvalitetsbilaga för Bröstpatologi (KVAST-bilaga). 2020. Available online: https://kunskapsbanken.cancercentrum.se/diagnoser/brostcancer/vardprogram/kvalitetsdokument-for--patologi/ (accessed on 5 October 2021).
- Goldhirsch, A.; Winer, E.P.; Coates, A.S.; Gelber, R.D.; Piccart-Gebhart, M.; Thürlimann, B.; Senn, H.J. Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann. Oncol. 2013, 24, 2206–2223. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, T.O.; Leung, S.C.Y.; Rimm, D.L.; Dodson, A.; Acs, B.; Badve, S.; Denkert, C.; Ellis, M.J.; Fineberg, S.; Flowers, M.; et al. Assessment of Ki67 in Breast Cancer: Updated Recommendations from the International Ki67 in Breast Cancer Working Group. J. Natl. Cancer Inst. 2021, 113, 808–819. [Google Scholar] [CrossRef] [PubMed]
- Tusher, V.G.; Tibshirani, R.; Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 2001, 98, 5116–5121. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Usami, S.; Moriya, T.; Kasajima, A.; Suzuki, A.; Ishida, T.; Sasano, H.; Ohuchi, N. Pathological aspects of core needle biopsy for non-palpable breast lesions. Breast Cancer 2005, 12, 272–278. [Google Scholar] [CrossRef]
- Cahill, R.A.; Walsh, D.; Landers, R.J.; Watson, R.G. Preoperative profiling of symptomatic breast cancer by diagnostic core biopsy. Ann. Surg. Oncol. 2006, 13, 45–51. [Google Scholar] [CrossRef]
- Usami, S.; Moriya, T.; Amari, M.; Suzuki, A.; Ishida, T.; Sasano, H.; Ohuchi, N. Reliability of prognostic factors in breast carcinoma determined by core needle biopsy. Jpn. J. Clin. Oncol. 2007, 37, 250–255. [Google Scholar] [CrossRef]
- Burge, C.N.; Chang, H.R.; Apple, S.K. Do the histologic features and results of breast cancer biomarker studies differ between core biopsy and surgical excision specimens? Breast 2006, 15, 167–172. [Google Scholar] [CrossRef]
- Connor, C.S.; Tawfik, O.W.; Joyce, A.J.; Davis, M.K.; Mayo, M.S.; Jewell, W.R. A comparison of prognostic tumor markers obtained on image-guided breast biopsies and final surgical specimens. Am. J. Surg. 2002, 184, 322–324. [Google Scholar] [CrossRef]
- Di Loreto, C.; Puglisi, F.; Rimondi, G.; Zuiani, C.; Anania, G.; Della Mea, V.; Beltrami, C.A. Large core biopsy for diagnostic and prognostic evaluation of invasive breast carcinomas. Eur. J. Cancer 1996, 32, 1693–1700. [Google Scholar] [CrossRef]
- Mann, G.B.; Fahey, V.D.; Feleppa, F.; Buchanan, M.R. Reliance on hormone receptor assays of surgical specimens may compromise outcome in patients with breast cancer. J. Clin. Oncol. 2005, 23, 5148–5154. [Google Scholar] [CrossRef] [PubMed]
- Romero, Q.; Bendahl, P.O.; Klintman, M.; Loman, N.; Ingvar, C.; Rydén, L.; Rose, C.; Grabau, D.; Borgquist, S. Ki67 proliferation in core biopsies versus surgical samples—A model for neo-adjuvant breast cancer studies. BMC Cancer 2011, 11, 341. [Google Scholar] [CrossRef] [PubMed]
- Hadad, S.M.; Jordan, L.B.; Roy, P.G.; Purdie, C.A.; Iwamoto, T.; Pusztai, L.; Moulder-Thompson, S.L.; Thompson, A.M. A prospective comparison of ER, PR, Ki67 and gene expression in paired sequential core biopsies of primary, untreated breast cancer. BMC Cancer 2016, 16, 745. [Google Scholar] [CrossRef]
- Orozco, J.I.J.; Chang, S.C.; Matsuba, C.; Ensenyat-Mendez, M.; Grunkemeier, G.L.; Marzese, D.M.; Grumley, J.G. Is the 21-Gene Recurrence Score on Core Needle Biopsy Equivalent to Surgical Specimen in Early-Stage Breast Cancer? A Comparison of Gene Expression Between Paired Core Needle Biopsy and Surgical Specimens. Ann. Surg. Oncol. 2021, 28, 5588–5596. [Google Scholar] [CrossRef] [PubMed]
- Qi, P.; Yang, Y.; Bai, Q.M.; Xue, T.; Ren, M.; Yao, Q.L.; Yang, W.T.; Zhou, X.Y. Concordance of the 21-gene assay between core needle biopsy and resection specimens in early breast cancer patients. Breast Cancer Res. Treat. 2021, 186, 327–342. [Google Scholar] [CrossRef]
- Valachis, A.; Mauri, D.; Polyzos, N.P.; Chlouverakis, G.; Mavroudis, D.; Georgoulias, V. Trastuzumab combined to neoadjuvant chemotherapy in patients with HER2-positive breast cancer: A systematic review and meta-analysis. Breast 2011, 20, 485–490. [Google Scholar] [CrossRef]
- Gianni, L.; Eiermann, W.; Semiglazov, V.; Manikhas, A.; Lluch, A.; Tjulandin, S.; Zambetti, M.; Vazquez, F.; Byakhow, M.; Lichinitser, M.; et al. Neoadjuvant chemotherapy with trastuzumab followed by adjuvant trastuzumab versus neoadjuvant chemotherapy alone, in patients with HER2-positive locally advanced breast cancer (the NOAH trial): A randomised controlled superiority trial with a parallel HER2-negative cohort. Lancet 2010, 375, 377–384. [Google Scholar]
- Korde, L.A.; Somerfield, M.R.; Carey, L.A.; Crews, J.R.; Denduluri, N.; Hwang, E.S.; Khan, S.A.; Loibl, S.; Morris, E.A.; Perez, A.; et al. Neoadjuvant Chemotherapy, Endocrine Therapy, and Targeted Therapy for Breast Cancer: ASCO Guideline. J. Clin. Oncol. 2021, 39, 1485–1505. [Google Scholar] [CrossRef]
- Cancercentrum R: Nationellt Vårdprogram Bröstcancer. 2020. Available online: https://cancercentrum.se/samverkan/cancerdiagnoser/brost/vardprogram/ (accessed on 7 October 2021).
- Acs, B.; Leung, S.C.Y.; Kidwell, K.M.; Arun, I.; Augulis, R.; Badve, S.S.; Bai, Y.; Bane, A.L.; Bartlett, J.M.S.; Bayani, J.; et al. Systematically higher Ki67 scores on core biopsy samples compared to corresponding resection specimen in breast cancer: A multi-operator and multi-institutional study. Mod. Pathol. 2022. [Google Scholar] [CrossRef]
- Smith, I.; Robertson, J.; Kilburn, L.; Wilcox, M.; Evans, A.; Holcombe, C.; Horgan, K.; Kirwan, C.; Mallon, E.; Sibbering, M.; et al. Long-term outcome and prognostic value of Ki67 after perioperative endocrine therapy in postmenopausal women with hormone-sensitive early breast cancer (POETIC): An open-label, multicentre, parallel-group, randomised, phase 3 trial. Lancet Oncol. 2020, 21, 1443–1454. [Google Scholar] [CrossRef]
- Vallon-Christersson, J. RNA Sequencing-Based Single Sample Predictors of Molecular Subtype and Risk of Recurrence for Clinical Assessment of Early-Stage Breast Cancer. Mendeley Data V1. 2022. Available online: https://data.mendeley.com/datasets/yzxtxn4nmd (accessed on 10 October 2021).
| Characteristic | N (%) | |
|---|---|---|
| Age | Years | 63 (27–88) * |
| Tumor Stage (T) | ||
| T1 | Tumor ≤ 20 mm | 120 (46%) |
| T2 | Tumor > 20 mm but ≤ 50 mm | 122 (47%) |
| T3 | Tumor > 50 mm | 12 (5%) |
| T4 | Tumor of any size with direct extension to chest wall or skin | 1 (0.4%) |
| Missing | 4 (2%) | |
| Nodal Status | ||
| N0 | No cancer in regional nodes | 210 (81%) |
| N+ | Cancer in regional nodes | 49 (19%) |
| Nottingham Histological Grade (NHG) | ||
| Grade 1 | Well differentiated | 31 (12%) |
| Grade 2 | Moderately differentiated | 128 (49%) |
| Grade 3 | Poorly differentiated | 91 (35%) |
| Missing | 9 (3%) |
| Biomarker (IHC) | CNB/Surgical Specimen | N | % Agreement | Kappa | McNemar’s p |
|---|---|---|---|---|---|
| ER (n = 183) | Pos/Pos | 169 | 98.91% | 0.917 | 1.000 |
| Neg/Neg | 12 | ||||
| Pos/Neg | 1 | ||||
| Neg/Pos | 1 | ||||
| PgR (n = 181) | Pos/Pos | 134 | 92.27% | 0.776 | 0.057 |
| Neg/Neg | 33 | ||||
| Pos/Neg | 3 | ||||
| Neg/Pos | 11 | ||||
| HER2 (n = 181) | Pos/Pos | 10 | 97.24% | 0.785 | 1.000 |
| Neg/Neg | 166 | ||||
| Pos/Neg | 3 | ||||
| Neg/Pos | 2 | ||||
| Ki67 (n = 181) | High/High | 129 | 82.87% | 0.474 | 0.011 |
| Low/Low | 21 | ||||
| High/Low | 8 | ||||
| Low/High | 23 | ||||
| St Gallen Subtype (n = 180) | LumA/LumA | 15 | 82.78% | 0.612 | 0.067 |
| LumB/LumB | 116 | ||||
| HER2/HER2 | 10 | ||||
| TNBC/TNBC | 8 | ||||
| Discordant subtype | 31 | ||||
| Biomarker (GEX) | CNB/Surgical Specimen | N | % Agreement | Kappa | McNemar’s p |
| ER (n = 176) | Pos/Pos | 133 | 88.07% | 0.606 | 0.027 |
| Neg/Neg | 22 | ||||
| Pos/Neg | 5 | ||||
| Neg/Pos | 16 | ||||
| PgR (n = 176) | Pos/Pos | 122 | 85.23% | 0.588 | 0.076 |
| Neg/Neg | 28 | ||||
| Pos/Neg | 8 | ||||
| Neg/Pos | 18 | ||||
| HER2 (n = 176) | Pos/Pos | 11 | 97.16% | 0.800 | 0.063 |
| Neg/Neg | 160 | ||||
| Pos/Neg | 0 | ||||
| Neg/Pos | 5 | ||||
| Ki67 (n = 176) | High/High | 48 | 86.36 | 0.699 | 0.002 |
| Low/Low | 104 | ||||
| High/Low | 4 | ||||
| Low/High | 20 | ||||
| Pam50 Subtype (n = 176) | LumA/LumA | 56 | 69.89% | 0.552 | 0.168 |
| LumB/LumB | 49 | ||||
| HER2/HER2 | 7 | ||||
| Basal/Basal | 9 | ||||
| Normal-like/Normal-like | 1 | ||||
| Unclassified/Unclassified | 1 | ||||
| Discordant subtype | 53 |
| Biomarker (CNBs) | IHC/GEX | N | % Agreement | Kappa |
|---|---|---|---|---|
| ER (n = 168) | Pos/Pos | 134 | 85.71% | 0.395 |
| Neg/Neg | 10 | |||
| Pos/Neg | 23 | |||
| Neg/Pos | 1 | |||
| PgR (n = 167) | Pos/Pos | 110 | 80.83% | 0.484 |
| Neg/Neg | 25 | |||
| Pos/Neg | 19 | |||
| Neg/Pos | 13 | |||
| HER2 (n = 166) | Pos/Pos | 8 | 96.64% | 0.708 |
| Neg/Neg | 152 | |||
| Pos/Neg | 3 | |||
| Neg/Pos | 3 | |||
| Ki67 (n = 166) | High/High | 43 | 47.59% | 0.152 |
| Low/Low | 36 | |||
| High/Low | 84 | |||
| Low/High | 3 | |||
| Subtype (n = 150) | LumA/LumA | 23 | 57.33% | 0.264 |
| LumB/LumB | 52 | |||
| HER2/HER2 | 6 | |||
| TNBC/Basal | 5 | |||
| Discordant subtype | 64 | |||
| Biomarker (Surgical) | IHC/GEX | N | % Agreement | Kappa |
| ER (n = 213) | Pos/Pos | 176 | 91.54% | 0.636 |
| Neg/Neg | 19 | |||
| Pos/Neg | 18 | |||
| Neg/Pos | 0 | |||
| PgR (n = 212) | Pos/Pos | 153 | 86.32% | 0.588 |
| Neg/Neg | 30 | |||
| Pos/Neg | 16 | |||
| Neg/Pos | 13 | |||
| HER2 (n = 216) | Pos/Pos | 14 | 93.52% | 0.632 |
| Neg/Neg | 188 | |||
| Pos/Neg | 3 | |||
| Neg/Pos | 11 | |||
| Ki67 (n = 215) | High/High | 83 | 55.34% | 0.205 |
| Low/Low | 36 | |||
| High/Low | 99 | |||
| Low/High | 0 | |||
| Subtype (n = 203) | LumA/LumA | 24 | 61.58% | 0.398 |
| LumB/LumB | 79 | |||
| HER2/HER2 | 11 | |||
| TNBC/Basal | 11 | |||
| Discordant subtype | 78 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saghir, H.; Veerla, S.; Malmberg, M.; Rydén, L.; Ehinger, A.; Saal, L.H.; Vallon-Christersson, J.; Borg, Å.; Hegardt, C.; Larsson, C.; et al. How Reliable Are Gene Expression-Based and Immunohistochemical Biomarkers Assessed on a Core-Needle Biopsy? A Study of Paired Core-Needle Biopsies and Surgical Specimens in Early Breast Cancer. Cancers 2022, 14, 4000. https://doi.org/10.3390/cancers14164000
Saghir H, Veerla S, Malmberg M, Rydén L, Ehinger A, Saal LH, Vallon-Christersson J, Borg Å, Hegardt C, Larsson C, et al. How Reliable Are Gene Expression-Based and Immunohistochemical Biomarkers Assessed on a Core-Needle Biopsy? A Study of Paired Core-Needle Biopsies and Surgical Specimens in Early Breast Cancer. Cancers. 2022; 14(16):4000. https://doi.org/10.3390/cancers14164000
Chicago/Turabian StyleSaghir, Hani, Srinivas Veerla, Martin Malmberg, Lisa Rydén, Anna Ehinger, Lao H. Saal, Johan Vallon-Christersson, Åke Borg, Cecilia Hegardt, Christer Larsson, and et al. 2022. "How Reliable Are Gene Expression-Based and Immunohistochemical Biomarkers Assessed on a Core-Needle Biopsy? A Study of Paired Core-Needle Biopsies and Surgical Specimens in Early Breast Cancer" Cancers 14, no. 16: 4000. https://doi.org/10.3390/cancers14164000
APA StyleSaghir, H., Veerla, S., Malmberg, M., Rydén, L., Ehinger, A., Saal, L. H., Vallon-Christersson, J., Borg, Å., Hegardt, C., Larsson, C., Haidar, A., Hedenfalk, I., Loman, N., & Kimbung, S. (2022). How Reliable Are Gene Expression-Based and Immunohistochemical Biomarkers Assessed on a Core-Needle Biopsy? A Study of Paired Core-Needle Biopsies and Surgical Specimens in Early Breast Cancer. Cancers, 14(16), 4000. https://doi.org/10.3390/cancers14164000







