A Novel Deep Learning-Based Mitosis Recognition Approach and Dataset for Uterine Leiomyosarcoma Histopathology
Abstract
:Simple Summary
Abstract
1. Introduction
- Release of 150 annotated bounding box dataset for uterine leiomyosarcoma histopathology and a baseline method for mitosis detection. To the best of our knowledge, this is the first study on leiomyosarcoma histopathology where a dataset and automated method for mitosis detection is provided. As stated earlier, the proposed method obviates the need for manually annotating that further reduces human errors.
- Benchmarks for the provided dataset using the YOLOv4 detection technique are provided. Moreover, standard computer vision metrics such as precision, recall, and F1-score are used for comparison with possible future work.
- An end-to-end framework for the detection of mitosis in ULMS, describing the data capturing, annotations, and detection, is provided.
- For research and development, the code and dataset are made publicly available.
2. Related Work
2.1. Handcrafted Features-Based
2.2. Deep Features-Based
3. Materials and Methods
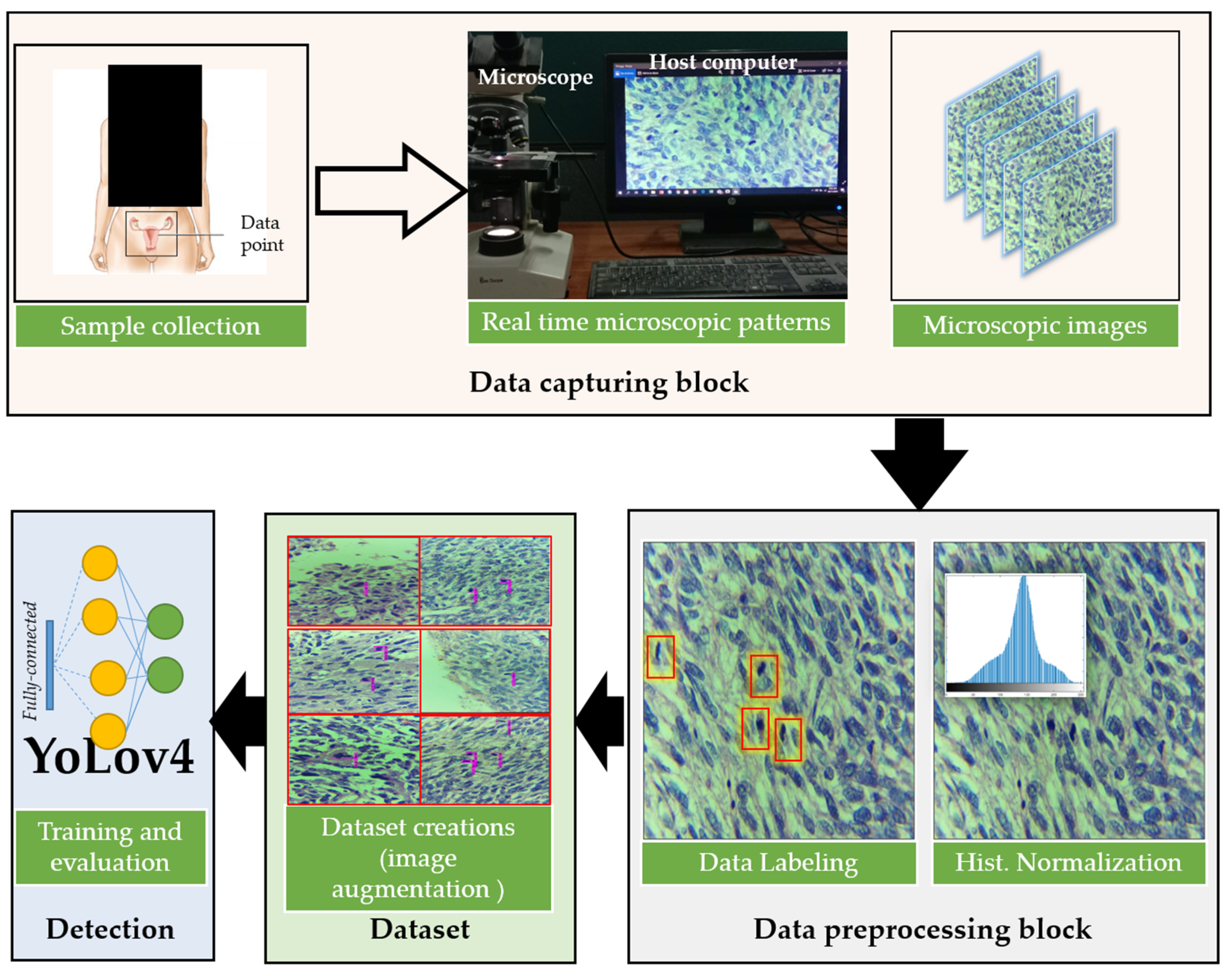
3.1. Overview of the Proposed Method
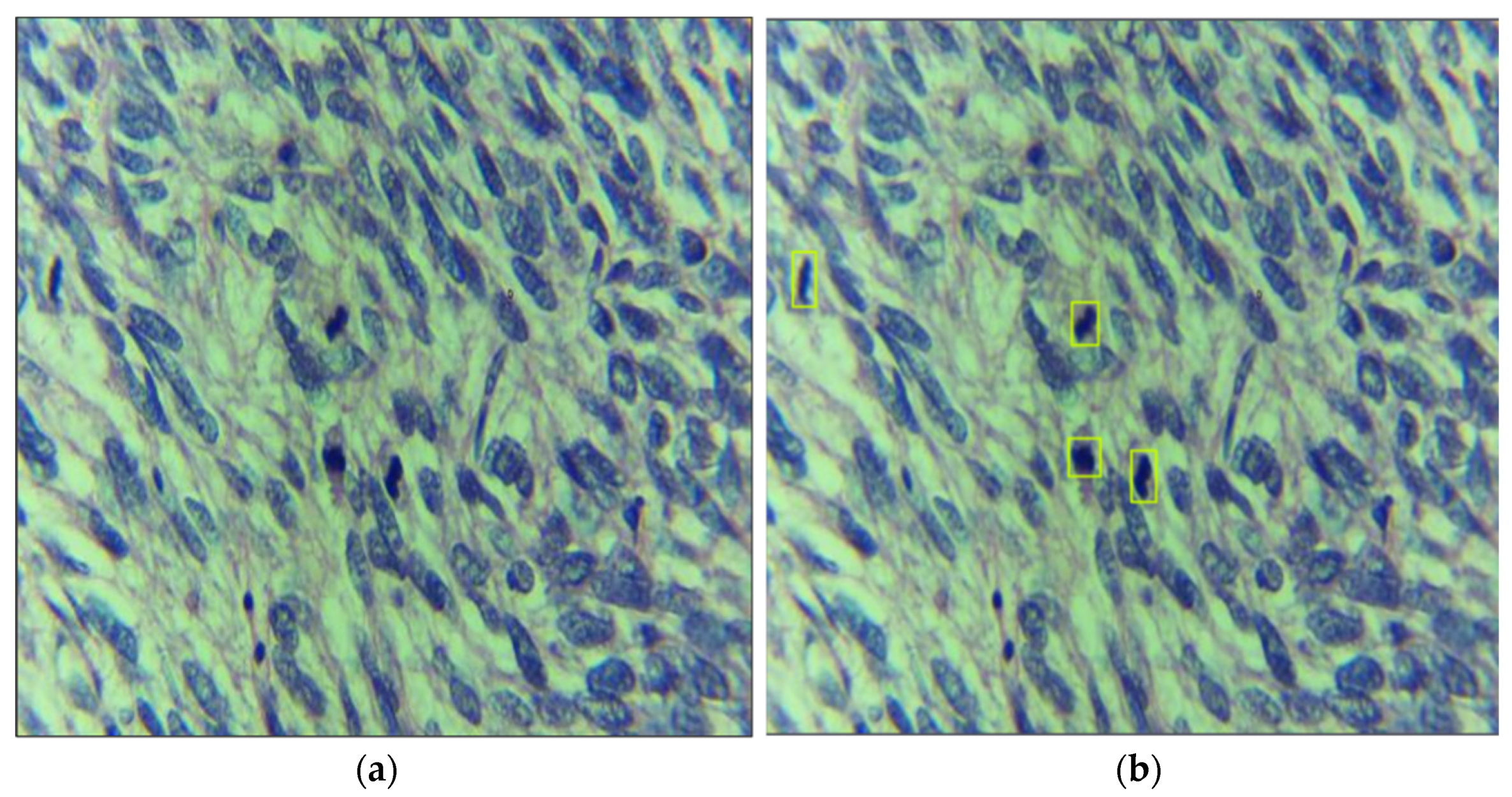
3.2. Dataset Acquisition, Preprocessing, and Labeling
3.3. Baseline Models
- Simple: YOLO is available in many libraries as an in-built example.
- Fast to set up: Unlike faster RCNN where first regions are divided and then classification is performed, YOLO performs the detection of region and classification all simultaneously. With CNN and RCNN, all of the potential regions need separate classification.
- Supports both GPUs and CPUs: In case the hardware does not have a GPU, only the CPU is capable of running the YOLO network.
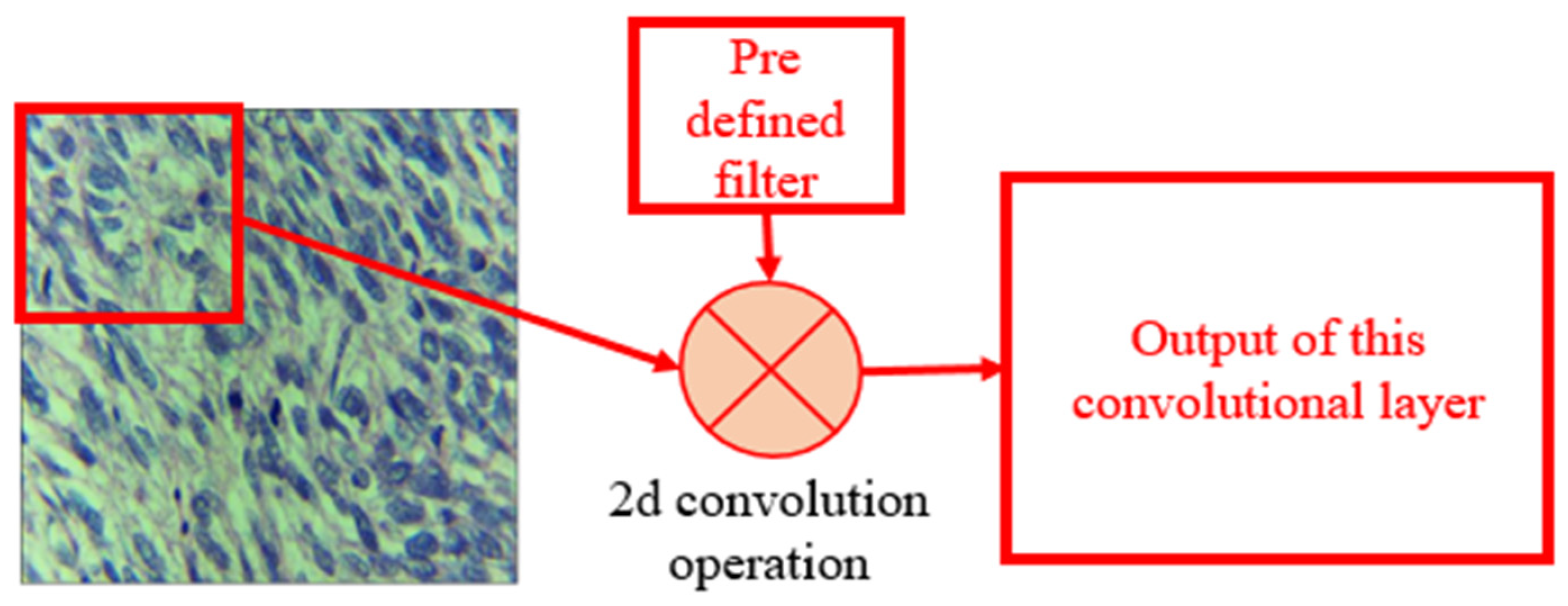
- CBL: Convolution, batch normalization, and leaky-ReLU module made up by combining the convolution layer, a batch normalization layer, and a leaky-ReLU activation function. The convolution layer was the main heart of this network where the input images convolved with a filter to find the output for the next convolution layer. The operation of the convolution layer is shown in Figure 5. The batch normalization block normalized the data to minimize the outlier’s effects on the data. A rectified linear unit (ReLU) was used as an activation function.
- CBM: As expressed in Figure 4b the CBM blocks shown in the figure performs convolution, batch normalization, and MISH, which serves as a non-monotonic activation function.
- Concatenation: This block simply concatenates the output of different intermediate layers to form the input for the next layer.
4. Results
4.1. Training
4.2. Performance Evaluations Metrics
- True Positive (TP): When our model correctly predicts mitosis.
- False Positive (FP): When there is no mitosis in the input image and the proposed algorithm still detects the mitosis.
- False Negative (FN): When there is mitosis in the input image and the proposed algorithm miss detects the mitosis.
- Confidence: The confidence score shows how confident the YOLO is regarding the presence of the mitosis region.
- Precision: It shows how much positive detection of the mitosis is actually correct. Equation (1) shows the precision:
- 6.
- Recall: From the correct mitosis, what portion is detected successfully. Equation (2), shows the recall:
- 7.
- F1-Score: The F1-score is calculated based on precision and recall. The higher the F-1 score the better the algorithm. Equation (3) represents the F1-score as follows:
4.3. Overall Performance Evaluation
4.4. Statistical Significance Tests
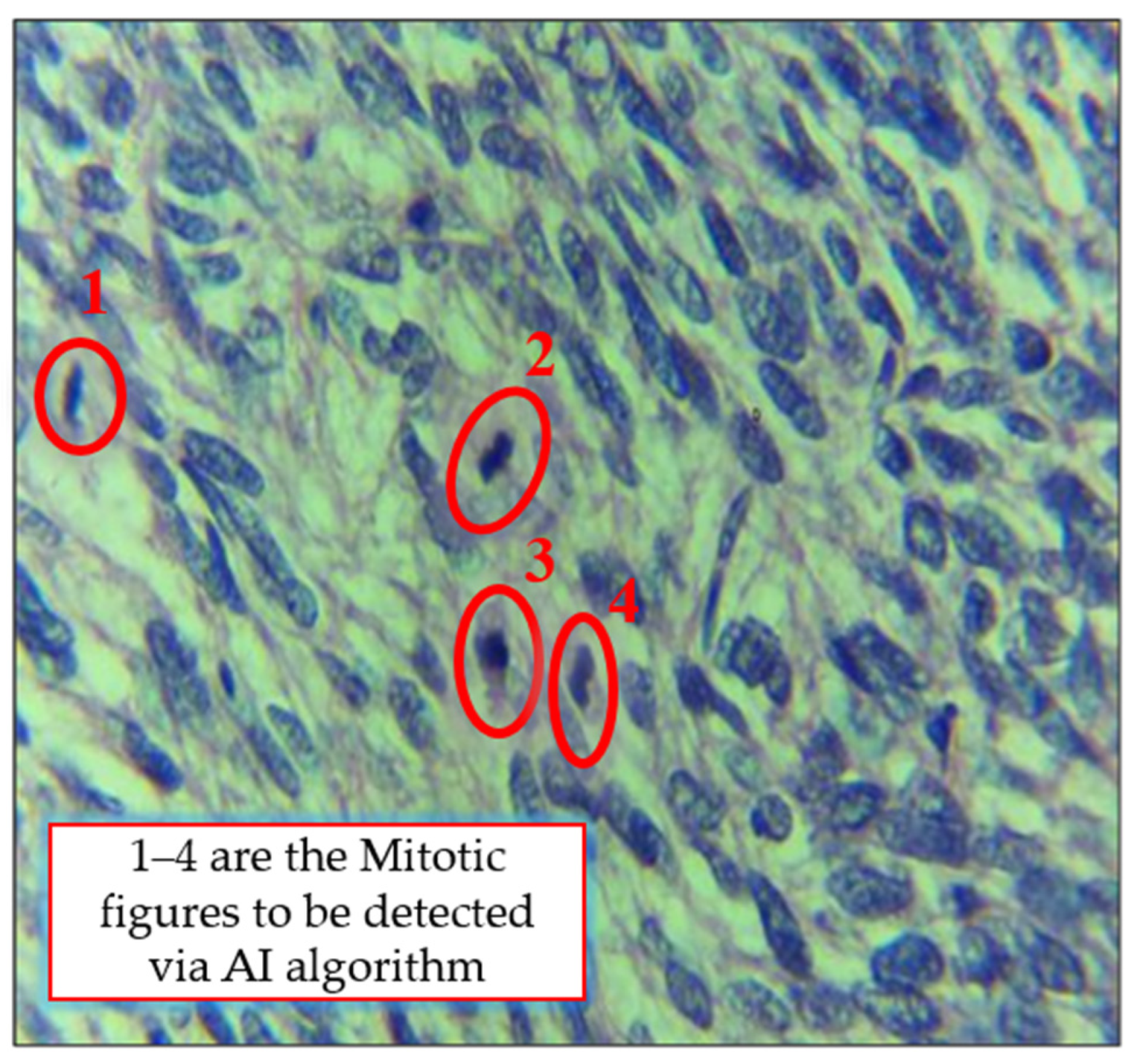
4.5. Visualization of Results
4.6. Comparison with the State-of-the-Art Methods
5. Discussion
- The development of AI-based methods is decreasing the gap between pathologists and computers. With the advent of technology, the trust level of stakeholders is increasing.
- In the case of ULMS, significant variations are observed among the mitosis objects as compared to other tumors. These variations increase the difficulty level of the mitosis detection task.
- It is crucial to carefully assess the morphological characteristics of the mitosis objects owing to the strong resemblance between leiomyoma, STUMP, and leiomyosarcoma. The leiomyomas are usually multiple and mostly do not show increased cellularity, significant nuclear atypia, or mitotic activity. However, cellular variants as well as variants with bizarre nuclei and those which are mitotically active are sometimes seen.
- Mitotically active leiomyomas often have more than 10 mitoses per 10 high power fields (HPF) but typically lack nuclear atypia or tumor necrosis. Uterine tumors labeled as STUMP may show focal or multifocal to diffuse, moderate to severe atypia, and mitotic count <10 (mean 3 to 4) per 10 HPF. Tumor necrosis is absent. Still, other cases show no atypia.
- AI-based techniques automatically detect mitotically active cells in the histopathology images (thereby accelerating the diagnosis) and are time efficient.
- This study opens a new door by providing a dataset and a baseline. However, its limitation is the size of the dataset used. In the future, the dataset size should be increased.
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Roberts, M.E.; Aynardi, J.T.; Chu, C.S. Uterine Leiomyosarcoma: A Review of the Literature and Update on Management Options. Gynecol. Oncol. 2018, 151, 562–572. [Google Scholar] [CrossRef] [PubMed]
- George, S.; Serrano, C.; Hensley, M.L.; Ray-Coquard, I. Soft Tissue and Uterine Leiomyosarcoma. J. Clin. Oncol. 2018, 36, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Kaur, K.; Kaur, P.; Kaur, A.; Singla, A. Uterine Leiomyosarcoma: A Case Report. J. Midlife Health 2014, 5, 202–204. [Google Scholar] [CrossRef] [PubMed]
- Maclean, A.; Kamal, A.; Adishesh, M.; Alnafakh, R.; Tempest, N.; Hapangama, D.K. Human Uterine Biopsy: Research Value and Common Pitfalls. Int. J. Reprod. Med. 2020, 2020, e9275360. [Google Scholar] [CrossRef] [PubMed]
- Selvanathan, S.; Acharya, N.; Singhal, S. Quality of Life after Hysterectomy and Uterus-Sparing Hysteroscopic Management of Abnormal Uterine Bleeding or Heavy Menstrual Bleeding. J. Midlife Health 2019, 10, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.W.; Kempson, R.L.; Hendrickson, M.R. Problematic Uterine Smooth Muscle Neoplasms: A Clinicopathologic Study of 213 Cases. Am. J. Surg. Pathol. 1994, 18, 535–558. [Google Scholar] [CrossRef]
- Chapel, D.B.; Sharma, A.; Lastra, R.R.; Maccio, L.; Bragantini, E.; Zannoni, G.F.; George, S.; Quade, B.J.; Parra-Herran, C.; Nucci, M.R. A Novel Morphology-Based Risk Stratification Model for Stage I Uterine Leiomyosarcoma: An Analysis of 203 Cases. Mod. Pathol. 2022, 35, 794–807. [Google Scholar] [CrossRef]
- Li, C.; Wang, X.; Liu, W.; Latecki, L.J. DeepMitosis: Mitosis Detection via Deep Detection, Verification and Segmentation Networks. Med. Image Anal. 2018, 45, 121–133. [Google Scholar] [CrossRef]
- Arsalan, M.; Owais, M.; Mahmood, T.; Cho, S.W.; Park, K.R. Aiding the Diagnosis of Diabetic and Hypertensive Retinopathy Using Artificial Intelligence-Based Semantic Segmentation. J. Clin. Med. 2019, 8, 1446. [Google Scholar] [CrossRef] [Green Version]
- Mahmood, T.; Arsalan, M.; Owais, M.; Lee, M.B.; Park, K.R. Artificial Intelligence-Based Mitosis Detection in Breast Cancer Histopathology Images Using Faster R-CNN and Deep CNNs. J. Clin. Med. 2020, 9, 749. [Google Scholar] [CrossRef] [Green Version]
- Sheikh, T.S.; Lee, Y.; Cho, M. Histopathological Classification of Breast Cancer Images Using a Multi-Scale Input and Multi-Feature Network. Cancers 2020, 12, 2031. [Google Scholar] [CrossRef] [PubMed]
- Deep Learning Assisted Mitotic Counting for Breast Cancer|Laboratory Investigation. Available online: https://www.nature.com/articles/s41374-019-0275-0 (accessed on 23 June 2022).
- Wang, M.; Aung, P.P.; Prieto, V.G. Standardized Method for Defining a 1-Mm2 Region of Interest for Calculation of Mitotic Rate on Melanoma Whole Slide Images. Arch. Pathol. Lab. Med. 2021, 145, 1255–1263. [Google Scholar] [CrossRef] [PubMed]
- Tabata, K.; Uraoka, N.; Benhamida, J.; Hanna, M.G.; Sirintrapun, S.J.; Gallas, B.D.; Gong, Q.; Aly, R.G.; Emoto, K.; Matsuda, K.M.; et al. Validation of Mitotic Cell Quantification via Microscopy and Multiple Whole-Slide Scanners. Diagn. Pathol. 2019, 14, 65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khalil, A.J.; Barhoom, A.M.; Abu-Nasser, B.S.; Musleh, M.M.; Abu-Naser, S.S. Energy Efficiency Prediction Using Artificial Neural Network. Int. J. Acad. Pedagog. Res. 2019, 3, 1–7. [Google Scholar]
- Mahmood, T.; Ziauddin, S.; Shahid, A.R.; Safi, A. Mitosis Detection in Breast Cancer Histopathology Images Using Statistical, Color and Shape-Based Features. J. Med. Imaging Health Inform. 2018, 8, 932–938. [Google Scholar] [CrossRef]
- Irshad, H. Automated Mitosis Detection in Histopathology Using Morphological and Multi-Channel Statistics Features. J. Pathol. Inform. 2013, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, S.; Rienties, B.; Khoja, S.A. The Role of Demographics in Online Learning; A Decision Tree Based Approach. Comput. Educ. 2019, 137, 32–47. [Google Scholar] [CrossRef]
- Chen, H.; Dou, Q.; Wang, X.; Qin, J.; Heng, P.-A. Mitosis Detection in Breast Cancer Histology Images via Deep Cascaded Networks. In Proceedings of the Thirtieth AAAI Conference on Artificial Intelligence, Phoenix, AZ, USA, 12 February 2016; pp. 1160–1166. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. In Advances in Neural Information Processing Systems; Curran Associates, Inc.: Red Hook, NY, USA, 2015; Volume 28. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Cai, D.; Sun, X.; Zhou, N.; Han, X.; Yao, J. Efficient Mitosis Detection in Breast Cancer Histology Images by RCNN. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 919–922. [Google Scholar]
- Dodballapur, V.; Song, Y.; Huang, H.; Chen, M.; Chrzanowski, W.; Cai, W. Mask-Driven Mitosis Detection In Histopathology Images. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 1855–1859. [Google Scholar]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), IEEE, Honolulu, HI, USA, 21–26 July 2017; pp. 1800–1807. [Google Scholar]
- Bochkovskiy, A.; Wang, C.-Y.; Liao, H.-Y.M. YOLOv4: Optimal Speed and Accuracy of Object Detection. arXiv 2020, arXiv:2004.10934. [Google Scholar]
- Roboflow: Give Your Software the Power to See Objects in Images and Video. Available online: https://roboflow.com/ (accessed on 23 June 2022).
- Ahmed, S.; Khan, F.; Ghaffar, A.; Hussain, F.; Cho, S.H. Finger-Counting-Based Gesture Recognition within Cars Using Impulse Radar with Convolutional Neural Network. Sensors 2019, 19, 1429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahmood, T.; Cho, S.W.; Park, K.R. DSRD-Net: Dual-Stream Residual Dense Network for Semantic Segmentation of Instruments in Robot-Assisted Surgery. Expert Syst. Appl. 2022, 202, 117420. [Google Scholar] [CrossRef]
- Wang, X.; Hua, X.; Xiao, F.; Li, Y.; Hu, X.; Sun, P. Multi-Object Detection in Traffic Scenes Based on Improved SSD. Electronics 2018, 7, 302. [Google Scholar] [CrossRef] [Green Version]
- Ge, Z.; Liu, S.; Wang, F.; Li, Z.; Sun, J. Yolox: Exceeding yolo series in 2021. arXiv 2021, arXiv:2107.08430. [Google Scholar]
- Kannadaguli, P. YOLO v4 Based Human Detection System Using Aerial Thermal Imaging for UAV Based Surveillance Applications. In Proceedings of the 2020 International Conference on Decision Aid Sciences and Application (DASA), Sakheer, Bahrain, 8–9 November 2020; pp. 1213–1219. [Google Scholar]
- Nersisson, R.; Iyer, T.J.; Joseph Raj, A.N.; Rajangam, V. A Dermoscopic Skin Lesion Classification Technique Using YOLO-CNN and Traditional Feature Model. Arab. J. Sci. Eng. 2021, 46, 9797–9808. [Google Scholar] [CrossRef]
- Yu, J.; Zhang, W. Face Mask Wearing Detection Algorithm Based on Improved YOLO-V4. Sensors 2021, 21, 3263. [Google Scholar] [CrossRef] [PubMed]
- Darknet: Open Source Neural Networks in C. Available online: https://pjreddie.com/darknet/ (accessed on 23 June 2022).
- CUDA by Example. Available online: https://developer.nvidia.com/cuda-example (accessed on 23 June 2022).


| Parameter | Value |
|---|---|
| True Positive (TP) | 97 |
| False Positive (FP) | 33 |
| False Negative (FN) | 11 |
| Precision | 0.7462 |
| Recall | 0.8981 |
| F1-Score | 0.8151 |
| Measure | Mean Value | Standard Deviation |
|---|---|---|
| Precision | 0.7462 | ±0.041 |
| Recall | 0.8981 | ±0.038 |
| F1-Score | 0.8151 | ±0.035 |
| Accuracy | 0.6879 | ±0.053 |
| Method | Precision | Recall | F1-Score |
|---|---|---|---|
| SSD | 0.7037 | 0.8796 | 0.7819 |
| Faster R-CNN | 0.7287 | 0.8704 | 0.7932 |
| YOLOv4 (Baseline method) | 0.7462 | 0.8981 | 0.8151 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zehra, T.; Anjum, S.; Mahmood, T.; Shams, M.; Sultan, B.A.; Ahmad, Z.; Alsubaie, N.; Ahmed, S. A Novel Deep Learning-Based Mitosis Recognition Approach and Dataset for Uterine Leiomyosarcoma Histopathology. Cancers 2022, 14, 3785. https://doi.org/10.3390/cancers14153785
Zehra T, Anjum S, Mahmood T, Shams M, Sultan BA, Ahmad Z, Alsubaie N, Ahmed S. A Novel Deep Learning-Based Mitosis Recognition Approach and Dataset for Uterine Leiomyosarcoma Histopathology. Cancers. 2022; 14(15):3785. https://doi.org/10.3390/cancers14153785
Chicago/Turabian StyleZehra, Talat, Sharjeel Anjum, Tahir Mahmood, Mahin Shams, Binish Arif Sultan, Zubair Ahmad, Najah Alsubaie, and Shahzad Ahmed. 2022. "A Novel Deep Learning-Based Mitosis Recognition Approach and Dataset for Uterine Leiomyosarcoma Histopathology" Cancers 14, no. 15: 3785. https://doi.org/10.3390/cancers14153785
APA StyleZehra, T., Anjum, S., Mahmood, T., Shams, M., Sultan, B. A., Ahmad, Z., Alsubaie, N., & Ahmed, S. (2022). A Novel Deep Learning-Based Mitosis Recognition Approach and Dataset for Uterine Leiomyosarcoma Histopathology. Cancers, 14(15), 3785. https://doi.org/10.3390/cancers14153785






