A Prospective Validation Study of Lung Cancer Gene Panel Testing Using Cytological Specimens
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Aim and Study Design
2.2. Patient Selection
2.3. Diagnostic Procedures
2.4. Sample Storage Conditions and Transport
2.5. Cytological Specimen Collection Using a GM-Tube
2.6. Sample Analysis
2.6.1. Sample Purification, Library Preparation, and NGS Sequencing
2.6.2. Pathological Diagnosis and Companion Diagnostic Test (CDx)
2.6.3. Assessment of Concordance of Variant Allele Frequency (VAF) between FFPE and Cytology
2.7. Statistical Analysis
3. Results
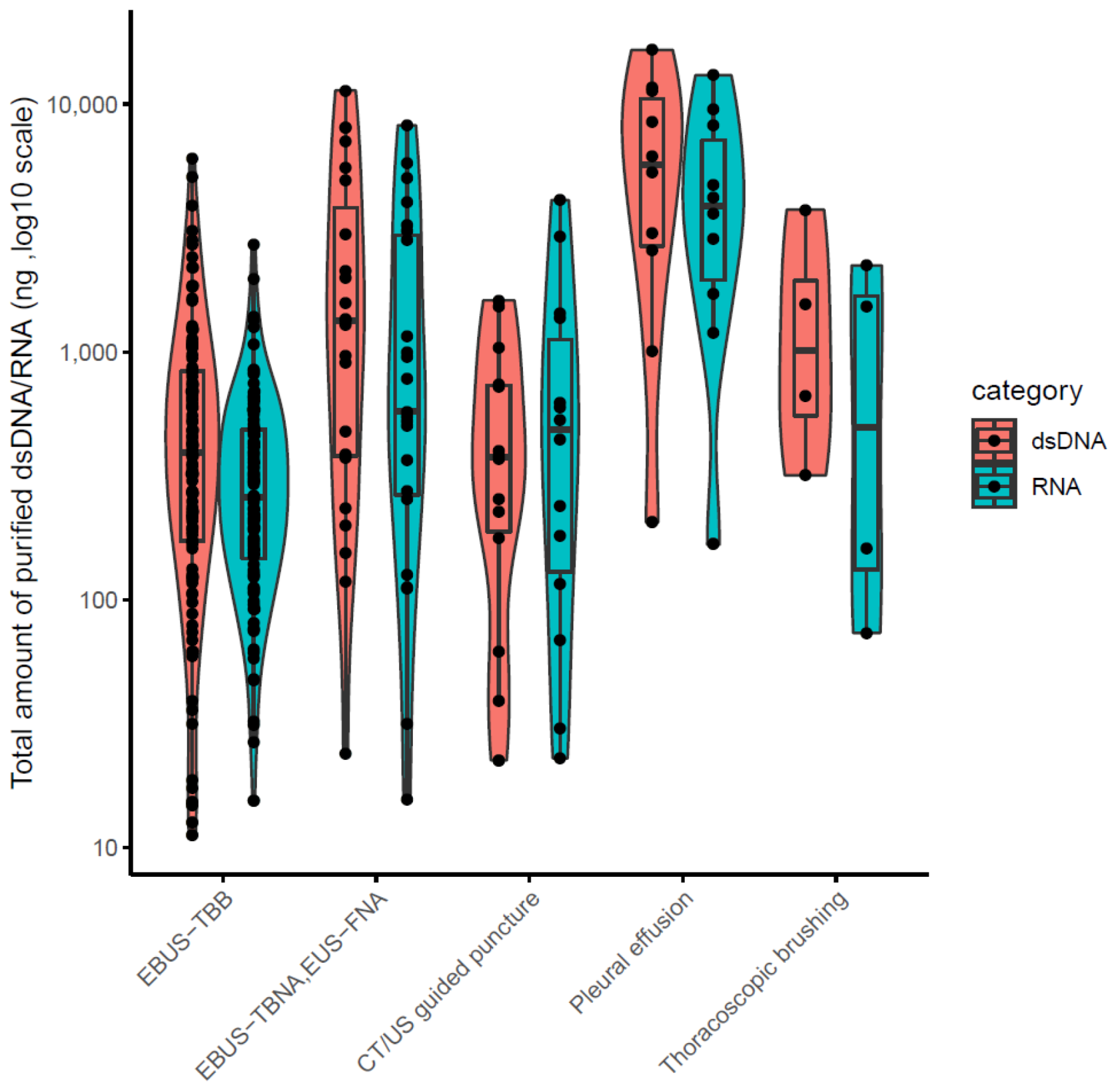
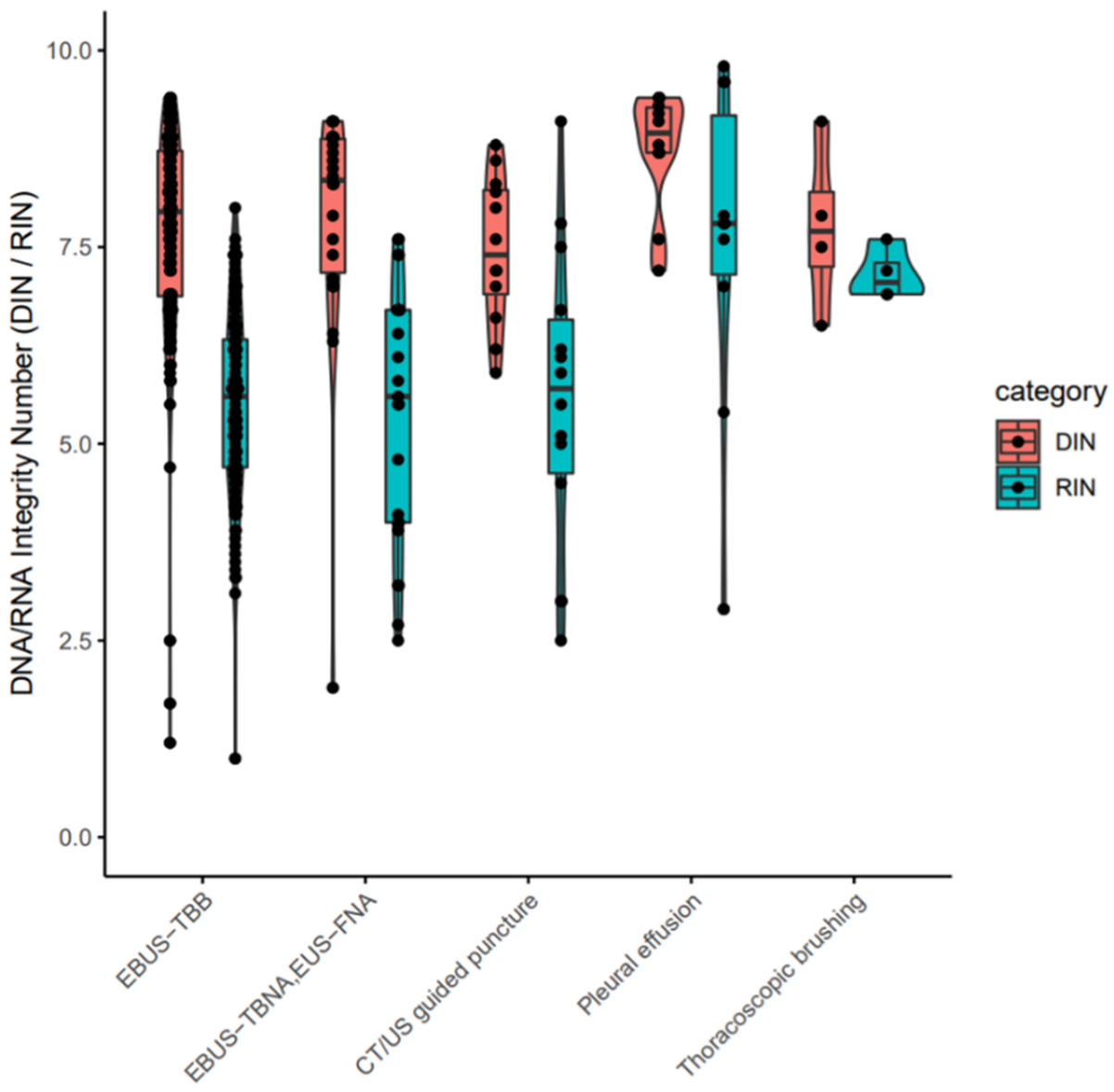
3.1. Patient Information, Quantity and Quality Evaluation of Purified Nucleotides
3.2. Assessment of Amplification Bias and Success Rate of LCCP Assay for Cytology Samples
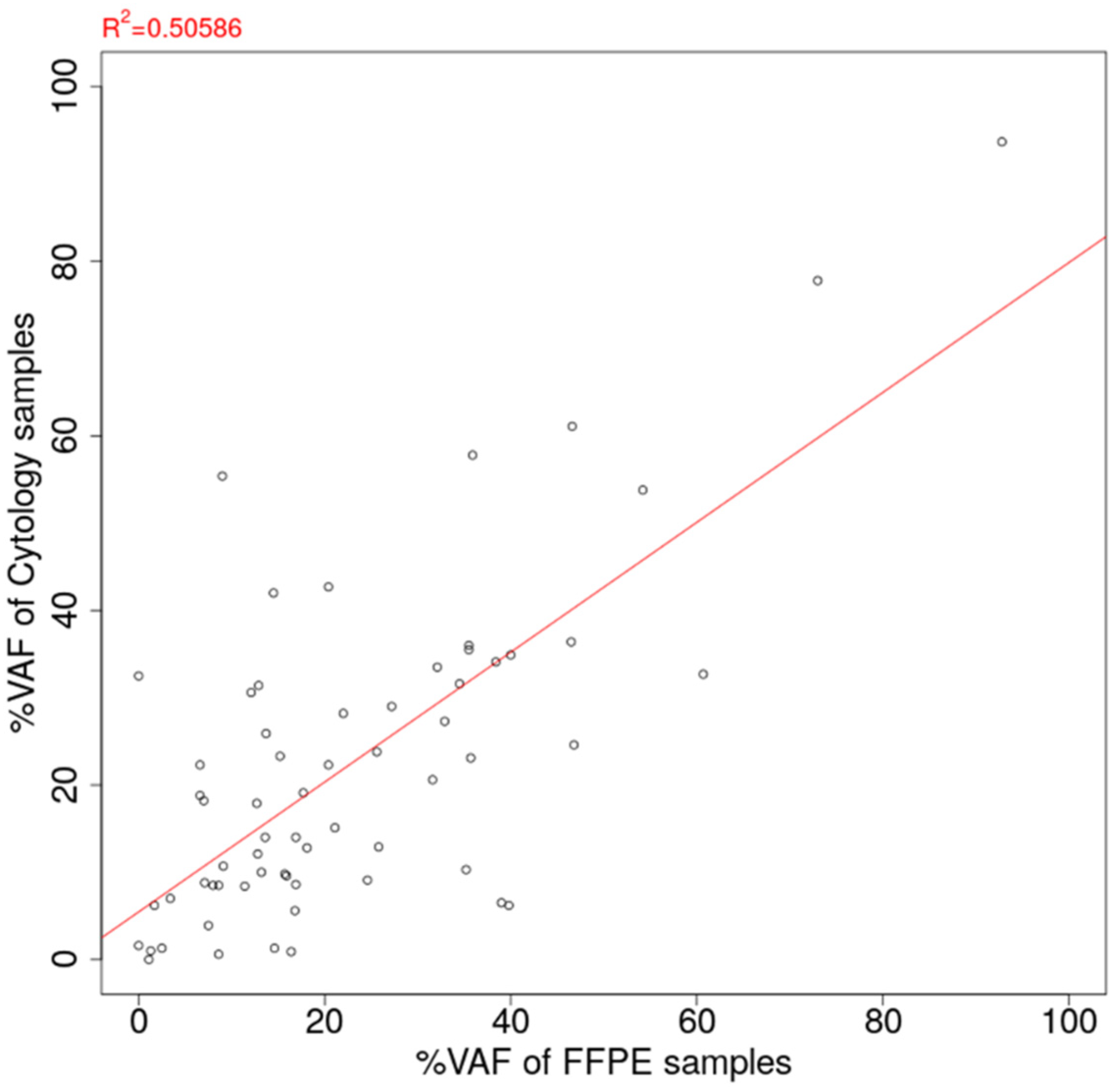
3.3. Comparison of VAF of Oncogenic Mutation Detected from Cytology-LCCP Panel Assay and FFPE-LCCP Panel Assay
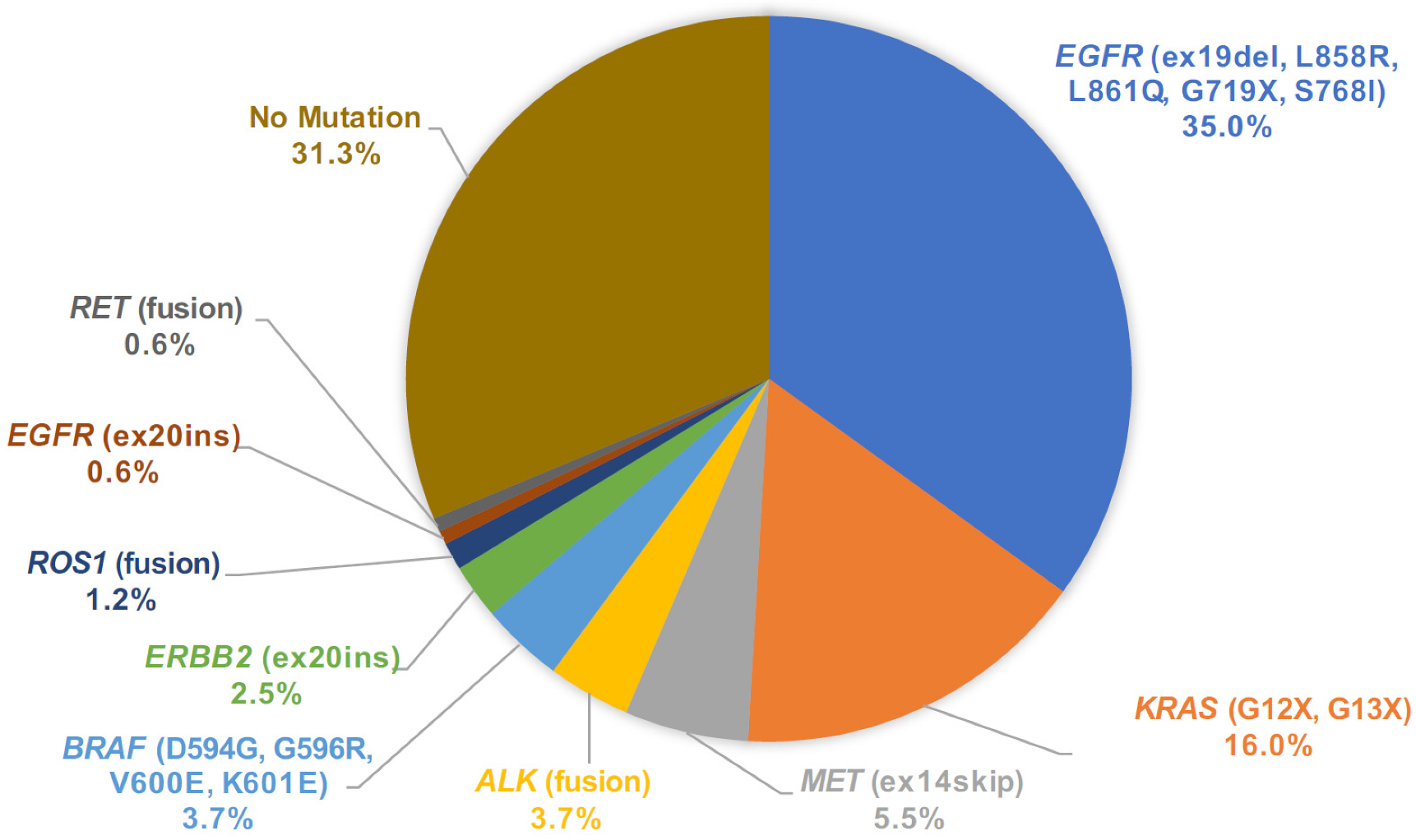
3.4. Mutation Call Profile from Cytology LCCP and Concordance with Companion Diagnostic Test
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kris, M.G.; Johnson, B.E.; Berry, L.D.; Kwiatkowski, D.J.; Iafrate, A.J.; Wistuba, I.I.; Varella-Garcia, M.; Franklin, W.A.; Aronson, S.L.; Su, P.F.; et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA 2014, 311, 1998–2006. [Google Scholar] [CrossRef] [PubMed]
- Gambardella, V.; Tarazona, N.; Cejalvo, J.M.; Lombardi, P.; Huerta, M.; Roselló, S.; Fleitas, T.; Roda, D.; Cervantes, A. Personalized Medicine: Recent Progress in Cancer Therapy. Cancers 2020, 12, 1009. [Google Scholar] [CrossRef] [Green Version]
- Mosele, F.; Remon, J.; Mateo, J.; Westphalen, C.; Barlesi, F.; Lolkema, M.; Normanno, N.; Scarpa, A.; Robson, M.; Meric-Bernstam, F.; et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2020, 31, 1491–1505. [Google Scholar] [CrossRef] [PubMed]
- Summary of Safety and Effectiveness Data for Oncomine Dx Target Test. 2017. Available online: https://www.accessdata.fda.gov/cdrh_docs/pdf16/P160045B.pdf (accessed on 27 March 2022).
- Murakami, S.; Yokose, T.; Nemoto, D.; Suzuki, M.; Usui, R.; Nakahara, Y.; Kondo, T.; Kato, T.; Saito, H. Suitability of Bronchoscopic Biopsy Tissue Samples for Next-Generation Sequencing. Diagnostics 2021, 11, 391. [Google Scholar] [CrossRef]
- Liam, C.K.; Mallawathantri, S.; Fong, K.M. Is tissue still the issue in detecting molecular alterations in lung cancer? Respirology 2020, 25, 933–943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kage, H.; Kohsaka, S.; Shinozaki-Ushiku, A.; Hiraishi, Y.; Sato, J.; Nagayama, K.; Ushiku, T.; Takai, D.; Nakajima, J.; Miyagawa, K.; et al. Small lung tumor biopsy samples are feasible for high quality targeted next generation sequencing. Cancer Sci. 2019, 110, 2652–2657. [Google Scholar] [CrossRef] [PubMed]
- Udagawa, H.; Kirita, K.; Naito, T.; Nomura, S.; Ishibashi, M.; Matsuzawa, R.; Hisakane, K.; Usui, Y.; Matsumoto, S.; Yoh, K.; et al. Feasibility and utility of transbronchial cryobiopsy in precision medicine for lung cancer: Prospective single-arm study. Cancer Sci. 2020, 111, 2488–2498. [Google Scholar] [CrossRef]
- Rolfo, C.; Mack, P.; Scagliotti, G.V.; Aggarwal, C.; Arcila, M.E.; Barlesi, F.; Bivona, T.; Diehn, M.; Dive, C.; Dziadziuszko, R.; et al. Liquid Biopsy for Advanced NSCLC: A Consensus Statement From the International Association for the Study of Lung Cancer. J. Thorac. Oncol. 2021, 16, 1647–1662. [Google Scholar] [CrossRef]
- Hofman, P. Next-Generation Sequencing with Liquid Biopsies from Treatment-Naïve Non-Small Cell Lung Carcinoma Patients. Cancers 2021, 13, 2049. [Google Scholar] [CrossRef]
- Kato, K.; Okami, J.; Nakamura, H.; Honma, K.; Sato, Y.; Nakamura, S.; Kukita, Y.; Nakatsuka, S.; Higashiyama, M. Analytical Performance of a Highly Sensitive System to Detect Gene Variants Using Next-Generation Sequencing for Lung Cancer Companion Diagnostics. medRxiv 2021. [Google Scholar] [CrossRef]
- Travis, W.D.; Brambilla, E.; Nicholson, A.G.; Yatabe, Y.; Austin, J.H.M.; Beasley, M.B.; Chirieac, L.R.; Dacic, S.; Duhig, E.; Flieder, D.B.; et al. The 2015 World Health Organization Classification of Lung Tumors: Impact of Genetic, Clinical and Radiologic Advances Since the 2004 Classification. J. Thorac. Oncol. 2015, 10, 1243–1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsunoda, A.; Morikawa, K.; Inoue, T.; Miyazawa, T.; Hoshikawa, M.; Takagi, M.; Mineshita, M. A prospective observational study to assess PD-L1 expression in small biopsy samples for non-small-cell lung cancer. BMC Cancer 2019, 19, 546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Wekken, A.J.; Pelgrim, R.; ’t Hart, N.; Werner, N.; Mastik, M.F.; Hendriks, L.; van der Heijden, E.H.F.M.; Looijen-Salamon, M.; de Langen, A.J.; Staal-van den Brekel, J.; et al. Dichotomous ALK-IHC Is a Better Predictor for ALK Inhibition Outcome than Traditional ALK-FISH in Advanced Non-Small Cell Lung Cancer. Clin. Cancer Res. 2017, 23, 4251–4258. [Google Scholar] [CrossRef] [Green Version]
- Morikawa, K.; Iinuma, M.; Shinozaki, Y.; Nishine, H.; Inoue, T.; Mineshita, M. A case of advanced adenocarcinoma genetically confirmed with EGFR/BRAF co-mutation in both primary and metastatic lesions. Ther. Adv. Med. Oncol. 2021, 13, 17588359211053420. [Google Scholar] [CrossRef]
- Kukita, Y.; Matoba, R.; Uchida, J.; Hamakawa, T.; Doki, Y.; Imamura, F.; Kato, K. High-fidelity target sequencing of individual molecules identified using barcode sequences: De novo detection and absolute quantitation of mutations in plasma cell-free DNA from cancer patients. DNA Res. 2015, 22, 269–277. [Google Scholar] [CrossRef] [Green Version]
- Kunimasa, K.; Kato, K.; Imamura, F.; Kukita, Y. Quantitative detection of ALK fusion breakpoints in plasma cell-free DNA from patients with non-small cell lung cancer using PCR-based target sequencing with a tiling primer set and two-step mapping/alignment. PLoS ONE 2019, 14, e0222233. [Google Scholar] [CrossRef]
- Kukita, Y.; Ohkawa, K.; Takada, R.; Uehara, H.; Katayama, K.; Kato, K. Selective identification of somatic mutations in pancreatic cancer cells through a combination of next-generation sequencing of plasma DNA using molecular barcodes and a bioinformatic variant filter. PLoS ONE 2018, 13, e0192611. [Google Scholar] [CrossRef]
- Von Ahlfen, S.; Missel, A.; Bendrat, K.; Schlumpberger, M. Determinants of RNA Quality from FFPE Samples. PLoS ONE 2007, 2, e1261. [Google Scholar] [CrossRef]
- Wang, S.; Kool, E.T. Origins of the Large Differences in Stability of DNA and RNA Helixes: C-5 Methyl and 2′-Hydroxyl Effects. Biochemistry 1995, 34, 4125–4132. [Google Scholar] [CrossRef]
- Fordyce, S.L.; Kampmann, M.-L.; van Doorn, N.L.; Gilbert, M.T.P. Long-term RNA persistence in postmortem contexts. Investig. Genet. 2013, 4, 7. [Google Scholar] [CrossRef] [Green Version]
- Jo, P.; Nietert, M.; Gusky, L.; Kitz, J.; Conradi, L.C.; Müller-Dornieden, A.; Schüler, P.; Wolff, H.A.; Rüschoff, J.; Ströbel, P.; et al. Neoadjuvant Therapy in Rectal Cancer—Biobanking of Preoperative Tumor Biopsies. Sci. Rep. 2016, 6, 35589. [Google Scholar] [CrossRef] [Green Version]
- Tu, H.-Y.; Li, Y.-S.; Bai, X.-Y.; Sun, Y.-L.; Zheng, M.-Y.; Ke, E.-E.; Liao, R.-Q.; Jiang, B.-Y.; Lin, J.-X.; Huang, J.; et al. Genetic Profiling of Cell-Free DNA From Pleural Effusion in Advanced Lung Cancer as a Surrogate for Tumor Tissue and Revealed Additional Clinical Actionable Targets. Clin. Lung Cancer 2021, 23, 135–142. [Google Scholar] [CrossRef]
- Vendrell, J.A.; Taviaux, S.; Béganton, B.; Godreuil, S.; Audran, P.; Grand, D.; Clermont, E.; Serre, I.; Szablewski, V.; Coopman, P.; et al. Detection of known and novel ALK fusion transcripts in lung cancer patients using next-generation sequencing approaches. Sci. Rep. 2017, 7, 12510. [Google Scholar] [CrossRef] [Green Version]
- Pinsolle, J.; Mondet, J.; Duruisseaux, M.; D’Alnoncourt, S.; Magnat, N.; de Fraipont, F.; Moro-Sibilot, D.; Toffart, A.-C.; Brambilla, E.; McLeer-Florin, A. A Rare Fusion of CLIP1 and ALK in a Case of Non–Small-Cell Lung Cancer With Neuroendocrine Features. Clin. Lung Cancer 2019, 20, e535–e540. [Google Scholar] [CrossRef]
- Ariyasu, R.; Nishikawa, S.; Uchibori, K.; Oh-Hara, T.; Yoshizawa, T.; Dotsu, Y.; Koyama, J.; Saiki, M.; Sonoda, T.; Kitazono, S.; et al. High ratio of T790M to EGFR activating mutations correlate with the osimertinib response in non-small-cell lung cancer. Lung Cancer 2018, 117, 1–6. [Google Scholar] [CrossRef]
- Kim, E.Y.; Cho, E.N.; Park, H.S.; Hong, J.Y.; Lim, S.; Youn, J.P.; Hwang, S.Y.; Chang, Y.S. Compound EGFR mutation is frequently detected with co-mutations of actionable genes and associated with poor clinical outcome in lung adenocarcinoma. Cancer Biol. Ther. 2016, 17, 237–245. [Google Scholar] [CrossRef] [Green Version]
- McLaughlin, C.W.; Skabelund, A.J.; Easterling, E.R.; Morris, M.J. The Safety and Utility of Fiberoptic Bronchoscopy in the Very Elderly. J. Bronchol. Interv. Pulmonol. 2018, 25, 300–304. [Google Scholar] [CrossRef]
- Mineshita, M.; Morikawa, K.; Furuya, N.; Kida, H.; Nishine, H.; Handa, H.; Inoue, T. Flexible bronchoscopy for lung cancer diagnosis in patients aged ≥ 85 years. Geriatr. Gerontol. Int. 2022, 22, 32–35. [Google Scholar] [CrossRef]
- Morikawa, K.; Kinoshita, K.; Kida, H.; Inoue, T.; Mineshita, M. Preliminary Results of NGS Gene Panel Test Using NSCLC Sputum Cytology and Therapeutic Effect Using Corresponding Molecular-Targeted Drugs. Genes 2022, 13, 812. [Google Scholar] [CrossRef]
- Sakaguchi, T.; Iketani, A.; Furuhashi, K.; Nakamura, Y.; Suzuki, Y.; Ito, K.; Fujiwara, K.; Nishii, Y.; Katsuta, K.; Taguchi, O.; et al. Comparison of the analytical performance between the Oncomine Dx Target Test and a conventional single gene test for epidermal growth factor receptor mutation in non-small cell lung cancer. Thorac. Cancer 2021, 12, 462–467. [Google Scholar] [CrossRef]


| Patient Characteristic | n = 163 |
|---|---|
| Male/female | 102/61 |
| Median age, years(range) | 72(44–90) |
| Clinical stage(I/II/III/IV) | 28/13/30/92 |
| Diagnosticprocedure | |
| EBUS-TBB | 112 |
| EBUS-TBNA, EUS-FNA | 23 |
| CT/US guided puncture | 14 |
| Pleural effusion | 10 |
| Thoracoscopic brushing | 4 |
| LCCP | ||||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Positive | Negative | Sensitivity | Specificity | PPV | NPV | ||
| CDx | Positive | EGFR | 57 | 0 | 1.000 (0.949–1.000) | - | 0.983 (0.908–1.000) | - |
| Negative | 1 | 94 | - | 0.989 (0.943–1.000) | - | 1.000 (0.969–1.000) | ||
| Positive | ALK | 6 | 1 | 0.857 (0.421–0.996) | - | 1.000 (0.607–1.000) | - | |
| Negative | 0 | 82 | - | 1.000 (0.964–1.000) | - | 0.988 (0.935–1.000) | ||
| Positive | BRAF V600E | 4 | 0 | 1.000 (0.473–1.000) | - | 1.000 (0.473–1.000) | - | |
| Negative | 0 | 48 | - | 1.000 (0.939–1.000) | - | 1.000 (0.939–1.000) | ||
| Positive | ROS1 | 2 | 0 | 1.000 (0.224–1.000) | - | 1.000 (0.224–1.000) | - | |
| Negative | 0 | 53 | - | 1.000 (0.945–1.000) | - | 1.000 (0.945–1.000) | ||
| Positive | MET | 6 | 0 | 1.000 (0.607–1.000) | - | 1.000 (0.607–1.000) | - | |
| Negative | 0 | 53 | - | 1.000 (0.945–1.000) | - | 1.000 (0.945–1.000) | ||
| Positive | ALL 5 Genes | 75 | 1 | 0.987 (0.929–1.000) | - | 0.987 (0.929–1.000) | - | |
| Negative | 1 | 330 | - | 0.997 (0.983–1.000) | - | 0.997 (0.983–1.000) | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morikawa, K.; Kida, H.; Handa, H.; Inoue, T.; Saji, H.; Koike, J.; Nakamura, S.; Sato, Y.; Ueda, Y.; Suzuki, F.; et al. A Prospective Validation Study of Lung Cancer Gene Panel Testing Using Cytological Specimens. Cancers 2022, 14, 3784. https://doi.org/10.3390/cancers14153784
Morikawa K, Kida H, Handa H, Inoue T, Saji H, Koike J, Nakamura S, Sato Y, Ueda Y, Suzuki F, et al. A Prospective Validation Study of Lung Cancer Gene Panel Testing Using Cytological Specimens. Cancers. 2022; 14(15):3784. https://doi.org/10.3390/cancers14153784
Chicago/Turabian StyleMorikawa, Kei, Hirotaka Kida, Hiroshi Handa, Takeo Inoue, Hisashi Saji, Junki Koike, Seiji Nakamura, Yoshiharu Sato, Yumi Ueda, Fumihiko Suzuki, and et al. 2022. "A Prospective Validation Study of Lung Cancer Gene Panel Testing Using Cytological Specimens" Cancers 14, no. 15: 3784. https://doi.org/10.3390/cancers14153784
APA StyleMorikawa, K., Kida, H., Handa, H., Inoue, T., Saji, H., Koike, J., Nakamura, S., Sato, Y., Ueda, Y., Suzuki, F., Matoba, R., & Mineshita, M. (2022). A Prospective Validation Study of Lung Cancer Gene Panel Testing Using Cytological Specimens. Cancers, 14(15), 3784. https://doi.org/10.3390/cancers14153784






