Recent Application of Artificial Intelligence in Non-Gynecological Cancer Cytopathology: A Systematic Review
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
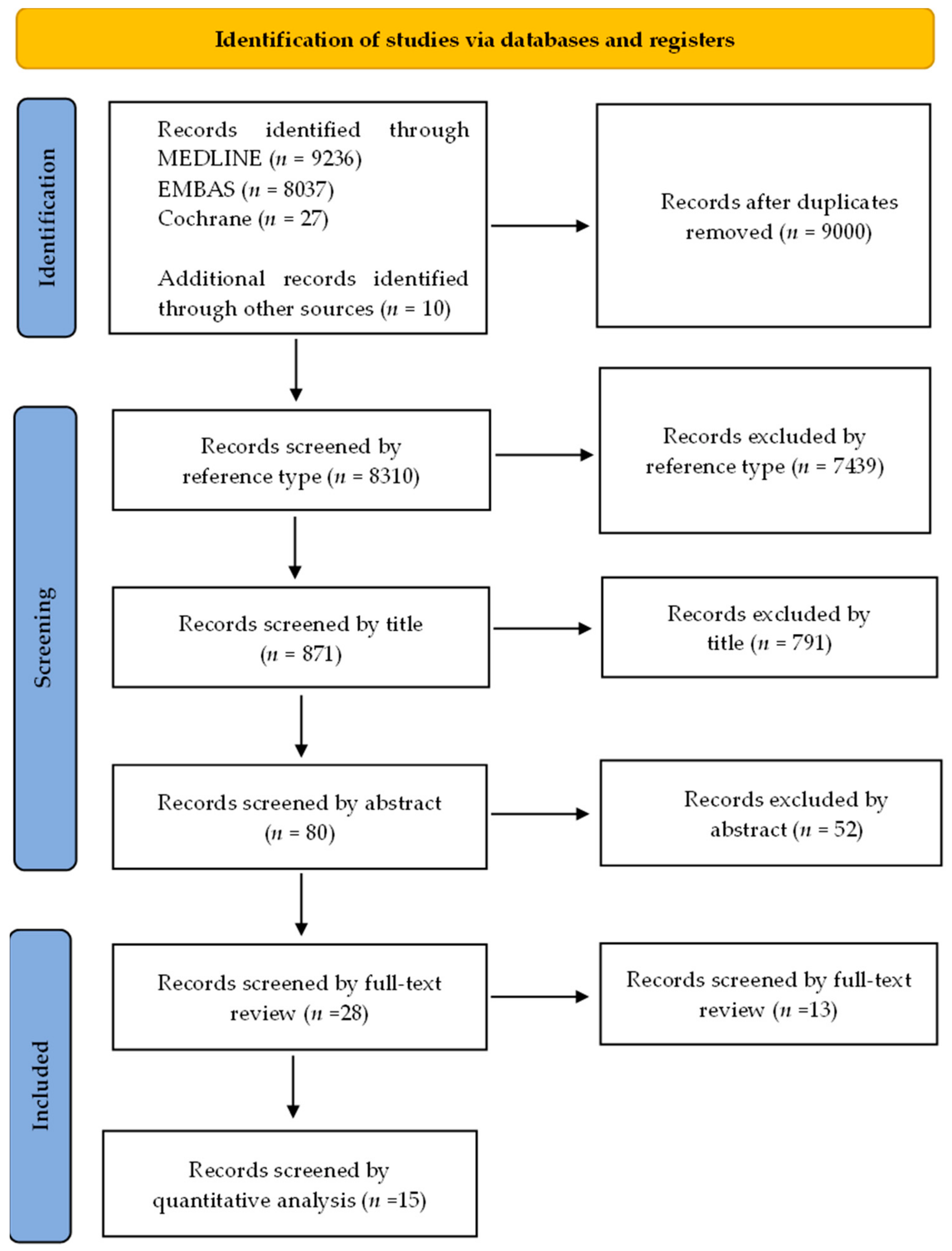
2.1. Literature Search
2.2. Study Selection, Reviewing, and Data Retrieval
3. Results
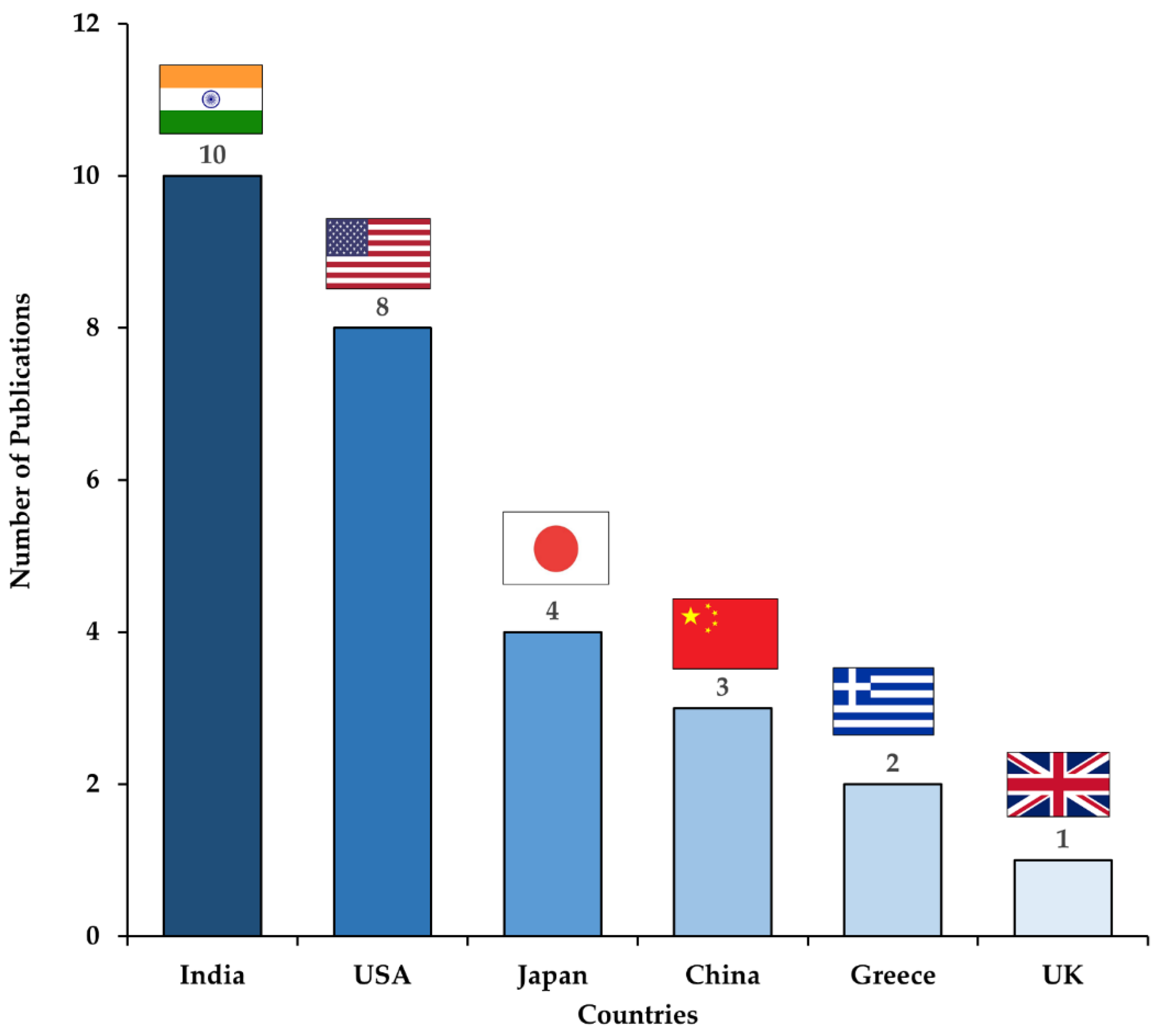
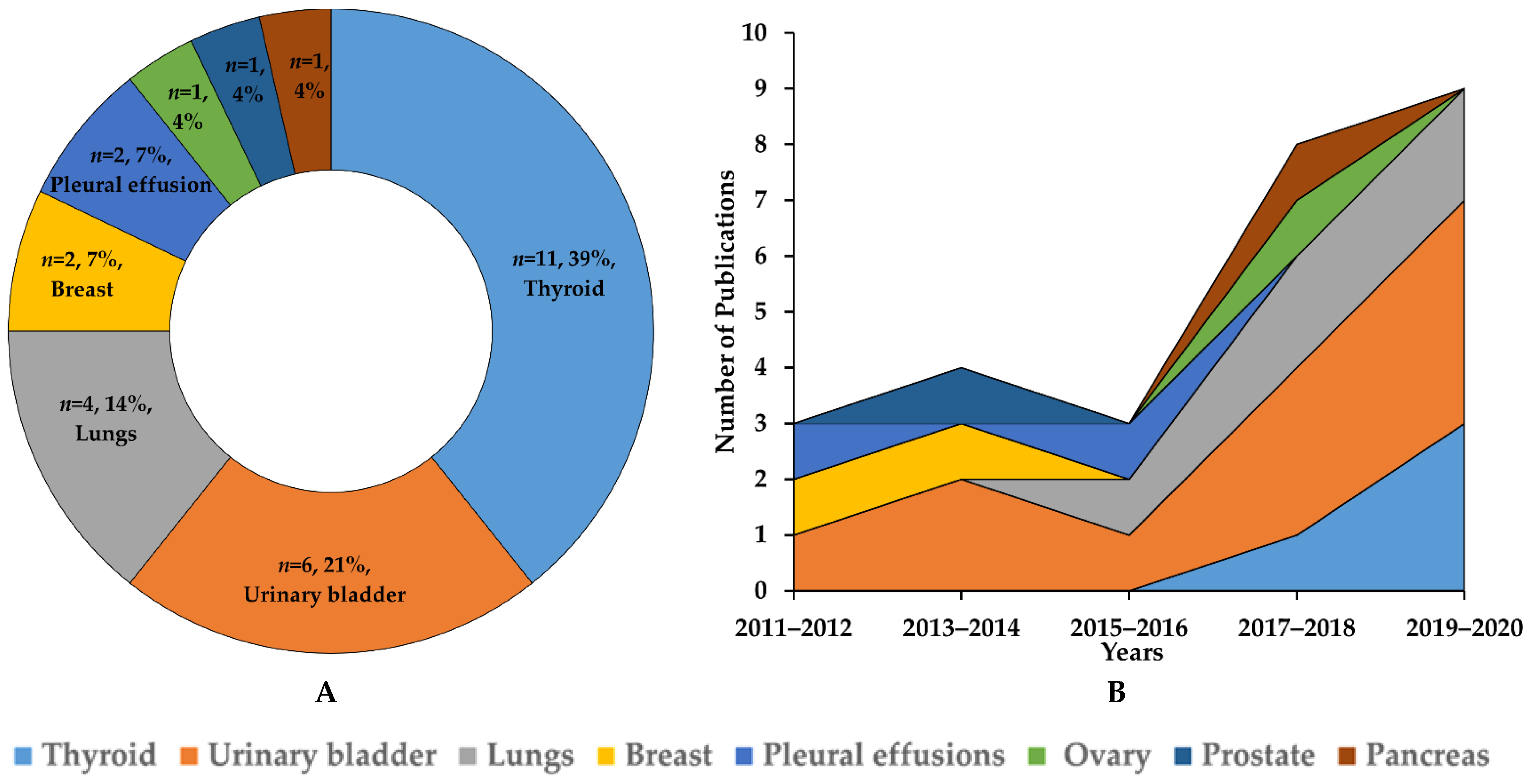
3.1. Study Selection and Characteristics
3.2. Applications of AI in Non-Gynecological Cancer Cytology Image Analysis
3.2.1. Classification for Thyroid FNAC
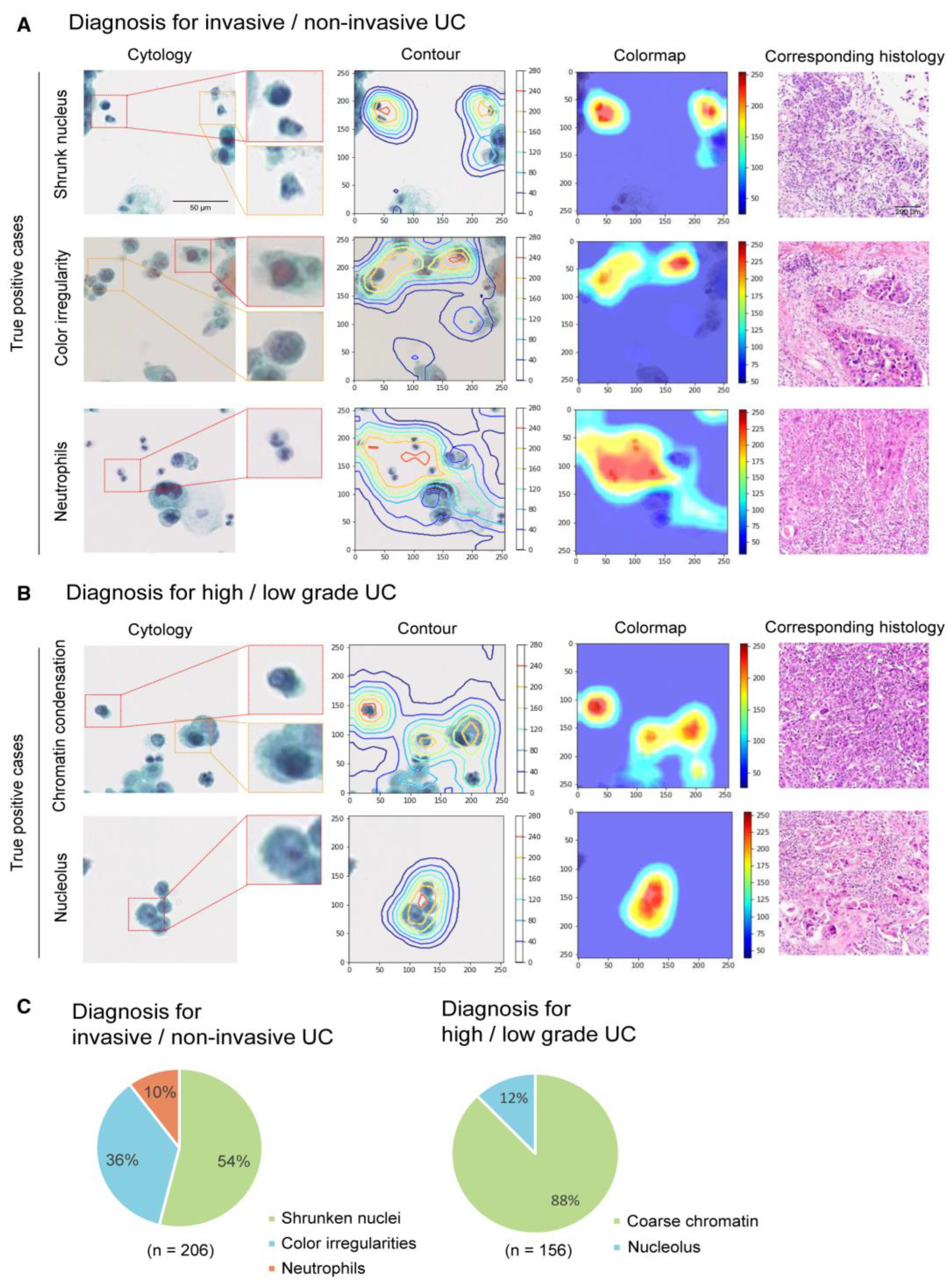
3.2.2. Classification for Urinary Tract Cytology
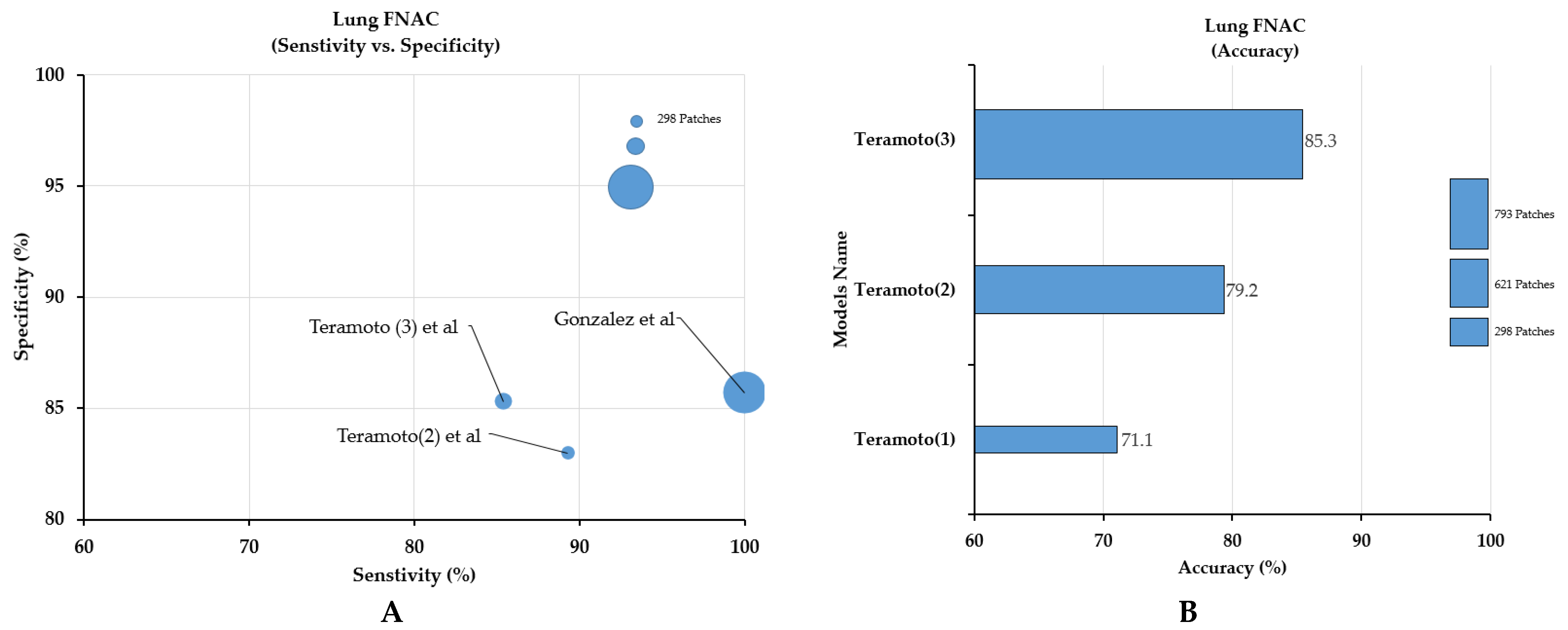
3.2.3. Classification for Lung FNAC or Bronchoscopic (Respiratory Tract) Aspirates
3.2.4. Classification for Breast FNAC
3.2.5. Classification for Pleural Fluids
3.2.6. Classification for Ovary FNAC
3.2.7. FNAC Classification for the Pancreas
3.2.8. FNAC Classification for the Prostate
4. Discussion
4.1. Challenges in Cytological Diagnosis
4.2. Challenges of Cytological Exams
4.2.1. Sampling Adequacy
4.2.2. Time-Consuming and Labor-Intensive Tasks
4.2.3. Intra-Examiner Variation in Sampling Procedures
4.2.4. Inter- and Intra-Observer Interpretational Variation among Cytologists
4.3. Challenges Related to the Application of AI in Cytology
4.3.1. The Larger Size of the Image
4.3.2. Difficulty in Annotation
4.3.3. Limited Z-Stacked Images
4.3.4. Lack of Well-Annotated Larger Datasets
4.3.5. Limited Publicly Available Datasets and Grand Challenges
4.3.6. Variation in the Annotation of Datasets and Image Quality
4.4. Evolving Trends of AI Models in Cytology
4.4.1. GYN Cytology
4.4.2. Application of AI in Non-GYN Cancer Cytology
4.5. Future Direction
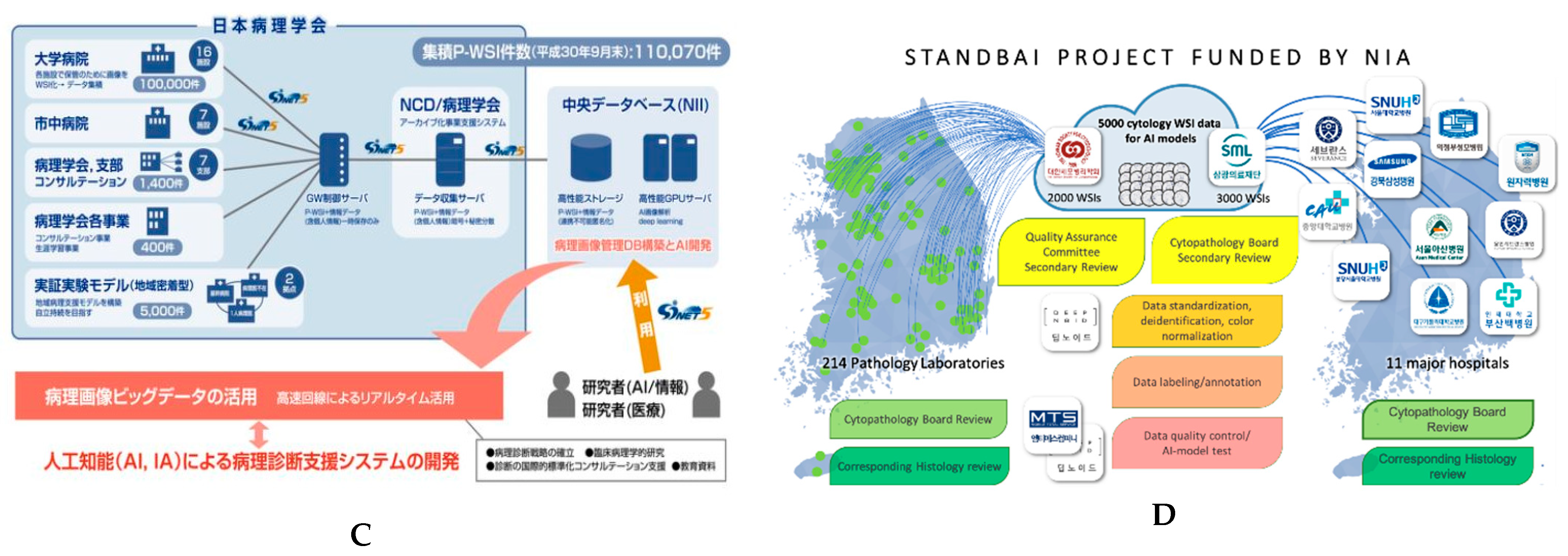
4.6. Recent Advancements in Digital Slide Repositories
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chong, Y.; Thakur, N.; Lee, J.Y.; Hwang, G.; Choi, M.; Kim, Y.; Yu, H.; Cho, M.Y. Diagnosis prediction of tumours of unknown origin using ImmunoGenius, a machine learning-based expert system for immunohistochemistry profile interpretation. Diagn. Pathol. 2021, 16, 19. [Google Scholar] [CrossRef]
- Chong, Y.; Lee, J.Y.; Kim, Y.; Choi, J.; Yu, H.; Park, G.; Cho, M.Y.; Thakur, N. A machine-learning expert-supporting system for diagnosis prediction of lymphoid neoplasms using a probabilistic decision-tree algorithm and immunohistochemistry profile database. J. Pathol. Transl. Med. 2020, 54, 462–470. [Google Scholar] [CrossRef]
- Bera, K.; Schalper, K.A.; Rimm, D.L.; Velcheti, V.; Madabhushi, A. Artificial intelligence in digital pathology—new tools for diagnosis and precision oncology. Nat. Rev. Clin. Oncol. 2019, 16, 703–715. [Google Scholar] [CrossRef]
- Ahmad, Z.; Rahim, S.; Zubair, M.; Abdul-Ghafar, J. Artificial intelligence (AI) in medicine, current applications and future role with special emphasis on its potential and promise in pathology: Present and future impact, obstacles including costs and acceptance among pathologists, practical and philosophical considerations. A comprehensive review. Diagn. Pathol. 2021, 16, 24. [Google Scholar] [PubMed]
- Ailia, M.J.; Thakur, N.; Abdul-Ghafar, J.; Jung, C.K.; Yim, K.; Chong, Y. Current Trend of Artificial Intelligence Patents in Digital Pathology: A Systematic Evaluation of the Patent Landscape. Cancers 2022, 14, 2400. [Google Scholar] [CrossRef] [PubMed]
- Abdar, M.; Yen, N.Y.; Hung, J.C.-S. Improving the Diagnosis of Liver Disease Using Multilayer Perceptron Neural Network and Boosted Decision Trees. J. Med. Biol. Eng. 2018, 38, 953–965. [Google Scholar] [CrossRef]
- Srinivasu, P.N.; SivaSai, J.G.; Ijaz, M.F.; Bhoi, A.K.; Kim, W.; Kang, J.J. Classification of Skin Disease Using Deep Learning Neural Networks with MobileNet V2 and LSTM. Sensors 2021, 21, 2852. [Google Scholar] [CrossRef]
- Vulli, A.; Srinivasu, P.N.; Sashank, M.S.K.; Shafi, J.; Choi, J.; Ijaz, M.F. Fine-Tuned DenseNet-169 for Breast Cancer Metastasis Prediction Using FastAI and 1-Cycle Policy. Sensors 2022, 22, 2988. [Google Scholar] [CrossRef]
- Hosny, A.; Parmar, C.; Quackenbush, J.; Schwartz, L.H.; Aerts, H.J. Artificial intelligence in radiology. Nat. Rev. Cancer 2018, 18, 500–510. [Google Scholar] [CrossRef]
- Li, H.; Tian, S.; Li, Y.; Fang, Q.; Tan, R.; Pan, Y.; Huang, C.; Xu, Y.; Gao, X. Modern deep learning in bioinformatics. J. Mol. Cell Biol. 2020, 12, 823–827. [Google Scholar] [CrossRef]
- Libbrecht, M.W.; Noble, W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015, 16, 321–332. [Google Scholar] [CrossRef] [Green Version]
- Lamberti, M.J.; Wilkinson, M.; Donzanti, B.A.; Wohlhieter, G.E.; Parikh, S.; Wilkins, R.G.; Getz, K. A study on the application and use of artificial intelligence to support drug development. Clin. Ther. 2019, 41, 1414–1426. [Google Scholar] [CrossRef] [Green Version]
- Stafford, I.; Kellermann, M.; Mossotto, E.; Beattie, R.; MacArthur, B.; Ennis, S. A systematic review of the applications of artificial intelligence and machine learning in autoimmune diseases. NPJ Digit. Med. 2020, 3, 30. [Google Scholar] [CrossRef]
- Kim, H.; Yoon, H.; Thakur, N.; Hwang, G.; Lee, E.J.; Kim, C.; Chong, Y. Deep learning-based histopathological segmentation for whole slide images of colorectal cancer in a compressed domain. Sci. Rep. 2021, 11, 22520. [Google Scholar] [CrossRef]
- Alam, M.R.; Abdul-Ghafar, J.; Yim, K.; Thakur, N.; Lee, S.H.; Jang, H.-J.; Jung, C.K.; Chong, Y. Recent Applications of Artificial Intelligence from Histopathologic Image-Based Prediction of Microsatellite Instability in Solid Cancers: A Systematic Review. Cancers 2022, 14, 2590. [Google Scholar] [CrossRef]
- Thakur, N.; Yoon, H.; Chong, Y. Current Trends of Artificial Intelligence for Colorectal Cancer Pathology Image Analysis: A Systematic Review. Cancers 2020, 12, 1884. [Google Scholar] [CrossRef]
- Liu, Y.; Gadepalli, K.; Norouzi, M.; Dahl, G.E.; Kohlberger, T.; Boyko, A.; Venugopalan, S.; Timofeev, A.; Nelson, P.Q.; Corrado, G.S. Detecting Cancer Metastases on Gigapixel Pathology Images. arXiv 2017, arXiv:1703.02442. [Google Scholar]
- Chong, Y.; Ji, S.-J.; Kang, C.S.; Lee, E.J. Can liquid-based preparation substitute for conventional smear in thyroid fine-needle aspiration? A systematic review based on meta-analysis. Endocr. Connect. 2017, 6, 817–829. [Google Scholar] [CrossRef] [Green Version]
- Al-Abbadi, M.A. Basics of cytology. Avicenna J. Med. 2011, 1, 18. [Google Scholar] [CrossRef]
- Wentzensen, N.; Lahrmann, B.; Clarke, M.A.; Kinney, W.; Tokugawa, D.; Poitras, N.; Locke, A.; Bartels, L.; Krauthoff, A.; Walker, J.; et al. Accuracy and Efficiency of Deep-Learning—Based Automation of Dual Stain Cytology in Cervical Cancer Screening. JNCI J. Natl. Cancer Inst. 2020, 113, 72–79. [Google Scholar] [CrossRef]
- Bao, H.; Bi, H.; Zhang, X.; Zhao, Y.; Dong, Y.; Luo, X.; Zhou, D.; You, Z.; Wu, Y.; Liu, Z.; et al. Artificial intelligence-assisted cytology for detection of cervical intraepithelial neoplasia or invasive cancer: A multicenter, clinical-based, observational study. Gynecol. Oncol. 2020, 159, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Elliott Range, D.D.; Dov, D.; Kovalsky, S.Z.; Henao, R.; Carin, L.; Cohen, J. Application of a machine learning algorithm to predict malignancy in thyroid cytopathology. Cancer Cytopatho. 2020, 128, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Teramoto, A.; Yamada, A.; Kiriyama, Y.; Tsukamoto, T.; Yan, K.; Zhang, L.; Imaizumi, K.; Saito, K.; Fujita, H. Automated classification of benign and malignant cells from lung cytological images using deep convolutional neural network. Inform. Med. Unlocked 2019, 16, 100205. [Google Scholar] [CrossRef]
- Dey, P.; Logasundaram, R.; Joshi, K. Artificial neural network in diagnosis of lobular carcinoma of breast in fine-needle aspiration cytology. Diagn. Cytopathol. 2013, 41, 102–106. [Google Scholar] [CrossRef]
- Subbaiah, R.M.; Dey, P.; Nijhawan, R. Artificial neural network in breast lesions from fine-needle aspiration cytology smear. Diagn. Cytopathol. 2014, 42, 218–224. [Google Scholar] [CrossRef]
- Teramoto, A.; Tsukamoto, T.; Kiriyama, Y.; Fujita, H. Automated Classification of Lung Cancer Types from Cytological Images Using Deep Convolutional Neural Networks. BioMed Res. Int. 2017, 2017, 4067832. [Google Scholar] [CrossRef] [Green Version]
- Teramoto, A.; Tsukamoto, T.; Yamada, A.; Kiriyama, Y.; Imaizumi, K.; Saito, K.; Fujita, H. Deep learning approach to classification of lung cytological images: Two-step training using actual and synthesized images by progressive growing of generative adversarial networks. PLoS ONE 2020, 15, e0229951. [Google Scholar] [CrossRef]
- Gonzalez, D.; Dietz, R.L.; Pantanowitz, L. Feasibility of a deep learning algorithm to distinguish large cell neuroendocrine from small cell lung carcinoma in cytology specimens. Cytopathology 2020, 31, 426–431. [Google Scholar] [CrossRef]
- Wu, M.; Yan, C.; Liu, H.; Liu, Q. Automatic classification of ovarian cancer types from cytological images using deep convolutional neural networks. Biosci. Rep. 2018, 38, BSR20180289. [Google Scholar] [CrossRef] [Green Version]
- Momeni-Boroujeni, A.; Yousefi, E.; Somma, J. Computer-assisted cytologic diagnosis in pancreatic FNA: An application of neural networks to image analysis. Cancer Cytopathol. 2017, 125, 926–933. [Google Scholar] [CrossRef] [Green Version]
- Barwad, A.; Dey, P.; Susheilia, S. Artificial neural network in diagnosis of metastatic carcinoma in effusion cytology. Cytom. Part B Clin. Cytom. 2012, 82, 107–111. [Google Scholar] [CrossRef]
- Tosun, A.B.; Yergiyev, O.; Kolouri, S.; Silverman, J.F.; Rohde, G.K. Detection of malignant mesothelioma using nuclear structure of mesothelial cells in effusion cytology specimens. Cytom. Part A 2015, 87, 326–333. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, K.; Jain, A.K.; Sabata, B. Prostate cancer detection: Fusion of cytological and textural features. J. Pathol. Inform. 2011, 2, 5464787. [Google Scholar] [CrossRef]
- Gopinath, B.; Shanthi, N. Support Vector Machine based diagnostic system for thyroid cancer using statistical texture features. Asian Pac. J. Cancer Prev. 2013, 14, 97–102. [Google Scholar] [CrossRef] [Green Version]
- Gopinath, B.; Shanthi, N. Computer-aided diagnosis system for classifying benign and malignant thyroid nodules in multi-stained FNAB cytological images. Australas. Phys. Eng. Sci. Med. 2013, 36, 219–230. [Google Scholar] [CrossRef]
- Gopinath, B.; Shanthi, N. Development of an automated medical diagnosis system for classifying thyroid tumor cells using multiple classifier fusion. Technol. Cancer Res. Treat. 2015, 14, 653–662. [Google Scholar] [CrossRef]
- Savala, R.; Dey, P.; Gupta, N. Artificial neural network model to distinguish follicular adenoma from follicular carcinoma on fine needle aspiration of thyroid. Diagn. Cytopathol. 2018, 46, 244–249. [Google Scholar] [CrossRef]
- Sanyal, P.; Mukherjee, T.; Barui, S.; Das, A.; Gangopadhyay, P. Artificial Intelligence in Cytopathology: A Neural Network to Identify Papillary Carcinoma on Thyroid Fine-Needle Aspiration Cytology Smears. J. Pathol. Inform. 2018, 9, 43. [Google Scholar] [CrossRef]
- Dov, D.; Kovalsky, S.Z.; Cohen, J.; Range, D.E.; Henao, R.; Carin, L. Thyroid cancer malignancy prediction from whole slide cytopathology images. In Proceedings of the Machine Learning for Healthcare Conference, Ann Arbor, MI, USA, 9–10 August 2019; pp. 553–570. [Google Scholar]
- Guan, Q.; Wang, Y.; Ping, B.; Li, D.; Du, J.; Qin, Y.; Lu, H.; Wan, X.; Xiang, J. Deep convolutional neural network VGG-16 model for differential diagnosing of papillary thyroid carcinomas in cytological images: A pilot study. J. Cancer 2019, 10, 4876. [Google Scholar] [CrossRef]
- Fragopoulos, C.; Pouliakis, A.; Meristoudis, C.; Mastorakis, E.; Margari, N.; Chroniaris, N.; Koufopoulos, N.; Delides, A.G.; Machairas, N.; Ntomi, V. Radial Basis Function Artificial Neural Network for the Investigation of Thyroid Cytological Lesions. J. Thyroid. Res. 2020, 2020, 5464787. [Google Scholar] [CrossRef]
- Muralidaran, C.; Dey, P.; Nijhawan, R.; Kakkar, N. Artificial neural network in diagnosis of urothelial cell carcinoma in urine cytology. Diagn. Cytopathol. 2015, 43, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Sanghvi, A.B.; Allen, E.Z.; Callenberg, K.M.; Pantanowitz, L. Performance of an artificial intelligence algorithm for reporting urine cytopathology. Cancer Cytopathol. 2019, 127, 658–666. [Google Scholar] [CrossRef] [PubMed]
- Vaickus, L.J.; Suriawinata, A.A.; Wei, J.W.; Liu, X. Automating the Paris System for urine cytopathology—A hybrid deep-learning and morphometric approach. Cancer Cytopathol. 2019, 127, 98–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Awan, R.; Benes, K.; Azam, A.; Song, T.-H.; Shaban, M.; Verrill, C.; Tsang, Y.W.; Snead, D.; Minhas, F.; Rajpoot, N. Deep learning based digital cell profiles for risk stratification of urine cytology images. Cytom. Part A 2021, 99, 732–742. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Fu, X.; Liu, J.; Huang, Z.; Liu, N.; Fang, F.; Rao, J. Developing a Machine Learning Algorithm for Identifying Abnormal Urothelial Cells: A Feasibility Study. Acta Cytol. 2020, 65, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Nojima, S.; Terayama, K.; Shimoura, S.; Hijiki, S.; Nonomura, N.; Morii, E.; Okuno, Y.; Fujita, K. A deep learning system to diagnose the malignant potential of urothelial carcinoma cells in cytology specimens. Cancer Cytopathol. 2021, 129, 984–995. [Google Scholar] [CrossRef]
- Varlatzidou, A.; Pouliakis, A.; Stamataki, M.; Meristoudis, C.; Margari, N.; Peros, G.; Panayiotides, J.G.; Karakitsos, P. Cascaded learning vector quantizer neural networks for the discrimination of thyroid lesions. Anal. Quant. Cytol. Histol. 2011, 33, 323–334. [Google Scholar]
- Gopinath, B. A benign and malignant pattern identification in cytopathological images of thyroid nodules using gabor filter and neural networks. Asian J. Converg. Technol. 2018, 4, 1–6. [Google Scholar]
- Ducatman, E.C.B. Cytology: Diagnostic Principles and Clinical Correlates, 5th ed.; Elsevier: Amsterdam, The Netherlands, 2019; Volume 5, p. 688. [Google Scholar]
- Chong, Y.; Jung, H.; Pyo, J.-S.; Hong, S.W.; Oh, H.K. Current status of cytopathology practices in Korea: Annual report on the Continuous Quality Improvement program of the Korean Society for Cytopathology for 2018. J. Pathol. Transl. Med. 2020, 54, 318. [Google Scholar] [CrossRef] [Green Version]
- Oh, E.J.; Jung, C.K.; Kim, D.-H.; Kim, H.K.; Kim, W.S.; Jin, S.-Y.; Yoon, H.K.; Fellowship Council; Committee of Quality Improvement of the Korean Society for Cytopathology. Current Cytology Practices in Korea: A Nationwide Survey by the Korean Society for Cytopathology. J. Pathol. Transl. Med. 2017, 51, 579–587. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.J.; Kim, D.W.; Jung, S.J.; Baek, H.J. Factors that Influence Sample Adequacy in Liquid-Based Cytology after Ultrasonography-Guided Fine-Needle Aspiration of Thyroid Nodules: A Single-Center Study. Acta Cytol. 2018, 62, 253–258. [Google Scholar] [CrossRef]
- Dey, P. Time for evidence-based cytology. Cytojournal 2007, 4. [Google Scholar] [CrossRef]
- Yamada, T.; Yoshimura, T.; Kitamura, N.; Sasabe, E.; Ohno, S.; Yamamoto, T. Low-grade myofibroblastic sarcoma of the palate. Int. J. Oral Sci. 2012, 4, 170–173. [Google Scholar] [CrossRef] [Green Version]
- Kuzan, T.Y.; Güzelbey, B.; Turan Güzel, N.; Kuzan, B.N.; Çakır, M.S.; Canbey, C. Analysis of intra-observer and inter-observer variability of pathologists for non-benign thyroid fine needle aspiration cytology according to Bethesda system categories. Diagn. Cytopathol. 2021, 49, 850–855. [Google Scholar] [CrossRef]
- Barkan, G.A.; Wojcik, E.M.; Nayar, R.; Savic-Prince, S.; Quek, M.L.; Kurtycz, D.F.; Rosenthal, D.L. The Paris System for Reporting Urinary Cytology: The Quest to Develop a Standardized Terminology. Acta Cytol. 2016, 60, 185–197. [Google Scholar] [CrossRef] [Green Version]
- Sahai, R.; Rajkumar, B.; Joshi, P.; Singh, A.; Kumar, A.; Durgapal, P.; Gupta, A.; Kishore, S.; Chowdhury, N. Interobserver reproducibility of The Paris System of Reporting Urine Cytology on cytocentrifuged samples. Diagn. Cytopathol. 2020, 48, 979–985. [Google Scholar] [CrossRef]
- Reid, M.D.; Osunkoya, A.O.; Siddiqui, M.T.; Looney, S.W. Accuracy of grading of urothelial carcinoma on urine cytology: An analysis of interobserver and intraobserver agreement. Int. J. Clin. Exp. Pathol. 2012, 5, 882. [Google Scholar]
- McAlpine, E.D.; Pantanowitz, L.; Michelow, P.M. Challenges Developing Deep Learning Algorithms in Cytology. Acta Cytol. 2020, 65, 301–309. [Google Scholar] [CrossRef]
- Thrall, M.; Pantanowitz, L.; Khalbuss, W. Telecytology: Clinical applications, current challenges, and future benefits. J. Pathol. Inform. 2011, 2, 51. [Google Scholar] [CrossRef]
- Tizhoosh, H.R.; Pantanowitz, L. Artificial Intelligence and Digital Pathology: Challenges and Opportunities. J. Pathol. Inform. 2018, 9, 38. [Google Scholar] [CrossRef]
- Donnelly, A.D.; Mukherjee, M.S.; Lyden, E.R.; Bridge, J.A.; Lele, S.M.; Wright, N.; McGaughey, M.F.; Culberson, A.M.; Horn, A.J.; Wedel, W.R.; et al. Optimal z-axis scanning parameters for gynecologic cytology specimens. J. Pathol. Inform. 2013, 4, 38. [Google Scholar] [CrossRef] [PubMed]
- Pinchaud, N.; Hedlund, M. Camelyon17 Grand Challenge. Submission Results Camelyon17 Challange. Available online: https://camelyon17.grand-challenge.org/evaluation/results/ (accessed on 1 December 2019).
- Cruz-Roa, A.; Gilmore, H.; Basavanhally, A.; Feldman, M.; Ganesan, S.; Shih, N.N.C.; Tomaszewski, J.; González, F.A.; Madabhushi, A. Accurate and reproducible invasive breast cancer detection in whole-slide images: A Deep Learning approach for quantifying tumor extent. Sci. Rep. 2017, 7, 46450. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, A.; Gamble, P.; Jaroensri, R.; Abdelsamea, M.M.; Mermel, C.H.; Chen, P.-H.C.; Rakha, E.A. Artificial intelligence in digital breast pathology: Techniques and applications. Breast 2020, 49, 267–273. [Google Scholar] [CrossRef] [Green Version]
- Vooijs, P.; Palcic, B.; Garner, D.M.; MacAulay, C.E.; Matisic, J.; Anderson, G.H.; Grohs, D.H.; Gombrich, P.P.; Domanik, R.A.; Kamentsky, L.A. The FDA review process: Obtaining premarket approval for the PAPNET Testing System. Acta Cytol. 1996, 40, 138–140. [Google Scholar]
- Thrall, M.J. Automated screening of Papanicolaou tests: A review of the literature. Diagn. Cytopathol. 2019, 47, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Landau, M.S.; Pantanowitz, L. Artificial intelligence in cytopathology: A review of the literature and overview of commercial landscape. J. Am. Soc. Cytopathol. 2019, 8, 230–241. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Lu, L.; Nogues, I.; Summers, R.M.; Liu, S.; Yao, J. DeepPap: Deep convolutional networks for cervical cell classification. IEEE J. Biomed. Health Inform. 2017, 21, 1633–1643. [Google Scholar] [CrossRef] [Green Version]
- McHenry, C.R.; Phitayakorn, R. Follicular adenoma and carcinoma of the thyroid gland. Oncologist 2011, 16, 585–593. [Google Scholar] [CrossRef] [Green Version]

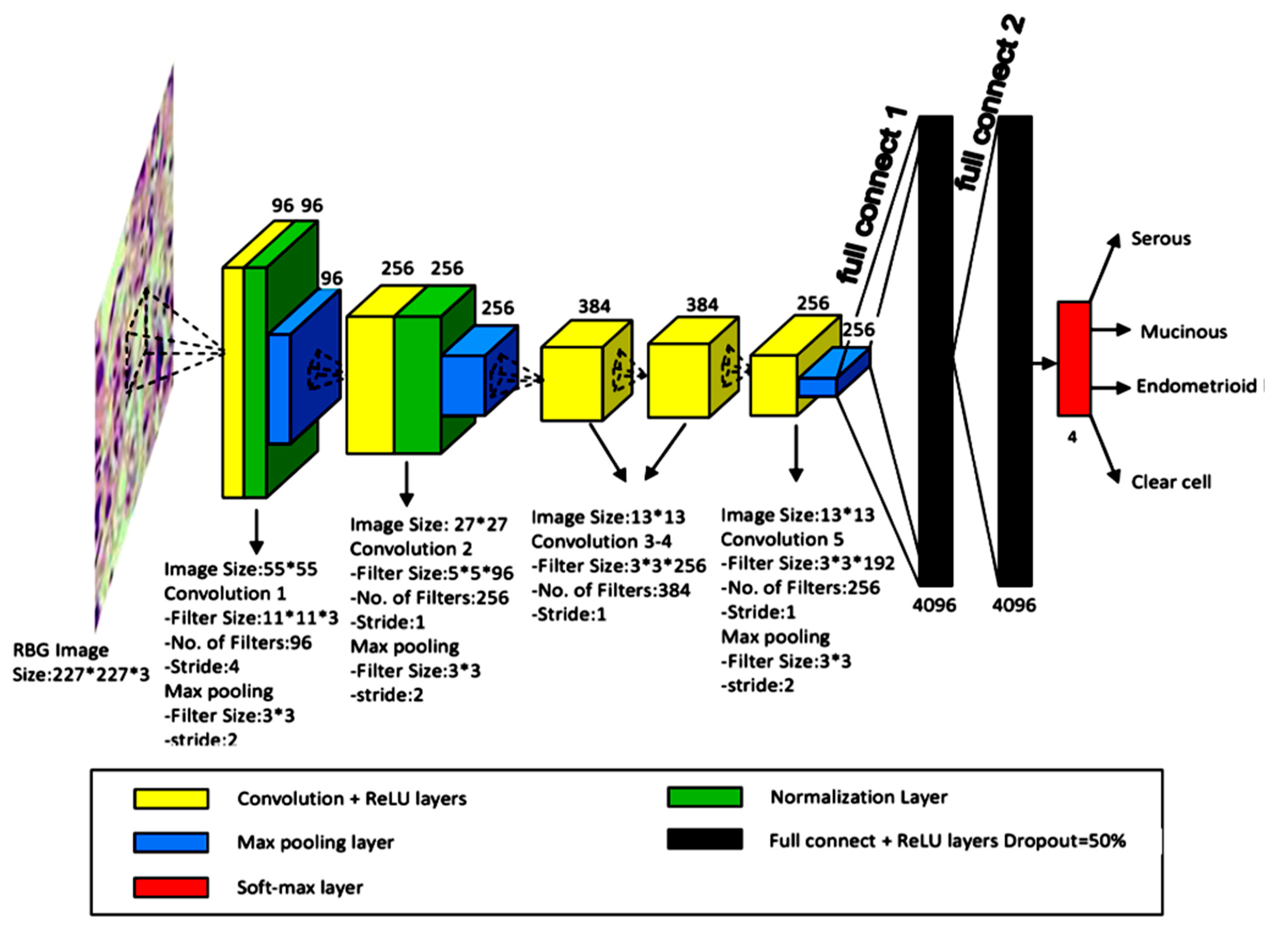
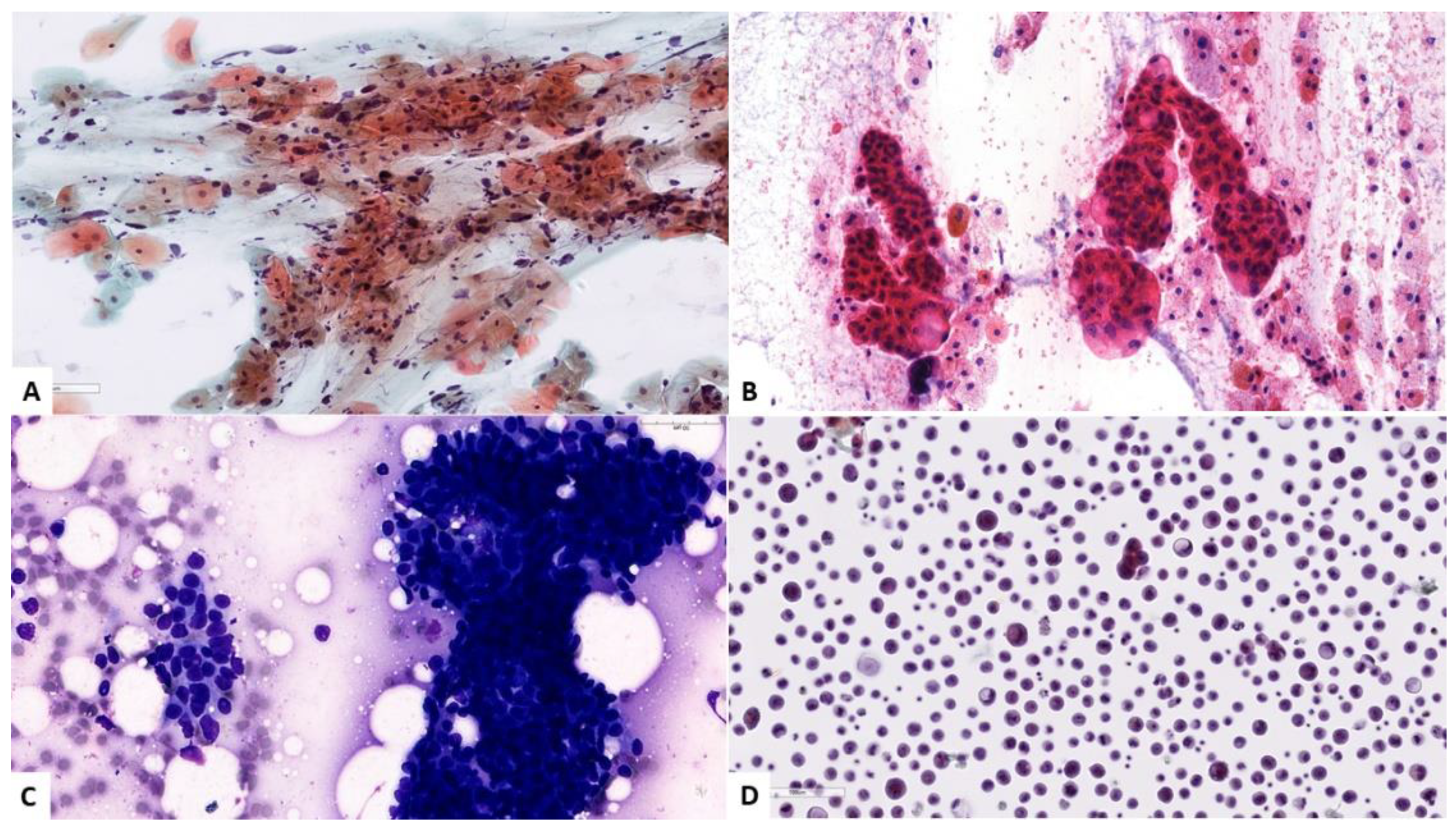
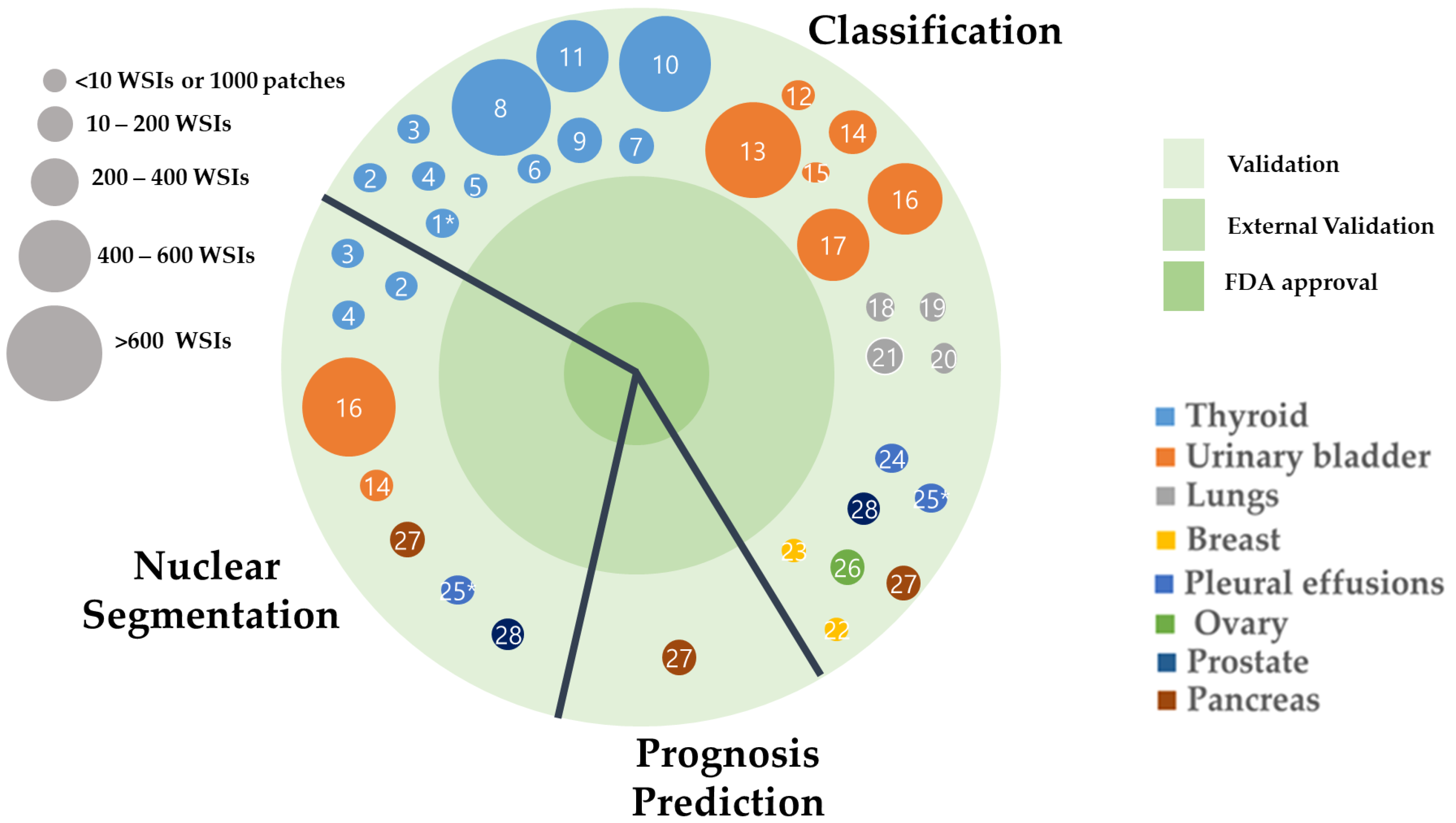
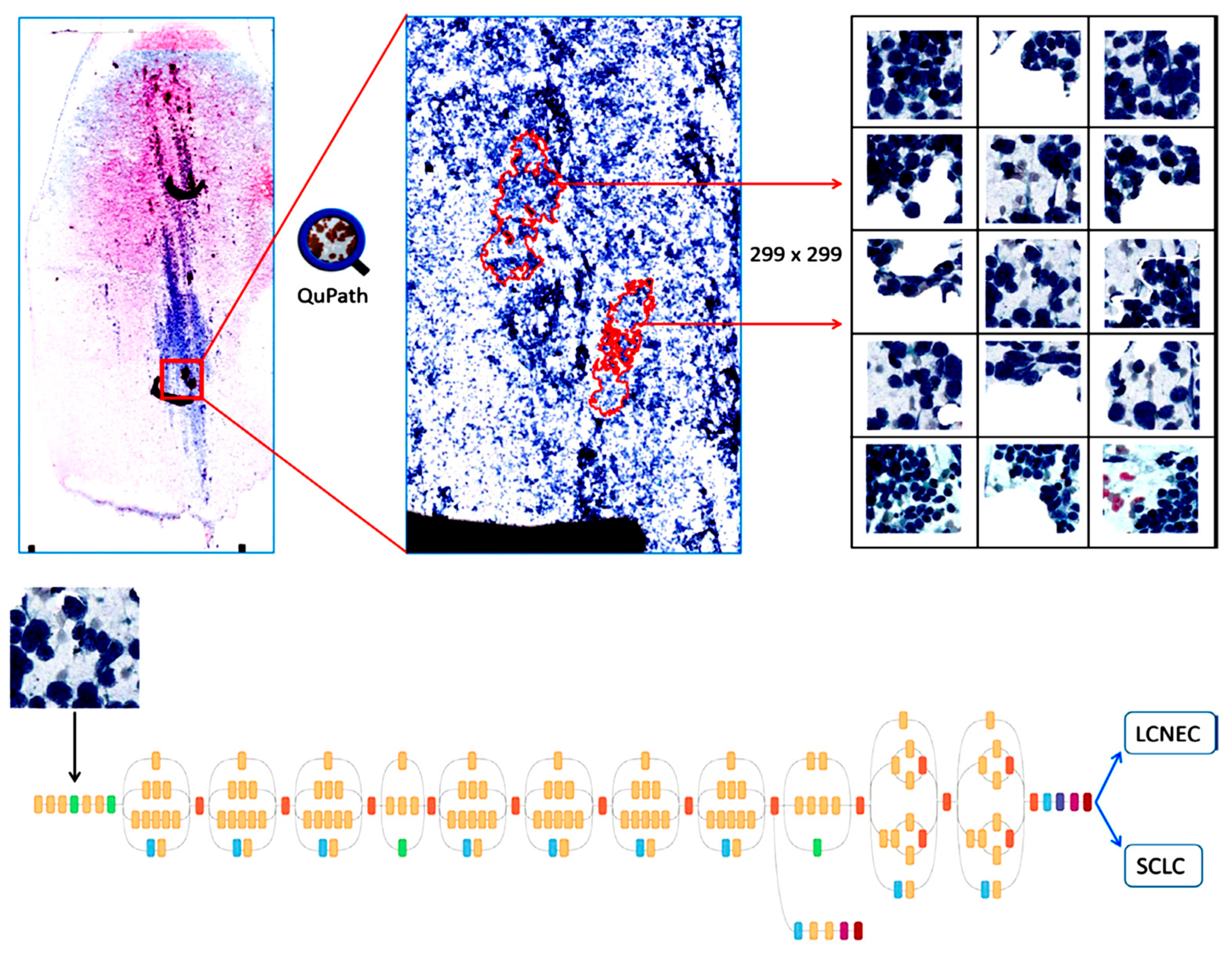

| No. | Organ | Author | Year | Country | Task | Staining and Preparation Method | Dataset | Pixel Level | Sampling | Z-Stacking Images | External Cross-Validation | Base Model | Performance | Pathologist Number |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Thyroid | Varlatzidou [48] | 2011 | Greece | Classification Benign/Malignant | Pap | 335 patients (32,887 nuclei) | 1024 × 768 | FNAC | ND | ND | ANN (LVQ) | Sens: 93.80% Spec: 94.11% Acc: 94.05% | NA |
| 2 | Gopinath (1) [34] | 2013 | India | Nuclear segmentation/ Classification Benign/Malignant | Pap | 110 patches | 256 × 256 | FNAC | ND | ND | SVM/ k-NN, | Sens: 95% Spec: 100% Acc: 96.7% | ATLAS committee | |
| 3 | Gopinath (2) [35] | 2013 | India | Nuclear segmentation/ Classification Benign/Malignant | Pap | 110 patches | 256 × 256 | FNAC | ND | ND | SVM/ ENN/ k-NN | Sens: 90% Spec: 100% Acc: 93.3% | ATLAS committee | |
| 4 | Gopinath (3) [36] | 2015 | India | Nuclear segmentation/ Classification Benign/Malignant | Pap | 110 patches | 256 × 256 | FNAC | ND | ND | SVM/ ENN/ k-NN/ DT | Sens: 100% Spec: 90% Acc: 96.6% | ATLAS committee | |
| 5 | Savala [37] | 2017 | India | Classification FA/FC | May Grunwald–Giemsa/H&E | 57 cases (57patches) | NA | FNAC | ND | ND | ANN | Acc: 100% AUC: 1.00% | 2 | |
| 6 | Gopinath (4) [49] | 2018 | India | Classification Benign/Malignant | Pap | 110 patches | 256 × 256 | FNAC | ND | ND | ANN/ ENN | Sens: 95% Spec: 100% Acc: 96.7% | ATLAS committee | |
| 7 | Sanyal [38] | 2018 | India | Classification PTC/non-PTC | Pap | 370 patches | 512 × 512 | FNAC | ND | ND | CNN | Sens: 90.48% Spec: 83.33% Acc: 85.1% | NA | |
| 8 | Dov [39] | 2019 | USA | Classification Benign/Malignant | Pap | 908 WSIs (5461 patches) | 150,000 × 100,000 | FNAC | ND | ND | CNN (VGG-11) | Sens: 92% Spec: 90.5% | 3 | |
| 9 | Guan [40] | 2019 | China | Classification Benign/PTC | LBC H&E | 279 WSI (887 patch images) | 224 × 224 | FNAC | ND | ND | VGG-16/ Inception-V3 | Sens 100% Spec 94.91% Acc: 97.6% | 1 | |
| 10 | Range [22] | 2020 | USA | Classification Benign/Malignant | Pap | 659 patients (908 WSIs) (4494 patches) | NA | FNAC | Yes | ND | Machine learning and CNNs | Sens: 92.0% Spec: 90.5% AUC: 0.93% | 1 | |
| 11 | Frago-poulos [41] | 2020 | Greece | Classification Benign/Malignant | LBC Pap-stained | 447 WSI (41,324 nuclei) | 1024 × 768 | FNAC | ND | ND | ANN (RBF) | Sens: 95.0%, Spec: 95.5% | NA | |
| 12 | Urinary bladder | Murali-daran [42] | 2015 | India | Classification Benign/Low-grade/ High-grade | Pap | 115 cases (115 patches) | NA | Urine sample | ND | ND | ANN | (1) All benign and malignant cases were diagnosed correctly (2) One of the low-grade cases was diagnosed as high-grade | 2 |
| 13 | Sanghvi [43] | 2019 | USA | Classification AU/HGUC/LGUN/SHGUC | NA | 2405 WSIs (26 million cells) | 150 × 150 | Urine sample | Yes | ND | CNN | Sens: 79.5% Spec: 84.5% AUC: 0.88% | 4 | |
| 14 | Vaickus [44] | 2018 | USA | Segmentation (Nucleus/Cytoplasm) Classification AU/BU/Sqc/Cry/Ery/Leu/BI/Deb | NA | 217 WSIs (1.42 × 107 million cells) | 40,000 × 40,000 | Urine sample | ND | ND | CNN (AlexNet/ResNet) | Acc: >95% | 2 | |
| 15 | Zhang [46] | 2020 | China | Classification UC/SqC/DC/ IC/AU/SHGUC | Pap | 49 cases 49 Images | NA | Urine sample | ND | ND | CNN | Identified abnormal urothelial cells | 1 | |
| 16 | Awan [45] | 2021 | UK | Segmentation, Detection Classification Bc/IC/AU/SqC/SHGUC | LBC Pap | 398 WSIs (9096 patches) | 256 × 256 500 × 500 5000 × 5000 | Urine sample | ND | ND | RetinaNet | AUC Atypical: 0.81 Malignant: 0.83 | NA | |
| 17 | Nojima [47] | 2021 | India | Classification Benign/Malignant Stromal invasion nuclear grading | LBC Pap | 232 cases 466 WSIs (61,512 patches) | 256 × 256 128 × 128 | Urine sample | ND | ND | VGG16 | AUC: 0.98, F1 score: 0.90 AUC: 0.86, F1 score: 0.82 AUC: 0.86, F1 score: 0.82 | NA | |
| 18 | Lungs | Teramoto (1) [26] | 2017 | Japan | Classification AdCC/SqCC/SCLC | Pap | 76 cases (298 patches) | 256 × 256 | FNAC /Bronchoscopy | ND | ND | CNN | Acc: 71.1% | NA |
| 19 | Teramoto (2) [23] | 2019 | Japan | Classification Benign/Malignant | Pap | 46 cases (621 patches) | 224 × 224 | FNAC /Bronchoscopy | ND | ND | CNN (VGG-16) | Sens: 89.3% Spec: 83.3% Acc: 79.2% | NA | |
| 20 | Teramoto (3) [27] | 2020 | Japan | Classification Benign/Malignant | Pap | 60 cases (793 patches) | 256 × 256 | FNAC /Bronchoscopy | ND | ND | CNN/ DCGAN/ PGGAN, | Sens: 85.4% Spec: 85.3% Acc: 85.3%% | NA | |
| 21 | Gonzalez [28] | 2020 | USA | Classification SCLC/LCNEC | Diff-Quik/ Pap/H&E | 40 cases (114 WSIs) (464,378 patches) | 299 × 299 | FNAC /Bronchoscopy | ND | ND | Inception V3 | For Diff-Quik Model Sens: 1.00%, Spec: 87.5%, AUC: 1.00% For the Pap-stained Model Sens: 1.00%, Spec: 85.7%, AUC: 1.00% For the H&E model Sens: 1.00% Spec: 87.5% AUC: 87.5% | NA | |
| 22 | Breast | Dey [24] | 2011 | India | Classification FAd/IDC/ILC | H&E | 64 cases (64 patches) | NA | FNAC | ND | ND | ANN | ANN classified all the FA and ILC cases and six out of seven IDC cases | 2 |
| 23 | Subbaiah [25] | 2013 | India | Classification FAd/IDC | H&E | 112 cases (112 patches) | NA | FNAC | ND | ND | ANN | Sens: 100% Spec: 100% | 2 | |
| 24 | Pleural effusions | Barwad [31] | 2011 | India | Classification (Benign/Metastatic Carcinoma) | Giemsa/ Pap | 114 cases (114 images) | NA | Pleural fluid | ND | ND | ANN | Acc: 100% | 2 |
| 25 | Tosun [32] | 2015 | USA | Nuclear segmentation/ Classification Benign/Malignant | Diff-Quik | 34 cases (1080 nuclei) | NA | Pleural fluid | ND | ND | OTBL/ k-nearest | Acc: 100% | 1 | |
| 26 | Ovary | Wu [29] | 2018 | China | Classification SC/MC/EC/CCC | H&E | 85 WSIs (7392 patches) | 227 × 227 | FNAC | ND | ND | CNN (AlexNet) | Acc: 78.20% | 2 |
| 27 | Pancreas | Boroujeni [30] | 2017 | USA | Nuclear segmentation/ Classification/ Survival (Benign/Malignant/Atypical) | Pap | 75 cases (277 images) | NA | FNAC | ND | ND | K-means clustering/MNN | Acc: 100% (Benign or malignant) Acc: 77% (Atypical cases classified as benign or malignant) | NA |
| 28 | Prostate | Nguyen [33] | 2012 | USA | Nuclear segmentation/ Classification Benign/Malignant | H&E | 17 WSIs | Training 4000 × 7000 Testing 5000 × 23,000 | NA | ND | ND | SVM/RBF kernel | Sens: 78% | NA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thakur, N.; Alam, M.R.; Abdul-Ghafar, J.; Chong, Y. Recent Application of Artificial Intelligence in Non-Gynecological Cancer Cytopathology: A Systematic Review. Cancers 2022, 14, 3529. https://doi.org/10.3390/cancers14143529
Thakur N, Alam MR, Abdul-Ghafar J, Chong Y. Recent Application of Artificial Intelligence in Non-Gynecological Cancer Cytopathology: A Systematic Review. Cancers. 2022; 14(14):3529. https://doi.org/10.3390/cancers14143529
Chicago/Turabian StyleThakur, Nishant, Mohammad Rizwan Alam, Jamshid Abdul-Ghafar, and Yosep Chong. 2022. "Recent Application of Artificial Intelligence in Non-Gynecological Cancer Cytopathology: A Systematic Review" Cancers 14, no. 14: 3529. https://doi.org/10.3390/cancers14143529
APA StyleThakur, N., Alam, M. R., Abdul-Ghafar, J., & Chong, Y. (2022). Recent Application of Artificial Intelligence in Non-Gynecological Cancer Cytopathology: A Systematic Review. Cancers, 14(14), 3529. https://doi.org/10.3390/cancers14143529







