Deep Learning Algorithms for Diagnosis of Lung Cancer: A Systematic Review and Meta-Analysis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Literature Search
2.2. Inclusion and Exclusion Criteria
2.3. Assessment of Methodologic Quality
2.4. Data Extraction
2.5. Data and Statistical Analysis
3. Results
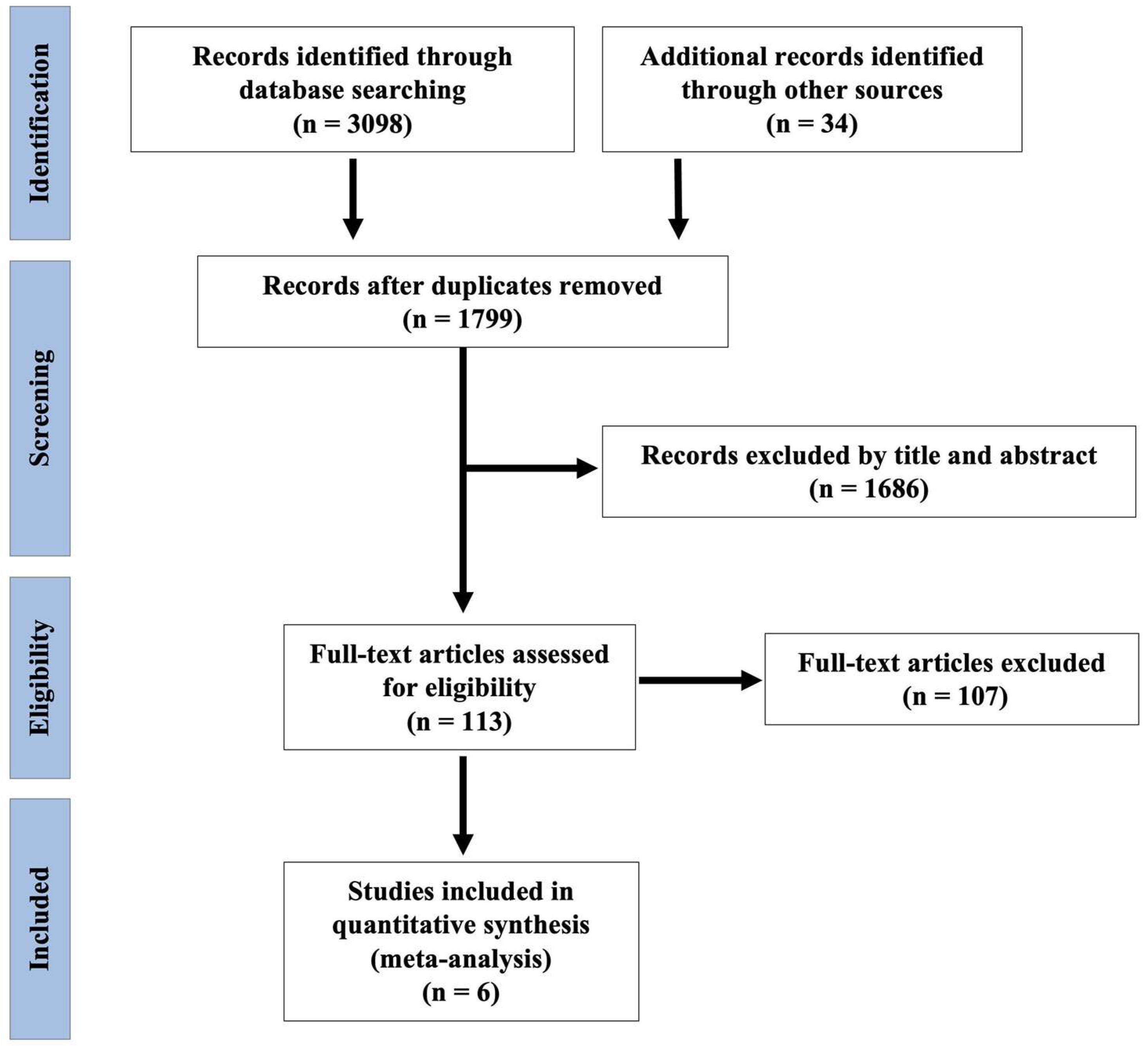
3.1. Search Results
3.2. Quality Appraisal
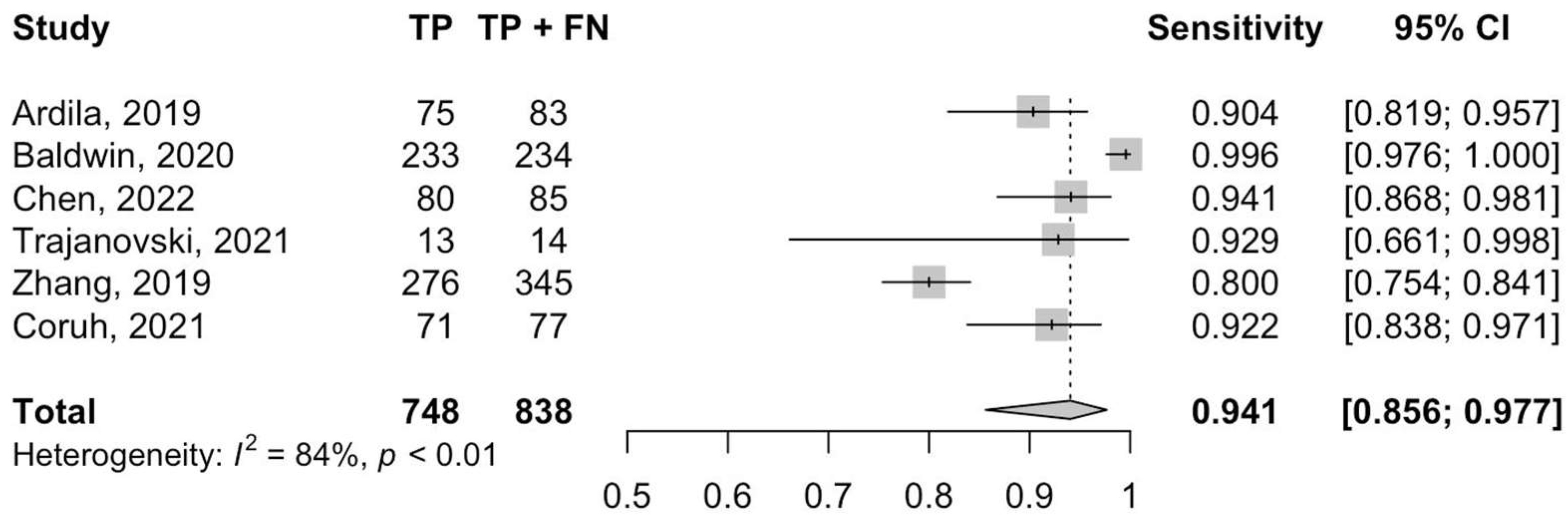
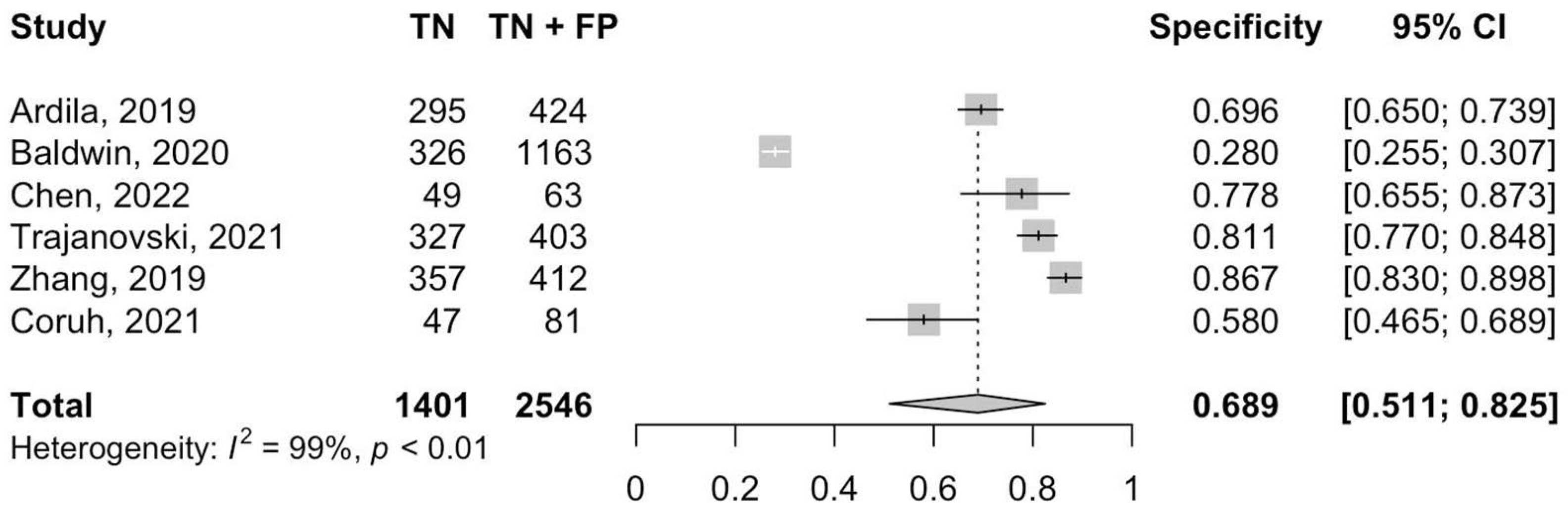
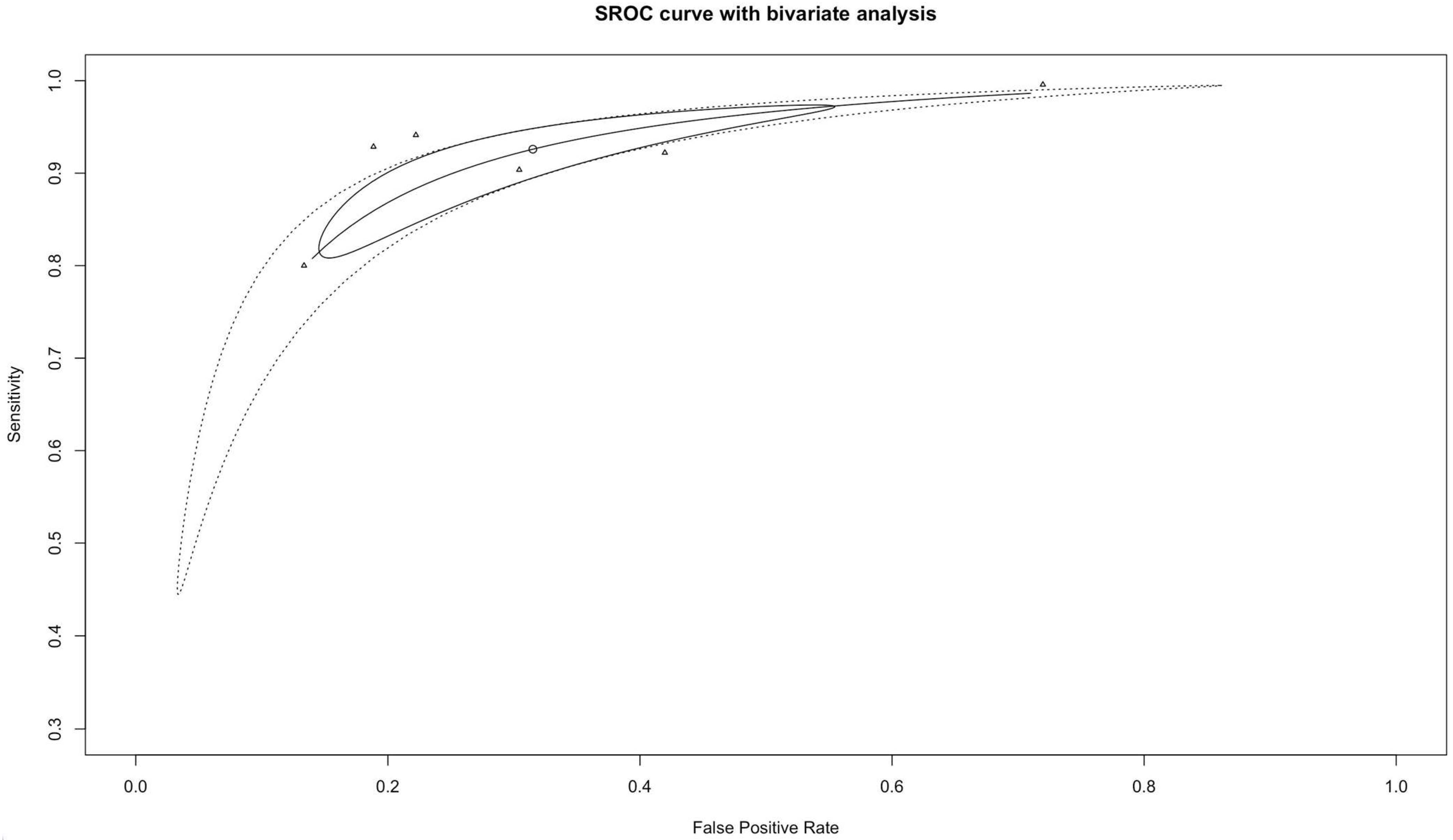
3.3. Diagnostic Accuracy and Heterogeneity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2022. CA A Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef]
- Siegel, R.; Ma, J.; Zou, Z.; Jemal, A. Cancer Statistics, 2014. CA A Cancer J. Clin. 2014, 64, 9–29. [Google Scholar] [CrossRef]
- Krist, A.H.; Davidson, K.W.; Mangione, C.M.; Barry, M.J.; Cabana, M.; Caughey, A.B.; Davis, E.M.; Donahue, K.E.; Doubeni, C.A.; Kubik, M.; et al. Screening for Lung Cancer: US Preventive Services Task Force Recommendation Statement. JAMA 2021, 325, 962. [Google Scholar] [CrossRef]
- Swensen, S.J.; Jett, J.R.; Hartman, T.E.; Midthun, D.E.; Sloan, J.A.; Sykes, A.-M.; Aughenbaugh, G.L.; Clemens, M.A. Lung Cancer Screening with CT: Mayo Clinic Experience. Radiology 2003, 226, 756–761. [Google Scholar] [CrossRef]
- Bach, P.B.; Mirkin, J.N.; Oliver, T.K.; Azzoli, C.G.; Berry, D.A.; Brawley, O.W.; Byers, T.; Colditz, G.A.; Gould, M.K.; Jett, J.R.; et al. Benefits and Harms of CT Screening for Lung Cancer. JAMA 2012, 307, 2418. [Google Scholar] [CrossRef]
- Larke, F.J.; Kruger, R.L.; Cagnon, C.H.; Flynn, M.J.; McNitt-Gray, M.M.; Wu, X.; Judy, P.F.; Cody, D.D. Estimated Radiation Dose Associated with Low-Dose Chest CT of Average-Size Participants in the National Lung Screening Trial. Am. J. Roentgenol. 2011, 197, 1165–1169. [Google Scholar] [CrossRef]
- Aberle, D.R.; Adams, A.M.; Berg, C.D. Reduced Lung-Cancer Mortality with Low-Dose Computed Tomographic Screening. Engl. J. Med. 2011, 365, 395–409. [Google Scholar] [CrossRef]
- de Koning, H.; van der Aalst, C.; ten Haaf, K.; Oudkerk, M. PL02.05 Effects of Volume CT Lung Cancer Screening: Mortality Results of the NELSON Randomised-Controlled Population Based Trial. J. Thorac. Oncol. 2018, 13, S185. [Google Scholar] [CrossRef]
- van Meerbeeck, J.P.; Franck, C. Lung Cancer Screening in Europe: Where Are We in 2021? Transl. Lung Cancer Res. 2021, 10, 2407–2417. [Google Scholar] [CrossRef]
- Wait, S.; Alvarez-Rosete, A.; Osama, T.; Bancroft, D.; Cornelissen, R.; Marušić, A.; Garrido, P.; Adamek, M.; van Meerbeeck, J.; Snoeckx, A.; et al. Implementing Lung Cancer Screening in Europe: Taking a Systems Approach. JTO Clin. Res. Rep. 2022, 3, 100329. [Google Scholar] [CrossRef]
- British Society of Thoracic Imaging and The Royal College of Radiologists. Considerations to Ensure Optimum Roll-Out of Targeted Lung Cancer Screening over the Next Five Years; British Society of Thoracic Imaging and The Royal College of Radiologists: London, UK, 2020. [Google Scholar]
- Kauczor, H.-U.; Bonomo, L.; Gaga, M.; Nackaerts, K.; Peled, N.; Prokop, M.; Remy-Jardin, M.; von Stackelberg, O.; Sculier, J.-P. ESR/ERS White Paper on Lung Cancer Screening. Eur. Radiol. 2015, 25, 2519–2531. [Google Scholar] [CrossRef]
- Kuo, E.; Bharat, A.; Bontumasi, N.; Sanchez, C.; Zoole, J.B.; Patterson, G.A.; Meyers, B.F. Impact of Video-Assisted Thoracoscopic Surgery on Benign Resections for Solitary Pulmonary Nodules. Ann. Thorac. Surg. 2012, 93, 266–273. [Google Scholar] [CrossRef]
- Deppen, S.A.; Blume, J.D.; Aldrich, M.C.; Fletcher, S.A.; Massion, P.P.; Walker, R.C.; Chen, H.C.; Speroff, T.; Degesys, C.A.; Pinkerman, R.; et al. Predicting Lung Cancer Prior to Surgical Resection in Patients with Lung Nodules. J. Thorac. Oncol. 2014, 9, 1477–1484. [Google Scholar] [CrossRef]
- El-Baz, A.; Beache, G.M.; Gimel’farb, G.; Suzuki, K.; Okada, K.; Elnakib, A.; Soliman, A.; Abdollahi, B. Computer-Aided Diagnosis Systems for Lung Cancer: Challenges and Methodologies. Int. J. Biomed. Imaging 2013, 2013, 942353. [Google Scholar] [CrossRef]
- American College of Radiology. Lung-RADS v1.1 Assessment Categories (2019 Release); American College of Radiology: Reston, VA, USA, 2019. [Google Scholar]
- Pedrosa, J.; Aresta, G.; Ferreira, C. Computer-Aided Lung Cancer Screening in Computed Tomography: State-of the-Art and Future Perspectives. In Detection Systems in Lung Cancer and Imaging; IOP Publishing: Bristol, UK, 2022; Volume 1, pp. 4–38. [Google Scholar]
- Aresta, G.; Araújo, T.; Jacobs, C.; van Ginneken, B.; Cunha, A.; Ramos, I.; Campilho, A. Towards an Automatic Lung Cancer Screening System in Low Dose Computed Tomography; Springer: Berlin/Heidelberg, Germany, 2018; pp. 310–318. [Google Scholar]
- Zheng, S.; Guo, J.; Cui, X.; Veldhuis, R.N.J.; Oudkerk, M.; van Ooijen, P.M.A. Automatic Pulmonary Nodule Detection in CT Scans Using Convolutional Neural Networks Based on Maximum Intensity Projection. IEEE Trans. Med. Imaging 2020, 39, 797–805. [Google Scholar] [CrossRef]
- Kaluva, K.C.; Vaidhya, K.; Chunduru, A.; Tarai, S.; Nadimpalli, S.P.P.; Vaidya, S. An Automated Workflow for Lung Nodule Follow-Up Recommendation Using Deep Learning; Springer: Berlin/Heidelberg, Germany, 2020; pp. 369–377. [Google Scholar]
- Katz, O.; Presil, D.; Cohen, L.; Schwartzbard, Y.; Hoch, S.; Kashani, S. Pulmonary-Nodule Detection Using an Ensemble of 3D SE-ResNet18 and DPN68 Models; Springer: Berlin/Heidelberg, Germany, 2020; pp. 378–385. [Google Scholar]
- Cao, H.; Liu, H.; Song, E.; Hung, C.-C.; Ma, G.; Xu, X.; Jin, R.; Lu, J. Dual-Branch Residual Network for Lung Nodule Segmentation. Appl. Soft Comput. 2019, 86, 105934. [Google Scholar] [CrossRef]
- Dong, X.; Xu, S.; Liu, Y.; Wang, A.; Saripan, M.I.; Li, L.; Zhang, X.; Lu, L. Multi-View Secondary Input Collaborative Deep Learning for Lung Nodule 3D Segmentation. Cancer Imaging 2020, 20, 53. [Google Scholar] [CrossRef]
- Usman, M.; Lee, B.-D.; Byon, S.-S.; Kim, S.-H.; Lee, B.; Shin, Y.-G. Volumetric Lung Nodule Segmentation Using Adaptive ROI with Multi-View Residual Learning. Sci. Rep. 2020, 10, 12839. [Google Scholar] [CrossRef]
- Wu, W.; Gao, L.; Duan, H.; Huang, G.; Ye, X.; Nie, S. Segmentation of Pulmonary Nodules in CT Images Based on 3D-UNET Combined with Three-dimensional Conditional Random Field Optimization. Med. Phys. 2020, 47, 4054–4063. [Google Scholar] [CrossRef]
- Liu, K.; Kang, G. Multiview Convolutional Neural Networks for Lung Nodule Classification. Int. J. Imaging Syst. Technol. 2017, 27, 12–22. [Google Scholar] [CrossRef]
- Kang, G.; Liu, K.; Hou, B.; Zhang, N. 3D Multi-View Convolutional Neural Networks for Lung Nodule Classification. PLoS ONE 2017, 12, e0188290. [Google Scholar] [CrossRef]
- da Nóbrega, R.V.M.; Rebouças Filho, P.P.; Rodrigues, M.B.; da Silva, S.P.P.; Dourado Júnior, C.M.J.M.; de Albuquerque, V.H.C. Lung Nodule Malignancy Classification in Chest Computed Tomography Images Using Transfer Learning and Convolutional Neural Networks. Neural Comput. Appl. 2020, 32, 11065–11082. [Google Scholar] [CrossRef]
- Dai, Y.; Yan, S.; Zheng, B.; Song, C. Incorporating Automatically Learned Pulmonary Nodule Attributes into a Convolutional Neural Network to Improve Accuracy of Benign-Malignant Nodule Classification. Phys. Med. Biol. 2018, 63, 245004. [Google Scholar] [CrossRef]
- Xiao, N.; Qiang, Y.; Bilal Zia, M.; Wang, S.; Lian, J. Ensemble Classification for Predicting the Malignancy Level of Pulmonary Nodules on Chest Computed Tomography Images. Oncol. Lett. 2020, 20, 401–408. [Google Scholar] [CrossRef]
- Liao, F.; Liang, M.; Li, Z.; Hu, X.; Song, S. Evaluate the Malignancy of Pulmonary Nodules Using the 3-D Deep Leaky Noisy-OR Network. IEEE Trans. Neural Netw. Learn. Syst. 2019, 30, 3484–3495. [Google Scholar] [CrossRef]
- Ozdemir, O.; Russell, R.L.; Berlin, A.A. A 3D Probabilistic Deep Learning System for Detection and Diagnosis of Lung Cancer Using Low-Dose CT Scans. IEEE Trans. Med. Imaging 2019, 39, 1419–1429. [Google Scholar] [CrossRef]
- Huang, P.; Lin, C.T.; Li, Y.; Tammemagi, M.C.; Brock, M.V.; Atkar-Khattra, S.; Xu, Y.; Hu, P.; Mayo, J.R.; Schmidt, H.; et al. Prediction of Lung Cancer Risk at Follow-up Screening with Low-Dose CT: A Training and Validation Study of a Deep Learning Method. Lancet Digit. Health 2019, 1, e353–e362. [Google Scholar] [CrossRef]
- Whiting, P.F.; Rutjes, A.W.S.; Westwood, M.E.; Mallet, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.G.; Sterne, J.A.C.; Bossuyt, P.M.M. QUADAS-2: A Revised Tool for the Quality Assessment of Diagnostic Accuracy Studies. Ann. Intern. Med. 2011, 155, 529–536. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. Int. J. Surg. 2010, 8, 336–341. [Google Scholar] [CrossRef]
- Devillé, W.L.; Buntinx, F.; Bouter, L.M.; Montori, V.M.; de Vet, H.C.; van der Windt, D.A.; Bezemer, P.D. Conducting Systematic Reviews of Diagnostic Studies: Didactic Guidelines. BMC Med. Res. Methodol. 2002, 2, 9. [Google Scholar] [CrossRef]
- Trajanovski, S.; Mavroeidis, D.; Swisher, C.L.; Gebre, B.G.; Veeling, B.S.; Wiemker, R.; Klinder, T.; Tahmasebi, A.; Regis, S.M.; Wald, C.; et al. Towards Radiologist-Level Cancer Risk Assessment in CT Lung Screening Using Deep Learning. Comput. Med. Imaging Graph. 2021, 90, 101883. [Google Scholar] [CrossRef] [PubMed]
- Ardila, D.; Kiraly, A.P.; Bharadwaj, S.; Choi, B.; Reicher, J.J.; Peng, L.; Tse, D.; Etemadi, M.; Ye, W.; Corrado, G.; et al. End-to-End Lung Cancer Screening with Three-Dimensional Deep Learning on Low-Dose Chest Computed Tomography. Nat. Med. 2019, 25, 954–961. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Sun, X.; Dang, K.; Li, K.; Guo, X.; Chang, J.; Yu, Z.; Huang, F.; Wu, Y.; Liang, Z.; et al. Toward an Expert Level of Lung Cancer Detection and Classification Using a Deep Convolutional Neural Network. Oncologist 2019, 24, 1159–1165. [Google Scholar] [CrossRef]
- Chen, Y.; Tian, X.; Fan, K.; Zheng, Y.; Tian, N.; Fan, K. The Value of Artificial Intelligence Film Reading System Based on Deep Learning in the Diagnosis of Non-Small-Cell Lung Cancer and the Significance of Efficacy Monitoring: A Retrospective, Clinical, Nonrandomized, Controlled Study. Comput. Math. Methods Med. 2022, 2022, 2864170. [Google Scholar] [CrossRef]
- Baldwin, D.R.; Gustafson, J.; Pickup, L.; Arteta, C.; Novotny, P.; Declerck, J.; Kadir, T.; Figueiras, C.; Sterba, A.; Exell, A.; et al. External Validation of a Convolutional Neural Network Artificial Intelligence Tool to Predict Malignancy in Pulmonary Nodules. Thorax 2020, 75, 306–312. [Google Scholar] [CrossRef]
- Çoruh, A.G.; Yenigün, B.; Uzun, C.; Kahya, Y.; Büyükceran, E.U.; Elhan, A.; Orhan, K.; Cangır, A.K. A Comparison of the Fusion Model of Deep Learning Neural Networks with Human Observation for Lung Nodule Detection and Classification. Br. J. Radiol. 2021, 94, 20210222. [Google Scholar] [CrossRef] [PubMed]
- Krupinski, E.A. Current Perspectives in Medical Image Perception. Atten. Percept. Psychophys. 2010, 72, 1205–1217. [Google Scholar] [CrossRef]
- Matsumoto, S.; Ohno, Y.; Aoki, T.; Yamagata, H.; Nogami, M.; Matsumoto, K.; Yamashita, Y.; Sugimura, K. Computer-Aided Detection of Lung Nodules on Multidetector CT in Concurrent-Reader and Second-Reader Modes: A Comparative Study. Eur. J. Radiol. 2013, 82, 1332–1337. [Google Scholar] [CrossRef]
- Brunyé, T.T.; Nallamothu, B.K.; Elmore, J.G. Eye-Tracking for Assessing Medical Image Interpretation: A Pilot Feasibility Study Comparing Novice vs. Expert Cardiologists. Perspect. Med. Educ. 2019, 8, 65–73. [Google Scholar] [CrossRef]
- Rampinelli, C.; Calloni, S.F.; Minotti, M.; Bellomi, M. Spectrum of Early Lung Cancer Presentation in Low-Dose Screening CT: A Pictorial Review. Insights Imaging 2016, 7, 449–459. [Google Scholar] [CrossRef]
- Setio, A.A.A.; Traverso, A.; de Bel, T.; Berens, M.S.N.; Bogaard, C.V.D.; Cerello, P.; Chen, H.; Dou, Q.; Fantacci, M.E.; Geurts, B.; et al. Validation, Comparison, and Combination of Algorithms for Automatic Detection of Pulmonary Nodules in Computed Tomography Images: The LUNA16 Challenge. Med. Image Anal. 2017, 42, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, C.; Setio, A.A.A.; Scholten, E.T.; Gerke, P.K.; Bhattacharya, H.; Hoesein, F.A.M.; Brink, M.; Ranschaert, E.; de Jong, P.A.; Silva, M.; et al. Deep Learning for Lung Cancer Detection on Screening CT Scans: Results of a Large-Scale Public Competition and an Observer Study with 11 Radiologists. Radiol. Artif. Intell. 2021, 3, e210027. [Google Scholar] [CrossRef] [PubMed]
- Ciompi, F.; Chung, K.; van Riel, S.J.; Setio, A.A.A.; Gerke, P.K.; Jacobs, C.; Scholten, E.T.; Schaefer-Prokop, C.; Wille, M.M.W.; Marchianò, A.; et al. Towards Automatic Pulmonary Nodule Management in Lung Cancer Screening with Deep Learning. Sci. Rep. 2017, 7, 46479. [Google Scholar] [CrossRef] [PubMed]
- Peikert, T.; Duan, F.; Rajagopalan, S.; Karwoski, R.A.; Clay, R.; Robb, R.A.; Qin, Z.; Sicks, J.; Bartholmai, B.J.; Maldonado, F. Novel High-Resolution Computed Tomography-Based Radiomic Classifier for Screen-Identified Pulmonary Nodules in the National Lung Screening Trial. PLoS ONE 2018, 13, e0196910. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.Z.; Huang, Y.L.; Wu, C.C.; Tang, E.K.; Chen, C.S.; Mar, G.Y.; Yen, Y.; Wu, M.T. Assessment of Selection Criteria for Low-Dose Lung Screening CT Among Asian Ethnic Groups in Taiwan: From Mass Screening to Specific Risk-Based Screening for Non-Smoker Lung Cancer. Clin. Lung Cancer 2016, 17, e45–e56. [Google Scholar] [CrossRef] [PubMed]
| Author | Year | Country | Study Design | Center | Artificial Intelligence | Source Validation Set | Threshold | Reference Standard Validation | Method Validation |
|---|---|---|---|---|---|---|---|---|---|
| Ardila et al. [38] | 2019 | USA | retrospective | multicenter | CNN | Lung cancer screening dataset | PPV = 0.11 | Histopathology | External validation |
| Baldwin et al. [41] | 2020 | UK | retrospective | multicenter | CNN | Private dataset | FN rate = 0% | Histopathology | External validation |
| Chen et al. [40] | 2022 | China | retrospective | single | CNN | Private dataset | Unknown (third party software) | Histopathology | External validation |
| Çoruh et al. [42] | 2021 | Turkey | retrospective | single | CNN | Private dataset | Youden index optimal cutoff | Histopathology | External validation |
| Trajanovski et al. [37] | 2021 | USA | retrospective | multicenter | CNN | Lung cancer screening dataset | Sensitivity = 93% | Histopathology | External validation |
| Zhang et al. [39] | 2019 | China | retrospective | multicenter | CNN | Private dataset | Probability of malignancy > 0.5 | Histopathology | Cross-validation |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Forte, G.C.; Altmayer, S.; Silva, R.F.; Stefani, M.T.; Libermann, L.L.; Cavion, C.C.; Youssef, A.; Forghani, R.; King, J.; Mohamed, T.-L.; et al. Deep Learning Algorithms for Diagnosis of Lung Cancer: A Systematic Review and Meta-Analysis. Cancers 2022, 14, 3856. https://doi.org/10.3390/cancers14163856
Forte GC, Altmayer S, Silva RF, Stefani MT, Libermann LL, Cavion CC, Youssef A, Forghani R, King J, Mohamed T-L, et al. Deep Learning Algorithms for Diagnosis of Lung Cancer: A Systematic Review and Meta-Analysis. Cancers. 2022; 14(16):3856. https://doi.org/10.3390/cancers14163856
Chicago/Turabian StyleForte, Gabriele C., Stephan Altmayer, Ricardo F. Silva, Mariana T. Stefani, Lucas L. Libermann, Cesar C. Cavion, Ali Youssef, Reza Forghani, Jeremy King, Tan-Lucien Mohamed, and et al. 2022. "Deep Learning Algorithms for Diagnosis of Lung Cancer: A Systematic Review and Meta-Analysis" Cancers 14, no. 16: 3856. https://doi.org/10.3390/cancers14163856
APA StyleForte, G. C., Altmayer, S., Silva, R. F., Stefani, M. T., Libermann, L. L., Cavion, C. C., Youssef, A., Forghani, R., King, J., Mohamed, T.-L., Andrade, R. G. F., & Hochhegger, B. (2022). Deep Learning Algorithms for Diagnosis of Lung Cancer: A Systematic Review and Meta-Analysis. Cancers, 14(16), 3856. https://doi.org/10.3390/cancers14163856






